Schlick T. Molecular Modeling and Simulation: An Interdisciplinary Guide
Подождите немного. Документ загружается.

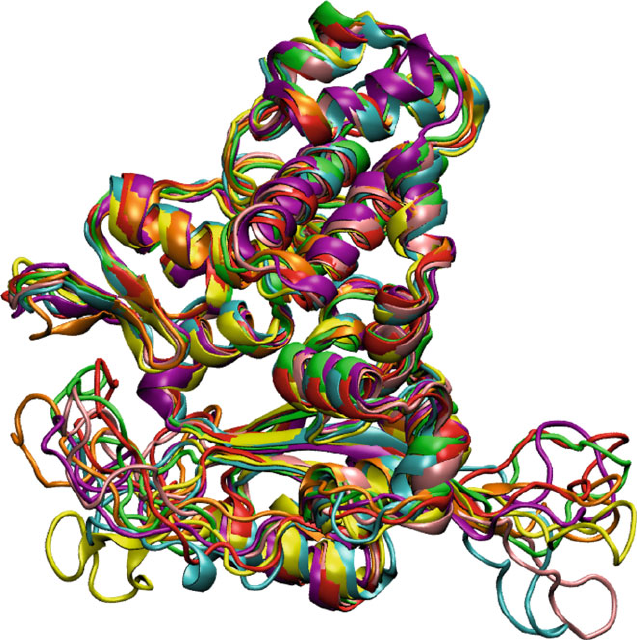
13.6. Various MD Ensembles 461
Algorithmic extensions to anisotropic systems, to allow changes in cell shape
in addition to size, have also been developed using the pressure tensor and matrix
analogs of the above approaches [123,835, for example].
14
Molecular Dynamics: Further Topics
Chapter 14 Notation
S
YMBOL DEFINITION
Matrices
C covariance matrix formulated for PCA
D
diffusion tensor (with sub-block matrices D
ij
)
H
local Hessian approximation
I
identity matrix
J
constant matrix used in defining symplectic
transformation
Λ
diagonal matrix with eigenvalues λ
1
, λ
2
, ...,3N
M
mass matrix
S
phase space transformation (S = DQD
−1
;also
Cholesky factor of D in BD)
T
hydrodynamic tensor
V
eigenvector matrix
Z
friction tensor (related to D via k
B
TD
−1
)
Ψ
J
Jacobian matrix of J
Vectors
F force
F
acceleration (
¨
X), or M
−1
F (X(t))
F
fast
,F
med
,F
slow
forces of ‘fast’, ‘medium’ and ‘slow’ components
P ,
˙
P
momentum and its first time derivative
R, R
B
random forces
V
velocity (
˙
X), components {v
i
} (also used for eigenvector)
X,
˙
X,
¨
X
position (components {x
i
}), and its first and second
time derivatives
X
r
reference position for MTS extrapolation
T. Schlick, Molecular Modeling and Simulation: An Interdisciplinary Guide, 463
Interdisciplinary Applied Mathematics 21, DOI 10.1007/978-1-4419-6351-2
14,
c
Springer Science+Business Media, LLC 2010
464 14. Molecular Dynamics: Further Topics
Chapter 14 Notation Table (continued)
S
YMBOL DEFINITION
Y ,
˙
Y vector and its first time derivative
∇E
energy gradient
Scalars & Functions
a hydrodynamic bead radius
k
1
,k
2
integers (used in MTS to relate timesteps; r = k
1
k
2
)
λ
n
eigenvalue n
n, m
integers (defining resonance condition); also m = mass
D
t
translational diffusion constant
E
k
kinetic energy
H
Hamiltonian
L
Liouville operator
N
number of atoms
N
b
number of beads (Brownian dynamics)
T
temperature
T
p
period (of characteristic frequency)
γ
Langevin damping constant
ζ
frictional coefficient (mγ for example)
η
solvent viscosity
θ
phase space rotation defined by integrator
θ
eff
effective phase space rotation
ω
natural frequency ( = ωΛt)
ω
eff
effective (scheme-dependent) frequency
Δτ, Δt
m
, Δt timesteps (inner, medium, and outer)
Φ(X)
‘dynamics function’ for implicit integration
Nature laughs at the difficulties of integration.
Pierre-Simon de Laplace (1749–1827).
Only by taking an infinitesimally small unit for observation (the dif-
ferential of history ...) and attaining to the art of integrating them
(that is, finding the sum of these infinitesimals) can we hope to arrive
at the laws of history.
Lev Tolstoy, in War and Peace (1828–1910).
14.1 Introduction
In this chapter we survey more advanced aspects of integration approaches for
biomolecular dynamics that are suitable for students interested in the mathe-
matical issues of numerical schemes. We begin in Section 14.2 by introducing
symplectic integrators in terms of an effective rotation in phase space and
illustrate basic concepts by simple harmonic analysis.
14.2. Symplectic Integrators 465
The multiple-timestep (MTS), or force splitting, methods are presented next
(Section 14.3), and the extreme variants of splitting by extrapolation versus split-
ting by impulses are contrasted with regard to resonance artifacts. Understanding
favorable and unfavorable features of MTS schemes with respect to stability, ac-
curacy, and resonance artifacts has led to important developments in recent years
of efficient, long-timestep methods for biomolecular dynamics, as well as to new
frameworks for method design and interpretation.
In this connection, we introduce in Sections 14.4 and 14.5, in turn, the stochas-
tic dynamics approaches of Langevin and Brownian dynamics; both are in fact
closely related, and the separation of terms may be arbitrary. These stochastic
formulations may be viewed as constructs that enhance sampling, or that, through
the introduction of random forces, make possible large timesteps by masking mild
instabilities resulting from Newtonian integration.
Stochastic approaches also constitute approximate models for following large-
scale motions in systems where random fluctuations (e.g., introduced by the bulk
solvent) are at least as important as the systematic forces. This is the case, for ex-
ample, in long DNA polymers that move with agility in solution, sampling many
equilibrium configurations, rather than remaining near a single state. Studies of
DNA supercoiling and of diffusion-controlled ligand gating events in enzyme
catalysis, for example, have relied on such Brownian dynamics approaches.
We also mention implicit integration schemes in Section 14.6. This class of
generally-more-expensive schemes has been explored as a possible way to in-
crease the timestep in biomolecular dynamics simulations. Section 14.7 describes
various enhanced sampling approaches for biomolecules, such as harmonic-
analysis based methods like essential dynamics, force biasing, altered protocols
(like replica-exchangeMD), and various innovative methods for exploringconfor-
mational space, deducing mechanisms and computing reaction rates. We conclude
in Section 14.8 with future perspectives on MD algorithm developments and with
a description of some promising integration alternatives.
The reader should have read Chapter 13, where basic aspects of molecular
dynamics and associated notation have been presented.
14.2 Symplectic Integrators
As introduced in the last chapter, symplectic or canonical integrators preserve
special properties associated with the Hamiltonian system of differential equa-
tions [731, 1090]. These properties include volume elements in phase space and
the Hamiltonian value (energy). In practice, the total energy is not preserved ex-
actly, but the energy error remains constant over long times. This is different from
nonsymplectic methods, which typically display a systematic energy drift in time
(usually damping). See Figure 13.6 for such an example of integration by the
symplectic Verlet versus the nonsymplectic Runge-Kutta method.

466 14. Molecular Dynamics: Further Topics
Such favorable properties can be explained by backward error analysis,
1
which
shows that the numerical trajectory of a symplectic integrator is the exact solu-
tion of a nearby Hamiltonian. The proximity measure depends on the timestep
of the integrator, Δt, and on the scheme’s order of accuracy, p. That is, it can
be shown that the Hamiltonian corresponding to the numerical trajectory of a
symplectic method remains order O(Δt)
p
away from the initial value of the true
Hamiltonian.
A comprehensive treatment of symplectic integration can be found in [1090].
Here only a few key concepts will be introduced, many of which are relevant to
future sections.
14.2.1 Symplectic Transformation
A Hamiltonian system is generally described by the motion of the collective po-
sition and momenta vectors X,P ∈R
3N
in time (here P = MV where M is the
diagonal mass matrix; see last chapter). The Hamiltonian H(X, P) is given as the
sum of the kinetic and potential energy of the system:
H(X, P)=
1
2
(P
T
M
−1
P )+E(X) . (14.1)
The evolution of this system is governed by the equations:
˙
P = −∂H/∂X ,
˙
X = ∂H/∂P , (14.2)
or, equivalently, in vector form as:
˙
X
˙
P
=
0 I
−I 0
∂H/∂X
∂H/∂P
≡ J ·∇H, (14.3)
where I is the 3N ×3N identity matrix.
An integrator defines a mapping Ψ that transforms coordinates and momenta
at time t, {X
n
,P
n
}, to coordinates and momenta at time t +Δt, {X
n+1
,P
n+1
}.
This transformation is symplectic if and only if its Jacobian matrix, Ψ
J
, satisfies:
Ψ
J
T
JΨ
J
= J , (14.4)
where Ψ
J
is the matrix
Ψ
J
≡
∂X
n+1
/∂X
n
∂X
n+1
/∂P
n
∂P
n+1
/∂X
n
∂P
n+1
/∂P
n
. (14.5)
1
See [280, Section 2.4.1] or [357, Section 2.7], for example. Roughly speaking, backward error
analysis transforms the problem of estimating computational errors (in our case due to the finite-
difference approximation) back to the problem of estimating the effect of changing the input data
slightly.

14.2. Symplectic Integrators 467
It can be shown that a composition of symplectic transformations is also
symplectic and that the inverse of a symplectic mapping is symplectic. These
properties are often used in practice to prove that an integrator is symplectic.
For a proof that the Verlet scheme is symplectic, see [1078]. Below we only
illustrate how the Verlet transformation can be interpreted for a linear system as
a rotation in phase space. Analysis of a harmonic oscillator is often a first step in
analyzing behavior of a numerical scheme and already sheds considerable insight
[821,866,967,1126,1199, 1437, for example].
14.2.2 Harmonic Oscillator Example
Consider the Verlet scheme of eq. (13.15) for linear forces F (X)=− ω
2
X,
where ω is the natural frequency of the oscillator. The transformation S can be
used to relate one phase point to the next [1199]. That is,
ωX
n+1
V
n+1
= S
ωX
n
V
n
, (14.6)
where S is defined as
S =
1 −
2
/2 (1 −
2
/4)
− 1 −
2
/2
, (14.7)
with the parameter as the frequency times the timestep:
= ωΔt. (14.8)
It is simple to show
2
that the product of the two eigenvalues is equal to
1(λ
1
λ
2
=1) and that the absolute value of their sum is less than or equal to
2(|λ
1
+ λ
2
|≤2). Thus, powers of S (which are matrices) are bounded if and
only if
2
< 4 , (14.9)
or equivalently
Δt<2/ω . (14.10)
14.2.3 Linear Stability
The above restriction on the timestep size is the stability condition for Verlet.
Thus, if the period of the oscillator is T
p
=2π/ω,thislinear stability condition
becomes
Δt<T
p
/π . (14.11)
2
Hint: For a 2 × 2 matrix, the matrix trace (sum of diagonals) is the sum of the eigenvalues, here
2 −
2
, and the matrix determinant is their product, here 1.
468 14. Molecular Dynamics: Further Topics
Table 14.1 gives corresponding linear stability limits on the timestep for
the high-frequency end of biomolecular vibrational modes. Clearly, in general,
we have:
T
p
(stretch) <T
p
(bend), and
T
p
(light atoms) <T
p
(heavy atoms).
We also note that the light-atom bends (e.g., H–O–H) and the heavy-atom bond
stretches (e.g., C=C) have very similar periods.
In general, the table emphasizes the lack of clear gaps in timescales among
these modes. This explains why constraining some high-frequency motions in
a dynamic formulation typically affects other vibrational modes and why the
computational benefit (larger permitted timestep) is relatively modest.
For example, if we constrain all bonds for a water model, thereby eliminating
the motion and stability limit corresponding to the O–H stretch (period ∼ 10 fs),
the next relevant period that limits the timestep is the H–O–H bend (period ∼ 21
fs), followed by water libration (∼ 42 fs). Thus, the effective timestep can at most
be doubled by constrained dynamics.
Table 14.1. The timestep limit for Verlet based on a harmonic oscillator analysis.
Vibrational Mode
a
Wave number Period T
p
T
p
/π
(1/λ)[cm
−1
] (λ/c) [fs]
b
[fs]
O–H, N–H stretch 3200-3600 9.8 3.1
C–H stretch 3000 11.1 3.5
O–C–O asymm. stretch 2400 13.9 4.5
C≡C, C≡N stretch 2100 15.9 5.1
C=O (carbonyl) stretch 1700 19.6 6.2
C=C stretch
H–O–H bend 1600 20.8 6.4
C–N–H, H–N–H bend 1500 22.2 7.1
C=C (aromatic) stretch
C–N stretch (amines) 1250 26.2 8.4
Water Libration (rocking) 800 41.7 13
O–C–O bending 700 47.6 15
C=C–H bending (alkenes)
C=C–H bending (aromatic)
a
All values are approximate.
b
The value of the speed of light is taken as c =3.00 × 10
10
cm s
−1
.

14.2. Symplectic Integrators 469
14.2.4 Timestep-Dependent Rotation in Phase Space
Under the linear stability assumption, the matrix S of eq. (14.7)hasthetwo
eigenvalues exp(±iθ) (i =
√
−1), where
θ =2sin
−1
(ω Δt/2) (14.12)
= ωΔt +
1
24
(ω Δt)
3
+ O(ω Δt)
5
. (14.13)
Thus, the angle θ depends on the timestep (and on ω). To see that this trans-
formation defines a rotation in phase space, we decompose the phase-space
transforming matrix S as
S = DQD
−1
. (14.14)
In this definition, the matrix
Q =
cos θ sin θ
−sin θ cos θ
(14.15)
defines a rotation of −θ radians in phase space, and D is the diagonal matrix
D =
10
01+tan
2
θ
2
. (14.16)
Behavior of the integrator in time can be interpreted through analysis of the
powers of S given by
S
n
= DQ
n
D
−1
,
where
Q =
cos nθ sin nθ
−sin nθ cos nθ
. (14.17)
The timestep-dependent behavior of the transformation S can be interpreted as
follows. Equations (14.12)and(14.13) show that the integrator is using θ as an
approximation to the exact rotation ωΔt. The smaller the timestep, the closer the
approximation. This is shown in Figure 14.1, where rapid divergence from the
target value (dashed diagonal line) is evident as Δt is increased.
The effective rotation θ
eff
can thus be defined as:
θ
eff
= ω
eff
Δt. (14.18)
For the Verlet method, the effective rotation is given by equation (14.12):
θ
verlet
eff
=2sin
−1
(ωΔt/2) . (14.19)

470 14. Molecular Dynamics: Further Topics
π/2 π
π/2
π
Verlet
θ
eff
θ
eff
, effevtive rotation
desired rotation,ω Δt
Figure 14.1. Theeffectiverotationθ
verlet
eff
(in radians), eq. (14.19), plotted against the
desired rotation, ωΔt, for the Verlet scheme.
14.2.5 Resonance Condition for Periodic Motion
For periodic motion with natural frequency ω, nonphysical resonance — an arti-
fact of the symplectic integrator — can occur when ω is related by integers n and
m to the forcing frequency (2π/Δt):
n
m
ω =
2π
Δt
. (14.20)
Here n is the resonance order, and the integers n and m are relatively prime.
Now recall that above we have shown that the Verlet method has the timestep-
dependent frequency ω
eff
given by θ
verlet
eff
/Δt where θ
verlet
eff
is defined in
eq. (14.19); this frequency thus depends on the timestep in a nonlinear way. Given
this frequency, the integrator-dependent resonance condition becomes
n
m
ω
eff
=
2π
Δt
. (14.21)
A “resonance of order n : m” means that n phase space points are sampled in m
revolutions:
nθ
eff
= nΔtω
eff
=2πm .
This special, finite-coverage of phase space can lead to incorrect, limited sampling
of configuration space. See Figure 14.2 for an illustration.
As was shown in [821], equation (14.19) can be used to formulate a condition
for a resonant timestep for the harmonic oscillator system. That is, using
ω
verlet
eff
=
2sin(ωΔt/2)
Δt
(14.22)

14.2. Symplectic Integrators 471
with the resonance condition of eq. (14.21), we have
ωΔt
2
=sin
mπ
n
. (14.23)
Equivalently,
Δt
verlet
n:m
=
2
ω
sin
mπ
n
=
T
p
π
sin
mπ
n
, (14.24)
where T
p
is the natural period of the oscillator.
Table 14.2 lists the values for low-order resonances (which are the most severe)
for m =1based on a period T
p
=10fs(orω =0.63 fs
−1
). This period corre-
sponds to the fastest period in biomolecular simulations, such as an O–H bond
stretch (see Table 14.1). For n =2,wehavelinear stability.
Clearly, since the limiting timesteps Δt
n:1
for resonance orders n>2 are
smaller than the linear stability limit (Δt
2:1
), resonance limits the timestep to
values lower than classical stability. Since the third-order resonance leads to in-
stability and the fourth-order resonance often leads to instability, in practice we
require Δt<Δt
4:1
. This implies for Verlet a stricter restriction than eq. (14.10),
which comes from the second-order resonance (or linear stability), to
Δt<
√
2/ω , (14.25)
which corresponds to the fourth-order resonance (Δt
2:1
and Δt
4:1
are listed in
Table 14.2).
14.2.6 Resonance Artifacts
For nonlinear systems, the effective forcing frequency for Verlet is not known, but
for relatively small energy values, it is reasonable to approximate this effective
frequency by expressions known for harmonic oscillators, as derived above. Such
approximations can be appropriate, as shown in [821] and Figure 14.2.
This figure from [821] shows the resonance artifacts of the Verlet scheme in
terms of peculiar phase-space diagrams (position versus momentum plot) ob-
tained for a Morse oscillator
3
integrated at Δt = 2 fs for increasing initial energies
(or corresponding temperatures). Around this timestep, resonances of order 6 and
higher are captured (see the reference resonant timesteps based on linear analy-
sis in Table 14.2). The 6:1 resonance shows the 6 islands (each a closed orbit;
see inset) covered in one revolution. As the initial energy increases, so does the
resonance order: see the 13:2 and 20:3 resonances near the box periphery. Anal-
ysis of stability and resonance for MTS schemes can be similarly performed by
extending the model to more than one frequency [94,1089].
3
The Morse bond potential and its relation to the harmonic potential is discussed in the Bond
Length Potentials section of Chapter 9 (see eq. (9.7)).
