Roff D.A. Modeling Evolution: An Introduction to Numerical Methods
Подождите немного. Документ загружается.

# Calculate new mean genetic value by applying fitness criterion
B <- SELECTION(X,N)
mu <- B[1] # New mu
N <- B[2] # New population size
} # End of Igen loop
par(mfrow¼c(2,2)) # Divide graphics page into quadrats
plot(Output[10:MaxGen,1], Output[10:MaxGen,2], type¼‘l’,
xlab¼‘Generation’, ylab¼‘Trait value’)
plot(Output[10:MaxGen,1], Output[10:MaxGen,3],type¼‘l’,
xlab¼‘Generation’,ylab¼‘Population Size, N’)
# Print out mean trait value and mean population size
c(mean(Output[2000:MaxGen,2]), mean(Output[1000:MaxGen,3]))
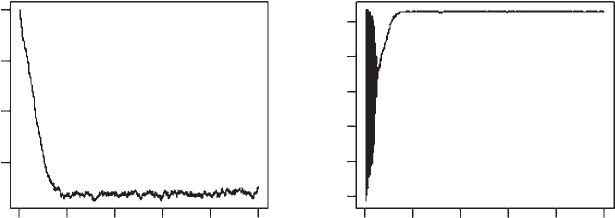
OUTPUT: (Figure 4.6)
> c(mean(Output[2000:MaxGen,2]), mean(Output[1000:MaxGen,3]))
[1] 2.752786 364.705144
After considerable fluctuation both the trait value (a) and population size settle
down to values more or less the same as those equilibria obtained in the invasi-
bility and elasticity analyses (2.73 and 368 [latter not previously given]).
4.8 Scenario 6: Evolution of two traits using an IVC model
The advantage of an IBM using a variance components approach is that it readily
accommodates scenarios that are difficult to incorporate in a population variance
components approach, particularly if one is interested in the effect of small popula-
tion sizes. The following scenario assumes selection on one trait that is negatively
correlated with another trait. The component that is difficult to incorporate in a
population variance components approach is that fitness is a truncated distribution.
Generation
0 2,000 4,000 6,000 8,000 10,000
Generation
0
100
46810
200
Population Size, N
Trait value
300
2,000 4,000 6,000 8,000 10,000
Figure 4.6 Results for Scenario 5.
258 MODELING EVOLUTION

4.8.1 General assumptions
1. Generations are non-overlapping.
2. There is a negative correlation (both genetic and phenotypic) between the two
traits X and Y .
3. Fitness is equal to the value of trait X for X > 0. If trait X is negative fitness is
zero. A simple example of this is if the realized manifestation of trait X is itself
fecundity.
4.8.2 Mathematical assumptions
1. The two traits follow the usual quantitative genetic assumptions.
2. Males and females have the same phenotypic values and subject to the same
selection. Thus the fitness of an individual, W
i
, with trait value X
i
is given by
if X
i
> 0z
i
¼ X
i
else z
i
¼ 0
W
i
¼
z
i
X
N
Pop
i¼1
z
i
ð4:39Þ
where N
Pop
is the number of individuals in the population.
3. Mating is random. Because males and females are identical in this scenario the
new genetic mean of trait X, m
GX
,is
m
GX
¼
P
N
Pop
i¼1
W
i
X
GXi
P
N
Pop
i¼1
W
i
ð4:40Þ
where X
GXi
is the genetic value of the ith individual. Because of the selection on
trait X, the mean genetic value of trait Y is given by
m
GY
¼
P
N
Pop
i¼1
W
i
X
GYi
P
N
Pop
i¼1
W
i
ð4:41Þ
4.8.3 Analysis
For the purpose of illustration the heritabilities are set at 0.4 and 0.5 for X and Y,
respectively. The genetic and phenotypic correlations are set at 0.8 and 0.7,
respectively. From these values plus the phenotypic variances (set at 1 and 0.5) the
genetic, and environmental variance–covariance matrices are calculated as de-
scribed in Section 1.2. Remember to load the library MASS, which is required for
the multivariate normal routine. The general flow of the program is
GENETIC MODELS 259
1. Selection function
2. Set parameter values
3. Enter loop for iterating over generations
4. Generate genetic, environmental, and phenotypic values using mvrnorm
5. Apply selection
6. Next generation
7. Plot results
R CODE:
rm(list¼ls()) # Clear workspace
library(MASS) # Load library MASS
SELECTION <- function(Trait.P, Trait.A)
{
# Determine fitness from trait X (¼trait 1)
# Traits with negative value have zero fitness, e.g. zero fecundity
Fec <- Trait.P[,1] # Preliminary fecundities
Fec[Trait.P[,1]<00] <- 0 # Adjust fecundity of individuals
Total.Fec <- sum(Fec) # Total fecundity of popn
NewX1 <- sum(Fec*Trait.A[,1])/Total.Fec # New mean X
NewX2 <- sum(Fec*Trait.A[,2])/Total.Fec # New mean Y
Mean.A <- c(NewX1, NewX2) # Combine
return(Mean.A)
} # End of function
####################### MAIN PROGRAM #######################
set.seed(100) # Initialize random number generator
Npop <- 1000 # Population size
MaxGen <- 10 # Nos of generations for simulation
Output <- matrix(0,MaxGen,3) # Allocate storage for trait
################### Create initial matrices ###################
# In this version the sexes are ignored
# This assumes that selection acts equally
# Give.Hmatrix is a dataframe with genetic correlations in upper
diagonal
# heritabilities along the diagonal and phenotypic correlations in
the lower diagonal
NX <- 2 # Number of traits
# Matrix of heritabilities and correlations
H2 <- matrix(c(0.4,-0.8,
-0.7, 0.5), 2,2, byrow¼TRUE)
Mean.A <- c(3, 3) # Initial additive genetic means
Mean.E <- c(0,0) # Environmental means
Var.P <- c(1, 0.5) # Phenotypic variances
# Phenotypic Covariance matrix
# Note that initial covariances set to 1 (arbitrary)
CovP <- matrix(1,NX,NX) # Phenotypic variances
260 MODELING EVOLUTION
diag(CovP) <- Var.P # Diagonal elements ¼ variances
# Establish CovA from h2 and CovP and CovE from CovA and CovP
CovA <- matrix(0,NX,NX) # Allocate memory for genetic matrix
CovE <- matrix(0,NX,NX) # Allocate memory for envir. matrix
for ( i in 1:NX) # Iterate over components of (co)variance matrix
{
CovA[i,i] <- CovP[i,i]*H2[i,i] # Genetic variance ¼ Vp*h2
CovE[i,i] <- CovP[i,i]-CovA[i,i] # Environmental covariances
if(CovE[i,i] < 0) stop (print(c(“CovE cannot be”,i,j,CovE[i,i])))
}
# Phenotypic and genetic covariances
N.minus.1 <- NX-1
for( i in 1:N.minus.1)
{
jj <-iþ1
for(j in jj:NX)
{
CovP[i,j] < CovP[j,i] <- CovP[i,j] # Matrix symmetrical
CovA[i,j] <- H2[i,j]*sqrt(CovA[i,i]*CovA[j,j])# Genetic covariance
CovA[j,i] <- CovA[i,j] # Matrix symmetrical
CovE[i,j] <- CovP[i,j]-CovA[i,j] # Environ. covariance
CovE[j,i] <- CovE[i,j]
} # End of j loop
} # End of i loop
###################### Start Simulation ######################
for (Igen in 1:MaxGen) # Iterate to MaxGen
{
# Generate additive and environmental values
Trait.E <- mvrnorm(Npop, mu ¼Mean.E, Sigma¼CovE) # Environ.
Trait.A <- mvrnorm(Npop, mu¼Mean.A, Sigma¼CovA) # Genetic
Trait.P <- Trait.A þ Trait.E # Phenotypic
# Store data
Output[Igen,1] <- Igen
Output[Igen,2] <- mean(Trait.P[,1]) # X
Output[Igen,3] <- mean(Trait.P[,2]) # Y
Mean.A <- SELECTION(Trait.P, Trait.A) # New X and Y
} # End of Igen loop
# Plot results
ymin <- min(Output[,2:3]); ymax <- max(Output[,2:3]) # Trait X
plot(Output[,1], Output[,2], xlab
¼“Generations”,
ylab¼“Trait
X (solid) and Y (dotted)”, type¼‘l’, ylim ¼ c(ymin, ymax)) # Trait
X on generation
lines(Output[,1], Output[,3], lty¼2) # Trait Y on generation
OUTPUT: (Figure 4.7)
GENETIC MODELS 261
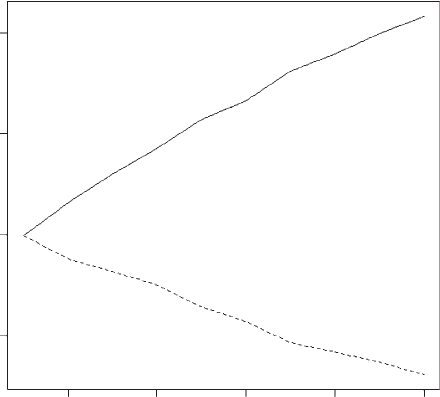
The figure shows that selection on trait X increases trait X but, because of the
negative correlation, trait Y declines in value.
4.9 Scenario 7: Evolution of two traits using an IL model
The general scenario is similar to Scenario 4 in which fitness is a threshold
function and to Scenario 6 in which two negatively correlated traits are modeled.
The basic coding follows which is similar to the one discussed in Section 1.3.
4.9.1 General assumptions
1. Generations are non-overlapping.
2. There is a negative correlation (both genetic and phenotypic) between the two
traits X and Y.
3. Fitness is a threshold value, such that individuals above some threshold
T0 contribute equally to the next generation whereas individuals below T0
have zero fitness.
4.9.2 Mathematical assumptions
1. Males and females do not differ in the expression of the trait. They are not
separately distinguished in this model.
2. Mating between selected individuals is at random and can include selfing.
3. Selected individuals do not differ in their fitness.
Generations
2
2.5 3.0
Trait X (solid) and Y (dotted)
3.5 4.0
46810
Figure 4.7 Results for Scenario 6.
262 MODELING EVOLUTION
4. The threshold for selection is set at the initial mean value of trait X
5. The phenotypic value is the sum of allelic values, which take values of 0 or 1,
plus an environmental deviation.
6. Parameter values are as follows:
a. Population size is 1,000.
b. Number of unique loci is 30.
c. Number of loci in common is 25.
d. Initial frequency at unique loci is 0.68.
e. Initial frequency at common loci is 0.47.
f. Initial genetic correlation given the forgoing values is 0.488.
g. Initial trait value of X is 64.3 and of Y is 17.3.
h. Initial heritabilities are 0.4 and 0.5.
i. Initial phenotypic correlation is 0.7.
4.9.3 Analysis
The general flow of the model is described in detail in Section 1.3. Simulation
proceeds in the following steps:
1. Assign parameter values as given above.
2. Calculate other required parameters (e.g., r
E
).
3. Set up initial genotypes using function ASSIGN.LOCI.
4. Set up selection threshold for required generations of selection (matrix T0). The
threshold is set for the first generations such that there is no selection, allowing
the stabilization of genotypic frequencies. After five generations the threshold
is set to the initial value of X (Trait.X).
5. Iterate over generations. Within this loop we
5a. Calculate genotypic values using rowSums
5b. Create environmental values using mvrnorm
5c. Add the environmental values to generate phenotypic values
5d. Apply function SELECTION
5e. Create the gamete pool using function GAMETE
5f. Get gametes from pool and create the next generation of genotypes
5g. Apply function MUTATION
6. Plot results.
R CODE:
rm(list¼ls()) # Remove all objects from memory
ASSIGN.LOCI <- function(G.loci, N.Pop, P)
{
GENETIC MODELS 263
Total.loci <- N.Pop*2*G.loci # Total number of loci in population
Alleles <- runif(Total.loci) # Generate random number 0-1
Alleles[Alleles < P] <- 0 # Allocate to 0s
Alleles[Alleles > P] <- 1 # Allocate to 1s
return(Alleles)
} # End of function
####################### SELECTION #######################
SELECTION <- function(Phenotype, Genotype, T0) # Selection
function
{
Selected <- Genotype[Phenotype >T0,] # Selection
return(Selected) # Return selected genotypes
} # End function
####################### GAMETE #######################
GAMETE <- function(X, G.loci) # Pick loci for gamete pool
{
Y <- sample(x¼X, size¼G.loci, replace¼FALSE) # Randomly select G.loci
return(Y)
}
################## Function Mutation ##################
MUTATION <- function(X, Pmut, G.loci, N.inds)
{
T.loci <- N.inds*2*G.loci
lambda <- Pmut*T.loci # Mean number of mutations in population
# Number of mutations using a Poisson distribution
N.mutations <- rpois(1,lambda)
# Randomly select N.mutations rows
Row <- ceiling(runif(N.mutations, min¼0, max¼T.loci))
Temp <- matrix(X) # Convert to a vector
Temp[Row] <- (abs(Temp[Row]-1)) # Convert mutated rows
X <- matrix(Temp, N.inds, 2*G.loci) # Convert back to matrix
return(X)
} # End function
####################### Main Program #######################
set.seed(10) # Initialize random number generator
N.Pop <- 1000 # Population size at each generation
X.loci <- Y.loci<- 30 # Loci per gamete unique to X
C.loci <- 25 # Loci per gamete common to X and Y
h2.X <- 0.4 # heritability of X
h2.Y <- 0.5 # Heritability of Y
S <
- -1 # Sign of genetic correlation
Rp <-
-0.7 # Phenotypic correlation
Pxy <- 0.68 # Proportion at x or y loci
Pc <- 0.47 # Proportion of c loci
TraitX <- 2*(Pxy*X.lociþPc*C.loci) # Initial mean trait value of X
264 MODELING EVOLUTION
TraitY <- 2*(Pxy*X.lociþS*Pc*C.loci) # Initial mean trait value of Y
VarGX <- 2*(X.loci*Pxy*(1-Pxy)þ C.loci*Pc*(1-Pc)) # Vg of X
VarGY <- VarGX # Vg of Y
CovGXY <- 2*C.loci*Pc*(1-Pc) # Genetic covariance
Rg <- S*CovGXY/sqrt(VarGX*VarGY) # Genetic correlation
print(c(Rg, TraitX, TraitY)) # Print values
# Calculate the environmental correlation
Re <- (Rp - Rg*sqrt(h2.X*h2.Y))/sqrt((1-h2.X)*(1-h2.Y))
# Check that this Re is possible
if (abs(Re)>1) stop (c(“Re not possible”))
# Environmental Variances and Standard deviations
Ve.X <- (1-h2.X)*VarGX/h2.X # Environmental variance for X
SDe.X <- sqrt(Ve.X) # Environmental SD for X
Ve.Y <- (1-h2.Y)*VarGY/h2.Y # Environmental variance for Y
SDe.Y <- sqrt(Ve.Y) # Environmental SD for Y
CovE <- Re*SDe.X*SDe.Y # Environmental covariance
Ematrix <- matrix(c(Ve.X,CovE,CovE,Ve.Y),2,2) # Covariance ma-
trix
# Nos of loci in each category
Nx.Alleles <- ASSIGN.LOCI(X.loci, N.Pop, Pxy)# Alleles unique to X
Ny.Alleles <- ASSIGN.LOCI(Y.loci, N.Pop, Pxy)# Alleles unique to Y
Nc.Alleles <- ASSIGN.LOCI(C.loci,N.Pop, Pc) # Alleles common toX&Y
# Now make three matrices for loci in individuals
G.Xmatrix <- matrix(Nx.Alleles, N.Pop, 2*X.loci) # X composition
G.Ymatrix <- matrix(Ny.Alleles, N.Pop, 2*Y.loci) # Y composition
G.Cmatrix <- matrix(Nc.Alleles, N.Pop, 2*C.loci) # C composition
################## Iterate over generations ##################
Maxgen <- 40 # Number of generations simulation runs
Output <- matrix(0, Maxgen,9) # Allocate space for output
T0 <- matrix(TraitX,Maxgen,1) # Set T0 for generations
T0[1:5] <- -100 # Set T0 so that 1st 5 gens there is no selection
for (Igen in 1:Maxgen) # Iterate over generations
{
# Get actual genotypic values
G.X <- rowSums(G.Xmatrix) þrowSums(G.Cmatrix) # X Genotypic values
VarGX <- var(G.X) # Vg for X
G.Y <- rowSums(G.Ymatrix) þ S*rowSums(G.Cma trix)# Y Genotypic values
VarGY <- var(G.Y) # Vg for Y
# Create phenotypic values
En
v <- mvrnorm(n¼N.Pop, mu¼c(0,0), Sigma¼Ematrix) # Environmental
values
P.X <- G.X þ Env[,1] # Vector of X phenotypes
P.Y <- G.Y þ Env[,2] # Vector of Y phenotypes
VarPX <- var(P.X) # Phenotypic variance of X
VarPY <- var(P.Y) # Phenotypic variance of Y
h2.X <- VarGX /VarPX # Heritability of X
GENETIC MODELS 265
h2.Y <- VarGY/VarPY # Heritability of Y
Rg <- cor(G.X, G.Y) # Genetic correlation
Rp <- cor(P.X, P.Y) # Phenotypic correlation
# Store results
Output[Igen,1:9] <- c(Igen, VarGX, VarGY, mean(P.X), mean(P.Y),
h2.X, h2.Y, Rg, Rp)
# Apply Selection. Note that selection here is only a function of X
ParentX <- SELECTION(P.X, G.Xmatrix, T0[Igen])
ParentY <- SELECTION(P.X, G.Ymatrix, T0[Igen])
ParentC <- SELECTION(P.X, G.Cmatrix, T0[Igen])
# Form Gamete pool
GameteX <- apply(ParentX, 1, GAMETE, X.loci)
GameteX <- t(GameteX) # Convert to proper matrix
GameteY <- apply(ParentY, 1, GAMETE, Y.loci)
GameteY <- t(GameteY) # Convert to proper matrix
GameteC <- apply(ParentC, 1, GAMETE, C.loci)
GameteC <- t(GameteC) # Convert to proper matrix
N.Parents <- nrow(ParentX) # Number of available parents
n <- seq(1, N.Parents) # sequence 1 to N.Parents
# Get 2*N.Pop random indices with replacement
G.Index <- sample(x¼n, size¼2*N.Pop, replace¼TRUE)
# Get gametes from gamete pool
S.GameteX <- GameteX[G.Index,]
S.GameteY <- GameteY[G.Index,]
S.GameteC <- GameteC[G.Index,]
# Form next generation
n1 <- N.Popþ1
n2 <- 2*N.Pop
G.Xmatrix <- cbind(S.GameteX[1:N.Pop,], S.GameteX[n1:n2,])
G.Ymatrix <- cbind(S.GameteY[1:N.Pop,], S.GameteY[n1:n2,])
G.Cmatrix <- cbind(S.GameteC[1:N.Pop,], S.GameteC[n1:n2,])
# Mutations
Pmut <- 0.0001 # Mutation probability
G.Xmatrix <- MUTATION(G.Xmatrix, Pmut, X.loci, N.Pop)
G.Ymatrix <- MUTATION(G.Ymatrix, Pmut, Y.loci, N.Pop)
G.Cmatrix <- MUTATION(G.Cmatrix, Pmut, C.loci, N.Pop)
} # Next generation
par(mfrow¼c(2,2))
#
Plot phenotypic value on generation
ymin <- min(Output[,4:5]); ymax <- max(Output[,4:5]) # Limits on y
plot( Output[,1], Output[,4], xlab¼‘Generation’, ylab¼‘Pheno-
types’, type¼‘l’, ylim¼c(ymin,ymax))
lines(Output[,1], Output[,5], lty¼2)
# Plot genetic variances on generation
ymin <- min(Output[,2:3]); ymax <- max(Output[,2:3]) # Limits on y
266 MODELING EVOLUTION

plot( Output[,1], Output[,2], xlab¼‘Generation’, ylab¼‘Genetic
variances’, type¼‘l’, ylim¼c(ymin,ymax))
lines(Output[,1], Output[,3], lty¼2)
# Plot heritabilities on generation
ymin <- min(Output[,6:7]); ymax <- max(Output[,6:7]) # Limits on y
plot( Output[,1], Output[,6], xlab¼‘Generation’, ylab-
¼‘Heritabilities’, type¼‘l’, ylim¼c(ymin,ymax))
lines(Output[,1], Output[,7], lty¼2)
# Plot correlations on generation
ymin <- min(Output[,8:9]); ymax <- max(Output[,8:9]) # Limits on y
plot( Output[,1], Output[,8], xlab¼‘Generation’, ylab-
¼‘Correlations’, type¼‘l’, ylim¼c(ymin,ymax))
lines(Output[,1], Output[,9], lty¼2)
OUTPUT: (Figure 4.8)
Generation
0
–0.7
–20
15 20 25
0 20406080
0.30 0.40 0.50
–0.5
Correlations
Genetic variances
Heritabilities
Phenotypes
–0.3
10 20 30 40
Generation
0 10203040
Generation
0 10203040
Generation
0 10203040
Figure 4.8 Results for Scenario 7. Solid lines shows results for trait X and dotted line shows
results for trait Y, except for the plot of correlations in which case the solid line gives
the genetic correlation and the dotted line the phenotypic correlation.
GENETIC MODELS 267
