Roff D.A. Modeling Evolution: An Introduction to Numerical Methods
Подождите немного. Документ загружается.

N.resident <- N.invader <- matrix(0,Tot.Gen) # Allocate space
N.resident[1] <- 1 # Initial pop size of resident
N.invader[Maxgen1] <- 1 # Initial pop size of invader
for (Igen in 2:Maxgen1) # Iterate over generations with resident
only
{
N <- N.resident[Igen-1]
N.resident[Igen] <- DD.FUNCTION(ALPHA.resident, N, N)
} # End of 1st Igen loop
# Now add invader
J <- Maxgen1þ1 # Starting generation
for (Igen in J:Tot.Gen) # Iterate over generations with invader
{
N <- N.resident[Igen-1]þ N.invader[Igen-1] # Total pop size
N.resident[Igen] <- DD.FUNCTION(ALPHA.resident, N.resident
[Igen-1], N)
N.invader[Igen] <- DD.FUNCTION(ALPHA.invader, N.invader[Igen-
1], N)
} # End of 2nd Igen loop
Generation <- seq(from¼1, to¼Tot.Gen) # Vector of generation
numbers
plot (Generation, N.resident, xlab¼’Generation’, ylab¼’Resident
N’, type¼’l’)
plot (Generation, N.invader, xlab¼’Generation’, ylab¼’Invader
N’, type¼’l’)
} # End of function
############## MAIN PROGRAM ##############
par(mfrow¼c(2,2)) # Divide graphics page into quadrats
Best.A <- 24.21635 # Best alpha from elasticity analysis
Invader.A <- 10 # Alpha for invader
Coeff <- Invader.A/Best.A # Calculate relevant coefficient
POP.DYNAMICS( Best.A,Coeff) # Call functiom
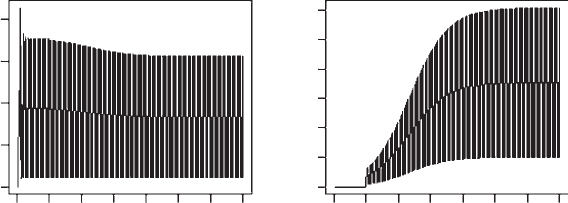
OUTPUT: (Figure 3.18)
Generation
Resident N
Invader N
0
0
0 102030405060
200 400 600 800
50 100 200 300
Generation
0 50 100 200 300
Figure 3.18 Temporal plots of resident and invader populations. Left‐hand plot shows the
population trajectory for the optimum a and the right‐hand plot shows the population
trajectory for an invader with a = 10.
218 MODELING EVOLUTION
It is evident from the output that an invader with ¼ 10 can invade the popula-
tion but not replace the resident population. This explains the relation between
the invasion coefficient and the optimum and emphasizes the importance of
elasticity analysis in conjunction with pairwise invasibility analysis.
3.8.5 Multiple invasibility analysis
The program is the same as in Scenario 6 except for: (a) it is run 10,000 generations
to ensure sufficient time for equilibrium to be attained (if possible), ( b) because the
elasticity analysis indicated that a single equilibrium is unlikely, frequency poly-
gons are plotted for the last three generations.
R CODE:
rm(list¼ls()) # Clear memory
DD.FUNCTION<- function(X, N.total) # Density-dependence function
{
# Set parameter values
ALPHA <- X[1]; N <- X[2]
Amin <-1;Amax<- 50; Bmin <-0;Bmax<-0.9;a<- 20; BETA <-0.01
AA <- (ALPHA-Amin)/(Amax-Amin) # For convenience
S <- (Bmax-Bmin)*(1-AA)/(1þa*AA) # Survival
N <- N*(ALPHA*exp(-BETA*N.total)þS) # new population
N <- max(0,N) # N cannot be negative
return(N)
} # End of function
############## MAIN PROGRAM ##############
set.seed(10) # Initialize the random number seed
Maxgen <- 10000 # Number of generations run
Stats <- matrix(0,Maxgen,3) # Allocate space for statistics
MaxAlpha <- 50 # maximum value of alpha
Ninc <- 50 # Number of classes for alpha
# Allocate space to store data for each generation
Store <- matrix(0,Maxgen, Ninc)
# Allocate space for alpha class and population size
Data <- matrix(0,Ninc,2)
Data[24,2] <- 1 # Initial population size and alpha class
ALPHA <- matrix(seq(from¼1, to ¼MaxAlpha, length¼Ninc),Ninc,1)
# Set Alpha
Data[,1] <- ALPHA # Place alpha in 1st column
for (Igen in 1:Maxgen) # Iterate over generations
{
N.total <- sum(Data[,2]) # Total population size
Data[,2] <- apply(Data,1,DD.FUNCTION, N.total) # New cohort
Store[Igen,] <
- Data[,2] # Store values for this generation
#
Keep track of population size, mean trait value and SD of trait value
Stats[Igen,2] <- sum(Data[,1]*Data[,2])/sum(Data[,2]) # Mean
S <- sum(Data[,2]) # Population size
Stats[Igen,1] <- S # Population size
INVASIBILITY ANALYSIS 219
SX1 <- sum(Data[,1]^2*Data[,2])
SX2 <- (sum(Data[,1]*Data[,2]))^2/S
Stats[Igen,3] <- sqrt((SX1-SX2)/(S-1)) # SD of trait
# Introduce a mutant by picking a random integer between 1 and 50
Mutant <- ceiling(runif(1, min¼0, max¼50))
Data[Mutant,2] <- Data[Mutant,2]þ1 # Add mutant to class
} # End of Igen loop
par(mfrow¼c(3,2)) # Split graphics page into 3 x 2 panels
for (Row in 9998:Maxgen) # Select rows to be plotted
{
plot(ALPHA, Store[Row,], type¼’l’, xlab¼’Alpha’, ylab¼ ’Number’)
} # End of frequency polygon plots
Generation <- seq(from¼1, to¼Maxgen) # Vector of generations
N0 <- 9900 # Starting value for plots
plot(Generation[N0:Maxgen], Stats[N0:Maxgen,1], ylab¼’Popula-
tion size’,xlab¼’Generat ion’ , type¼’l’)
plot(Generation[N0:Maxgen], Stats[N0:Maxgen,2], ylab¼’Mean’,
xlab¼’Generation’, type¼’l’)
plot(Generation[N0:Maxgen], Stats[N0:Maxgen,3], ylab¼’SD’,
xlab¼’Generation’, type¼’l’)
print(c(’Mean alpha in last gen ¼ ’,Stats[Maxgen,2]))
print(c(’SD of alpha in last gen ¼ ’,Stats[Maxgen,3]))
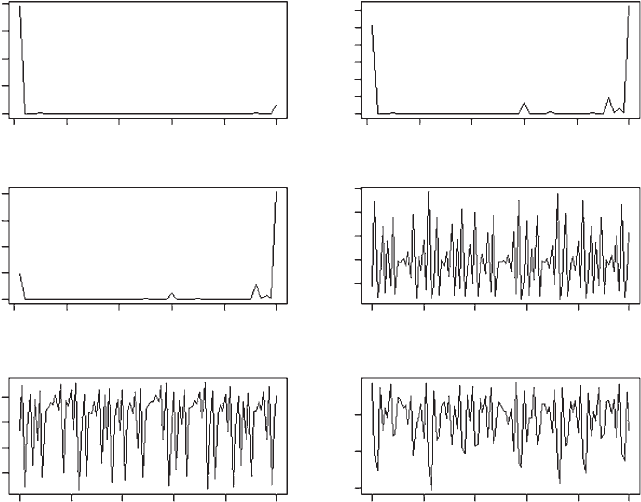
OUTPUT: (Figure 3.19)
Generation
9,900
0
00
0 40 80 120
20 40 60 80
100 300
10 20
Alpha
30 40 50
01020
Alpha
30 40 50 0 10 20
Alpha
30 40 50
10
10
200 600 1,000
15 20
SD
20
Mean
Number
Number
Number
Population size
30 40
9,920 9,940 9,960 9,980 10,000
Generation
9,900 9,920 9,940 9,960 9,980 10,000
Generation
9,900 9,920 9,940 9,960 9,980 10,000
Figure 3.19 Results of multiple invasibility analysis of Scenario 6. The three frequency
polygons show results for generation 9,998, 9,999 and 10,000.
220 MODELING EVOLUTION
> print(c(’Mean alpha in last gen ¼ ’,Stats[Maxgen,2]))
[1] “Mean alpha in last gen ¼ ” “40.6067676012847”
> print(c(’SD of alpha in last gen ¼ ’,Stats[Maxgen,3]))
[1] “SD of alpha in last gen ¼ ” “17.8941418836399”
The frequency polygon plots for the last three generations show dramatic shifts in
the mean and modal values of . As indicated by the previous analyses, the
equilibrium from the pairwise invasibility analysis is highly unstable and is un-
likely to be ever attained, since any small deviations set the population into
extreme fluctuations. The population consists of different clones, with their
frequencies changing through time.
3.9 Some exemplary papers
Katsukawa, Y., T. Katsukawa, and H. Matsuda. 2002. Indeterminate growth is
selected by a trade-off between high fecundity and risk avoidance in stochas-
tic environments. Population Ecology 44:265–272.
Problem: Optimal allocation of energy to growth, survival, and reproduction when
survival is temporally variable
Model type: Two age/stage
Density-dependence: None
Trade-off: Total allocation of energy to survival, growth, and reproduction fixed
Analysis: Maximization of
l
Reference scenario: Scenario 1
Ebenman, B., A. Johansson, T. Jonsson, and U. Wennergren. 1996. Evolution
of stable population dynamics through natural selection. Proceedings of the
Royal Society of London – Series B: Biological Sciences 263:1145–1151.
Problem: Optimal combination of density-dependent parameters when population
dynamics can range from a fixed equilibrium to chaotic
Model type: Two-stage
Density-dependence: Ricker function applied to maturation rate and juvenile
survival
Trade-off: Maturation rate and juvenile survival
Analysis: Invasibility analysis
Reference scenario: Scenarios 3 and 5
Lalonde, R. G. and B. D. Roitberg. 2006. Chaotic dynamics can select for long-
term dormancy. American Naturalist 168:127–131.
Problem: The optimal dormancy when population dynamics are chaotic
Model type: Up to 13 generations of dormancy
Density-dependence: Ricker function
Trade-off: No explicit trade-off
INVASIBILITY ANALYSIS 221
Analysis: Multiple invasibility analysis
Reference scenario: Scenarios 3 and 6
Johst, K., M. Doebeli, and R. Brandl . 1999. Evolution of complex dynamics in
spatially structured populations. Proceedings of the Royal Society of London –
Series B: Biological Sciences 266:1147–1154.
Problem: The optimal density-dependent and density-independent dispersal rates
Model type: One generation per time step
Density-dependence: Fecundity varies with density using an equation that can take a
variety of shapes
Trade-off: No explicit trade-offs
Analysis: Multiple invasibility analysis
Reference scenario: Scenarios 3 and 6
Benton, R. A. and A. Grant. 1999. Optimal reproductive effort in stochastic,
density-dependent environments. Evolution 53:677–688.
Problem: Optimal reproductive effort
Model type: Two age/stage class
Density-dependence: Various
Trade-off: Various combinations of reproductive effort and survival at age with or
without temporal variation
Analysis: Elasticity analysis
Reference scenario: Scenario 4
Wilbur, H. M. and V. H. W. Rudolf. 2006. Life-history evolution in uncertain
environments: Bet hedging in time. American Naturalist 168:398–411.
Problem: Optimal iteroparity and delayed maturity in a stochastic environment
Model type: Two-stage
Density-dependence: Ricker function applied to fertility with parameters selected to
ensure a stable population in a deterministic environment
Trade-off: Adult survival and clutch size in some model variants
Analysis: Invasibility analysis
Reference scenario: Scenarios 4 and 5
White, A., J. V. Greenman, T. G. Benton, and M. Boots. 2006. Evolutionary
behaviour in ecological systems with trade-offs and non-equilibrium popula-
tion dynamics. Evolutionary Ecology Research 8:387–398.
Problem: Optimal reproductive effort when population dynamics are complex
Model type: No explicit age structure
Density-dependence: Ricker function
Trade-off: Density-independent fecundity and survival
Analysis: Invasibility analysis
Reference scenario: Scenario 6
222 MODELING EVOLUTION

CHAPTER 4
Genetic Models
4.1 Introduction
The “Fisherian” optimality approach determines the optimal combination of trait
values but not the evolutionary trajectory nor, in general, the variation to be
expected about the optimal combination. Invasibility analysis is also primarily
concerned with the Evolutionarily Stable Strategy (ESS) values; though assuming
clonal inheritance, it can give insight into the temporal evolution of trait values.
Whether such analyses can be extended to organisms with other modes of inheri-
tance is not clear, though the analysis presented in Scenario 5 of this chapter
suggests that caution is warranted in applying the results of invasibility analysis to
sexually reproducing organisms.
There are basically three ways to implement a genetic model, which I shall refer
to as (a) the population variance components approach, ( b) the individual variance
components approach, and (c) the individual locus approach. The first two are
primarily for quantitative genetic modeling whereas the last can be used for both
quantitative genetic modeling and modeling simple Mendelian inheritance (e.g.,
single locus). The advantages of an individual-based model (IBM) are that it is easy
to incorporate complex population dynamics, frequency-dependence, and func-
tional constraints. IBMs are, however, relatively computer-intensive (particularly
individual locus models).
4.1.1 Population variance components (PVC) models
Rather than focus upon individual loci this method tackles the problem by focus-
ing at the population level upon the variances and covariances of the traits. The
phenotypic value, X, of a trait can be decomposed into the sum of genetic, G, and
environmental, E, components:
X ¼ G þ E ð4:1Þ
where it is assumed that G and E are uncorrelated and normally distributed, the
latter with a zero mean. The phenotypic variance, s
2
P
is thus also composed of the
sum of the genetic, s
2
G
, and the environmental, s
2
E
, variances
s
2
P
¼ s
2
G
þ s
2
E
ð4:2Þ

The genetic variance can itself be broken down into additive, dominance, and
epistatic components, but I shall ignore the latter two, as is typically done.
Heritability is a measure of the extent to which the resemblance between rela-
tives is due to additive genetic effects. A simple way to estimate heritability is to
regress the average trait value of offspring on the mid-parent value. Provided that
there are no maternal (or paternal) effects or environmental effects due to simi-
larity in the rearing environment within but not among families, the slope of this
regression is equal to the heritability of the trait. The general definition of herita-
bility, h
2
, is the ratio of additive genetic variance to phenotypic variance:
h
2
¼
s
2
G
s
2
P
ð4:3Þ
Heritability measures the proportion of the total variance that can be ascribed to the
additive effect of genes. A zero heritability means that variation is entirely environ-
mental whereas a heritability of one means that the variation is entirely due to
the additive effects of genes. The general relationship between offspring and par-
ents is
m
Offspring
¼ð1 h
2
Þm
Population
þ h
2
m
Parents
ð4:4Þ
where m refers to the mean values. Under directional selection the mean value of
parents is greater or less that of the population, depending on the direction of
selection, and by rearranging equation 4.4 we get the response to the selection as
m
Offspring
m
Population
¼ h
2
ðm
Parents
m
Offspring
Þ
R ¼ h
2
S ¼
s
2
G
s
2
P
S
ð4:5Þ
where S is the selection differential. The above equation is referred to as the
Breeder’s equation. It is frequently more convenient to write S in terms of
phenotypic standard deviations:
S ¼ is
P
ð4:6Þ
where i is called the selection intensity,
i ¼
S
s
P
¼
z
P
ð4:7Þ
P is the proportion selected for each generation and z is the ordinate at the point of
truncation. Suppose we wish to apply truncation selection by selecting the top P
proportion of the population, the selection intensity can be calculated using the
following R code:
Vp <- 1 # Phenotypic variance
P <- 0.2 # Proportion selected
x <- -qnorm(P, mean ¼ 0, sd ¼ 1) # x value
z <- exp(-0.5*x^2)/(sqrt(2*pi)) # z
224 MODELING EVOLUTION

i <- z/P # Selection intensity
S <- i*sqrt(Vp) # Selection differential
The change per generation is a constant and hence a plot of mean trait value on
generation will give a straight line (i.e., m
tþ1
¼ m
t
þ h
2
S).
In nature, selection is likely to be generally stabilizing rather than directional.
Two functions often used for such a scenario are a quadratic fitness function and a
Gaussian fitness function. Under the latter function the fitness of a trait, W, can be
written as
W ¼ e
1
2
ðxyÞ
2
o
ð4:8Þ
where y is the value at which fitness is maximized and o is a measure of the
strength of stabilizing selection, the strength of selection decreasing with increas-
ing o. Stabilizing selection can be incorporated into the single trait model as
R
t
¼
s
2
G
o þ s
2
P
ðy m
Population;t
Þð4:9Þ
Note that the response to selection is now a function of the distance from the
optimum value and hence is a function of generation. Over time the population
moves smoothly to the optimum value.
Extension of the above model to multiple traits requires the introduction of the
correlation between traits. Correlations can arise because genes may affect several
traits (called pleiotropy), be associated because of linkage or because of environ-
mental factors. Assuming, as is generally done, that the environmental and genet-
ic covariances are independent we can break the phenotypic covariance between
traits X and Y, s
PXY
, into the sum of the two covariances:
s
PXY
¼ s
GXY
þ s
EXY
ð4:10Þ
Noting that a correlation, r, is equal to
r ¼
s
XY
s
X
s
Y
ð4:11Þ
the phenotypic correlation can be obtained from the above relationship as
r
PXY
¼ r
G
s
GX
s
GY
s
PX
s
PY
þ r
E
s
EX
s
EY
s
PX
s
PY
¼ r
G
ffiffiffiffiffiffiffiffiffiffi
h
2
X
h
2
Y
q
þ r
E
ffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffiffi
ð1 h
2
X
Þð1 h
2
Y
Þ
q
ð4:12Þ
For multiple traits the Breeder’s equation can be expanded using matrix nota-
tion as
D z
¼ GP
1
S ð4:13Þ
where is D
z the vector of changes in trait means, G is the additive genetic variance–
covariance matrix, P is the phenotypic variance–covariance matrix, S is the vector of
GENETIC MODELS 225
selection differentials, and the 1 means the matrix inverse. For two traits, labeled 1
and 2, equation (4.13) can be written in explicit matrix format as
D
z
1
D
z
2
¼
s
2
G1
s
G12
s
G21
s
2
G2
s
2
P1
s
P12
s
P21
s
2
P2
1
S
1
S
2
ð4:14Þ
The diagonal elements of the genetic and phenotypic matrices hold the variances
and the off-diagonals the covariances (note that the matrices are symmetrical and
hence s
ij
¼ s
ji
). The response to directional truncation selection is the same as in
the single trait model, in that both traits change by a fixed amount per generation,
though in this case the amount is a function of the selection differential and the
correlation between the traits.
To set the values in equation (4.14) it is generally easier to commence with the
heritabilities, phenotypic variances, and correlations, and then make use of
equations (4.3) and (4.12). For two traits the coding is simple: for example,
h2 <- c(0.2,0.4) # Set heritabilities
Vp <- c(1,2) # Set phenotypic variances
Rp <- 0.4 # Set phenotypic correlation
Ra <- 0.15 # Set genetic correlation
Va <- h2*Vp # Using h2 ¼Va/Vp
Covp <- Rp*sqrt(Vp[1]*Vp[2]) # Using r ¼ Cov/SD1SD2
Cova <- Ra*sqrt(Va[1]*Va[2]) # Using r ¼ Cov/SD1SD2
Gmatrix <- matrix( c(Va[1],Cova, Cova, Va[2]),2,2) # G matrix
Pmatrix <- matrix(c(Vp[1],Covp, Covp, Vp[2]),2,2) # P matrix
The environmental correlation must lie between 1andþ1, which is not
guaranteed by the above coding. It is therefore necessary to put in a check,
using equation (4.12) and stop the program if r
E
falls outside the permissible
limits:
# Check that Re is possible
Re <- (Rp-Ra*sqrt(h2[1]*h2[2]))/sqrt((1-h2[1])*(1-h2[2]))
if(abs(Re) >1 ) stop (c(“problem with Re”))
The value of the heritability is unlikely to matter in terms of the final equilibrium
but will certainly affect the time taken to reach this value, should it exist, and the
variation about the value.
With more than two traits the above coding can be tedious but it can be
replaced by the following more general code. Suppose we have three traits. First
we construct a matrix, which I have called H2, which holds the heritabilities on
the diagonal, the genetic correlations above the diagonal and the phenotypic
correlations below the diagonal. We then use this matrix in conjunction with
the phenotypic variances (assumed to be 1.5, 1.0, and 0.5 in the example below) to
construct the necessary covariance matrices:
226 MODELING EVOLUTION
NX <- 3 # Number of traits
# Matrix of heritabilities and correlations.
# Values in bold are the heritabilities
# Genetic correlations are above the diagonal, phenotypic correl-
ations below
H2 <- matrix (c (0.4, 0.7, 0.3,
0.6, 0.5, 0.1,
0.4, 0.6, 0.3), NX, NX, byrow¼TRUE)
# Construct the Phenotypic Covariance matrix
# Note that initial covariances are set to 1 (arbitrary)
CovP <- matrix (1, NX, NX) # Phenotypic variances
diag(CovP) <- c (1.5,1.0,0.5)# Sets diagonal elements ¼variances
# Establish CovA from h2 and CovP and CovE from CovA and CovP
CovA <- matrix (0, NX, NX) # Allocate space
CovE <- matrix (0, NX, NX) # Allocate space
for ( i in 1:NX) # Iterate over cells
{
CovA[i,i] <- CovP[i,i]*H2[i,i] # ¼ Vp*h2
CovE[i,i] <- CovP[i,i]-CovA[i,i] # Variances
# Check that environmental variance is positive
if (CovE[i,i] < 0) stop (c(“Problem with CovE”))
}
# Phenotypic and genetic covariances
N. minus. 1 <- NX-1
for ( i in 1:N.minus.1)
{
jj <-iþ1
for (j in jj:NX)
{
# Phenotypic covariances
CovP [i,j] <- H2[j,i]*sqrt (CovP[i,i]*CovP[j,j])
CovP[j,i] <- CovP[i,j]
CovA[i,j] <- H2[i,j]*sqrt (CovA[i,i]*CovA[j,j]) # Ra*sqrt (VaxVay)
CovA j,i] <- CovA [i,j]
CovE[i,j] <- CovP [i,j]-CovA[i,j] # By difference
CovE[j,i] <- CovE [i,j]
}
}
Stabilizing selection can be incorporated using a third matrix (Lande 1980),
D
z¼ GðW þ PÞ
1
Sðm uÞð4:15Þ
where W is a positive semidefinite matrix, m is a vector of trait means, and y is a
vector of optimal trait values. The diagonal elements of W are measures of the
strength of stabilizing selection acting directly on the character (e.g., o in
GENETIC MODELS 227
