Nielsen H. RNA: Methods and Protocols
Подождите немного. Документ загружается.

104 Josefsen and Nielsen
20 min with gentle shaking. Extensive alkaline treatment
will fragment the RNA to the extent that it is no longer
hybridization competent. Before transfer, the gel is neu-
tralized in 0.1 M Tris-HCl, pH 7.6, for 10–15 min.
9. Membranes should be handled carefully. Finger grease on
the membrane will reduce its performance and contami-
nation with finger RNases are detrimental. Membranes are
supplied in a sandwich between two sheets of protective
paper. It is a good idea to keep the protective paper in place
while cutting out a gel-sized piece of membrane.
10. A wetted membrane appears gray. Patches of white indicate
areas that have been damaged. If these are in critical parts,
the membrane should be discarded.
11. RNA on nylon membranes can be stained by the non-toxic
methylene blue. The staining solution is 0.02% methylene
blue in 0.3 M sodium acetate, p H 5.5. The RNA will stain
in a matter of few minutes. De-staining is by incubation in
1× SSPE (10 mM phosphate buffer, pH 7.4, containing
150 mM NaCl and 1 mM EDTA).
12. Filters that have not been used for hybridization experi-
ments are stored dry between sheets of Whatman 3MM
paper. Filters that are stored after a hybridization experi-
ments are stored damp in vacuo or frozen to avoid oppor-
tunistic growth. Storage can be for several months.
13. In formaldehyde gels, RNases are inhibited due to the pres-
ence of formaldehyde in the gel. In glyoxal gels, inhibi-
tion of RNases can be achieved by addition of solid sodium
iodoacetate to 10 mM to the melted agarose.
References
1. Alwine, J. C., Kemp, D. J., Stark, G.
R. (1977) Method for detection of spe-
cific RNAs in agarose gels by transfer to
diazobenzyloxymethyl-paper and hybridiza-
tion with DNA probes. Proc Natl Acad Sci
USA 74, 5350–5354.
2. Alwine, J. C., Kemp, D. J., Parker, B. A.,
Reiser, J., Renart, J., Stark, G. R., Wahl, G.
M. (1979) Detection of specific RNAs or
specific fragments of DNA by fractionation
in gels and transfer to diazobenzyloxymethyl
paper. Methods Enzymol 68, 220–242.
3. Southern, E. M. (1975) Detection of spe-
cific sequences among DNA fragments sep-
arated by gel electrophoresis. J M ol Biol 98,
503–517.
4. Lamond, A. I., Sproat, B. S. (1994) Isola-
tion and characterization of Ribonucleopro-
tein complexes, in (Higgins, S. J. and Hames,
B. D., eds.) RNA Processing. A Practical
Approach. IRL Press, Oxford, Vol. 1, pp.
103–140.
5. Sambrook, J., Fritsch, E. F., Maniatis, T.
(1989) Molecular Cloning: A Laboratory
Manual. Cold Spring Harbor Laboratory
Press, Cold Spring Harbor, NY.
6. Darling, D. C., Brickell, P. M. (1994) Nucleic
Acid Blotting. The Basics.. IRL Press, Oxford.
7. Farrell, R. E.,Jr. (1993) RNA Methodologies.
A Laboratory Guide for Isolation and Charac-
terization. Academic, San Diego, CA.
8. Chomczynski, P., Sacchi, N. (1987)
Single-step method of RNA isolation
by acid guanidinium thiocyanate-phenol-
chloroform extraction. Anal Biochem 162,
156–159.
9. Aviv, H., Leder, P. (1972) Purification
of biologically active globin messenger
Northern Blotting Analysis 105
RNA by chromatography on oligothymidylic
acid-cellulose. Proc Natl Acad Sci USA 69,
1408–1412.
10. Lehrach, H., Diamond, D., Wozney, J. M.,
Boedtker, H. (1977) RNA molecular weight
determinations by gel electrophoresis under
denaturing conditions, a critical reexamina-
tion. Biochemistry 16, 4743–4751.
11. McMaster, G. K., Carmichael, G. G. (1977)
Analysis of single- and double-stranded
nucleic acids on polyacrylamide and agarose
gels by using glyoxal and acridine orange.
Proc Natl Acad Sci USA 74, 4835–4838.
12. Reijnders, L., Sloof, P., Sival, J., Borst, P.
(1973) Gel electrophoresis of RNA under
denaturing conditions. Biochim Biophys Acta
324, 320–333.
13. Chomczynski, P., Mackey, K. (1994) One-
hour downward capillary blotting of RNA at
neutral pH. Anal Biochem 221, 303–305.
14. Bittner, M., Kupferer, P., Morris, C. F.
(1980) Electrophoretic transfer of proteins
and nucleic acids from slab gels to dia-
zobenzyloxymethyl cellulose or nitrocellu-
lose sheets. Anal Biochem 102, 459–471.
15. Herrin, D. L., Schmidt, G. W. (1988)
Rapid, reversible staining of northern blots
prior to hybridization. Biotechniques 6,
196–200.
16. Church, G. M., Gilbert, W. (1984) Genomic
sequencing. Proc Natl Acad Sci USA 81,
1991–1995.
17. Saito, I., Sugiyama, H., Furukawa, N., Mat-
suura, T.. (1981) Photoreaction of thymidine
with primary amines. Application to specific
modification of DNA. Nucleic Acids Symp Ser
10, 61–64.
18. Feinberg, A. P., Vogelstein, B.. (1983) A
technique for radiolabeling DNA restric-
tion endonuclease fragments to high specific
activity. Anal Biochem 132, 6–13.
19. Casey, J., Davidson, N. (1977) Rates of for-
mation and thermal stabilities of RNA:DNA
and DNA:DNA duplexes at high concen-
trations of formamide. Nucleic Acids Res 4,
1539–1552.
20. Bonner, J., Kung, G., Bekhor, I. (1967) A
method for the hybridization of nucleic acid
molecules at low t emperature. Biochemistry 6,
3650–3653.

Chapter 8
Rapid Amplification of cDNA Ends (RACE)
Oladapo Yeku and Michael A. Frohman
Abstract
Rapid Amplification of cDNA ends (RACE) provides an inexpensive and powerful tool to quickly obtain
full-length cDNA when the sequence is only partially known. Starting with an mRNA mixture, gene-
specific primers generated from the known regions of the gene and non-specific anchors, full-length
sequences can be identified in as little as 3 days. RACE can also be used to identify alternative transcripts
of a gene when the partial or complete sequence of only one transcript is known. In the following sections,
we outline details for rapid amplification of 5
and 3
cDNA ends using the “new RACE” technique.
Key words: RACE, new RACE, alternative transcripts.
1. Introduction
The advent of microarrays and powerful bioinformatics analysis
has not only led to the discovery of new genes but has also pro-
vided tools for analysis of existing genes (1–3). Full or partial
sequences of newly discovered genes can be aligned against entire
organism genomes in search of homology or any other clue that
can yield insight into the identity or function of the gene. There
are instances, however, where bioinformatics data is unavailable or
incomplete. In these instances, rapid amplification of cDNA ends
(RACE) can be used to identify full-length sequences of a gene
if only part of its sequence is known (4–6). RACE can be used
to amplify both the 5
and the 3
ends of genes yielding valu-
able infor mation such as the location of transcription initiation
sites, cis-acting elements, and the localization and stability of the
transcript. By using 3
and 5
RACE, full-length cDNA sequences
can be cloned from partial sequences in as little as 3 days.
H. Nielsen (ed.), RNA, Methods in Molecular Biology 703,
DOI 10.1007/978-1-59745-248-9_8, © Springer Science+Business Media, LLC 2011
107
108 Yeku and Frohman
Since the initial description of RACE (7), many labs and
commercial companies have adapted and modified the protocol
to increase specificity and user friendliness (8–18)(see Note 1).
Amplification of the 5
end can generally be divided into two clas-
sifications; classic RACE and “new RACE.” The same principles
underlie all RACE protocols and they differ only in terms of speci-
ficity, ease of use and cost. While the principles for classic RACE
will be touched on, it has been thoroughly described elsewhere
for both 5
(19)and3
(20) end amplification. This protocol will
address the identification of alternative transcripts using the “new
RACE” protocol (21).
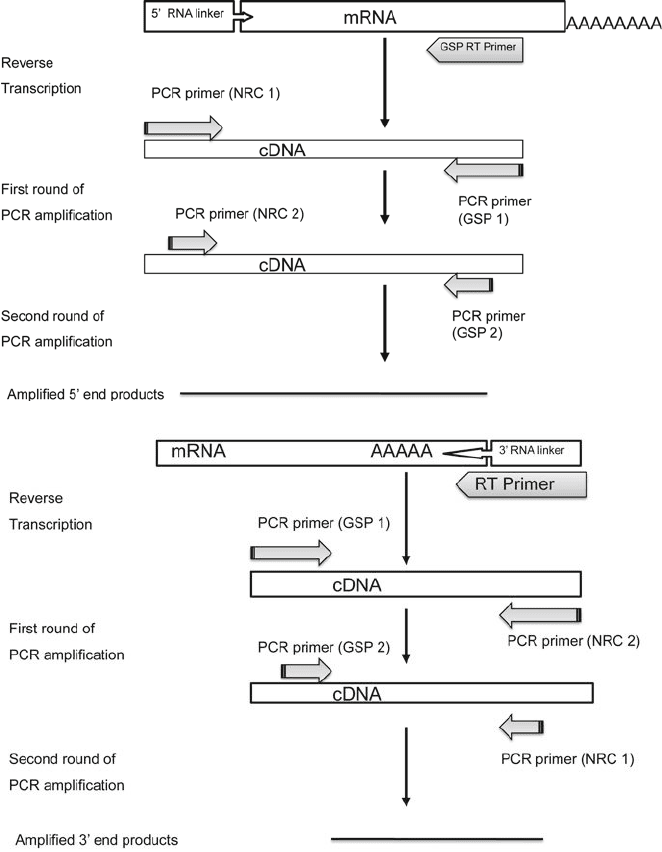
RACE primarily utilizes RT-PCR (Reverse Transcriptase-
Polymerase Chain Reaction) and PCR to amplify the ends of tran-
scripts starting with mRNA and cDNA, respectively (Fig. 8.1).
To perform RACE, the partial or a complete sequence of the
mRNA of interest has to be known. This is required to gener-
ate the Gene Specific Primers (GSPs) that will be used for the
amplifications. A second set of p rimers that extend from the
unknown end of the of the message back to the known region
of the 3
end is provided by the poly (A) tail (or an appended
homopolymer), while an appended homopolymer tail is used for
the 5
end. Herein lies the first and perhaps most important dif-
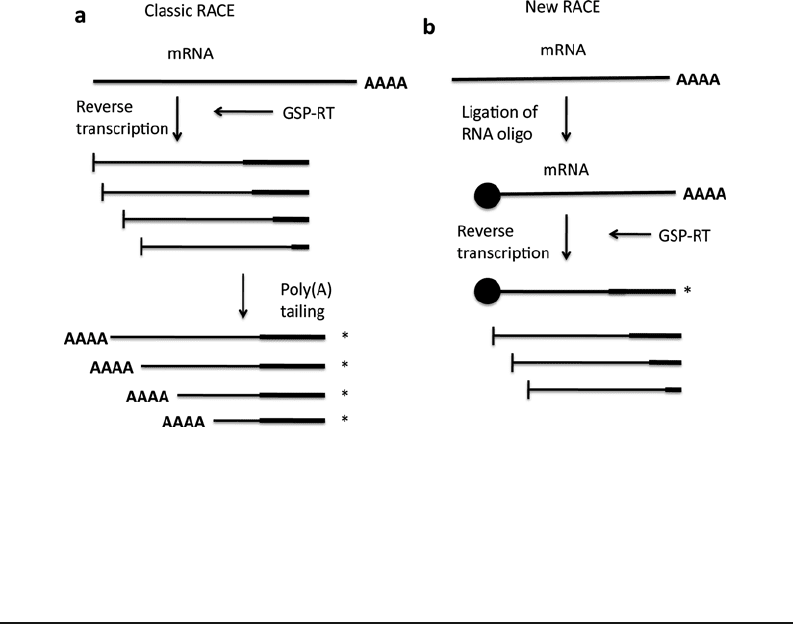
ference between “classic” and “new” RACE (22, 23). In classic
RACE (19), the homopolymer is appended after the mRNA is
reverse transcribed, whereas in “new” RACE, the homopolymer is
appended before the reverse transcriptase reaction (see Fig. 8.2).
This simple difference eliminates the amplification of non-full
length products, which greatly impr oves the ability to identify
transcription start sites. After the mRNA is reverse transcribed
to cDNA, the 5
and 3
ends are amplified using two nested PCR
reactions using gene-specific primers and primers derived from
the sequence of the RNA oligonucleotide (appended homopoly-
mer). The use of sequential nested PCR reactions is important
to reduce the amplification of unwanted products, since in each
reaction, only one of the primers is specific for the gene of inter-
est and the other binds to all cDNAs present in the starting mix-
ture (by comparison, standard PCR reactions employ two gene-
specific primers).
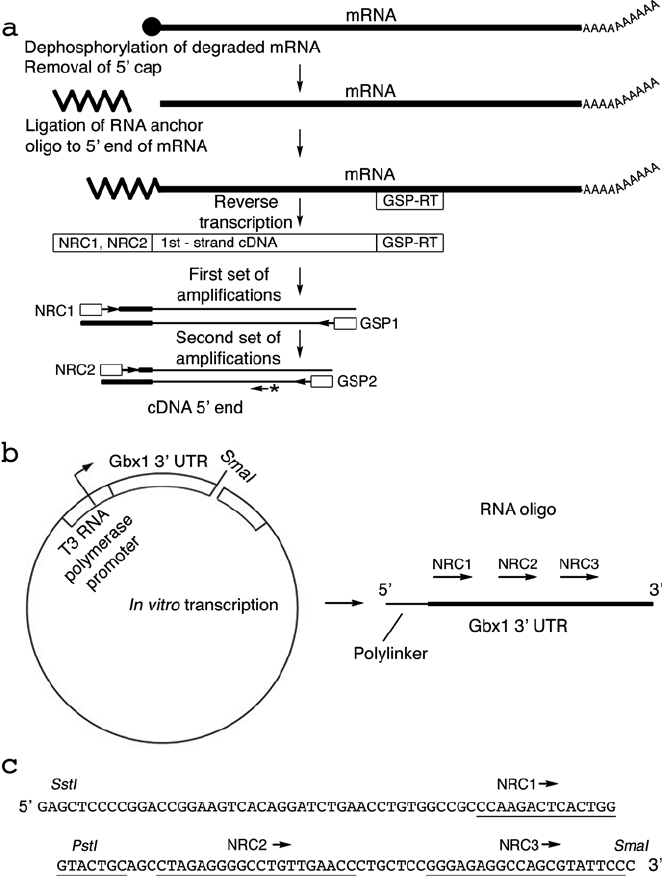
The starting mRNA mixture is dephosphorylated with shrimp
alkaline phosphatase (SAP). This dephosphorylates degraded
and uncapped (non full-length) mRNAs, leaving the full length
mRNAs with methylated “G” caps intact (24). The methylated
“G” cap is then removed with tobacco acid pyrophosphatase
(TAP). Treatment with TAP exposes the phosphorylated 5
end of
the mRNA and prepares it for ligation to the linker or homopoly-
mer. A short synthetic RNA, prepared by in vitro transcription
of a linearized plasmid (25), is ligated to the uncapped 5
and
3
end using T4 RNA ligase (see Fig. 8.3). Degraded or other
Rapid Amplification of cDNA Ends (RACE) 109
a
b
Fig. 8.1. Figure showing the general schematic for Rapid Amplification of cDNA Ends (RACE) starting with an mRNA pool.
a Amplification of the 5
end. See text for full details. b Amplification of the 3
end. Note that the NRC 1, NRC 2 and NRC
3 primers are the reverse compliments of sequences described in text. RT: Reverse Transcriptase. GSP: Gene Specific
Primer. NRC: New RACE Complement.
non-full length mRNA will not be ligated with the synthetic
oligonucleotide, since they are dephosphorylated. The mRNA-
RNA oligonucleotide hybrids are then reverse-transcribed using
a GSP. Full-length cDNA generated from the pr evious step then
contains specific sequences at the 5
and 3
ends that can be used
to amplify the full-length cDNA ends. Two nested PCR reactions
are used to amplify the full-length cDNA product with high speci-
ficity (see Fig. 8.3).
110 Yeku and Frohman
Fig. 8.2. Comparison between “classic RACE” and “new RACE.” a In “classic RACE,” the homopolymer (PolyA
tail) is added after the reverse transcription process. This anchor provides specificity for the amplifications down-
stream of the GSP-reverse transcription step. b In “new RACE,” the homopolymer is appended before the reverse
transcription takes place. This ensures full length products from the onset. Figure reproduced from citation (21).
http://www.nature.com/nprot/journal/v1/n6/images/nprot.2006.480-F3.jpg.
2. Materials
2.1.
Dephosphorylation of
Degraded RNA
1. RNA sample (TAP-treated and untre ated). All reagents must
be RNase fr ee.
2. Phosphatase buffer (10×): 0.1 M Tris-HCl (pH 7.5 at
37
◦
C), 0.1 M MgCl
2
, and 1 mg/mL BSA.
3. DTT (0.1 M).
4. RNasin (40 U/μL).
5. Shrimp Alkaline Phosphatase SAP (1 U/μL; Fermentas).
2.2. Decapping of
Intact RNA
1. Tobacco Acid Pyrophosphatase (TAP) buffer (10×):
0.5 M sodium acetate (pH 6.0), 10 mM EDTA, 1%
β-mercaptoethanol, and 0.1% Triton X-100
2. TAP (5 U/μl; Epicentre)
3.TEbuffer:10mMTris-HCl,pH7.5,and1mMEDTA,
pH 8.0
4. RNA spin column (Qiagen)
5. DTT (0.1 M)
6. RNasin (40 U/μL).
Rapid Amplification of cDNA Ends (RACE) 111
2.3. Preparation of
RNA Oligonucleotide
1.TEbuffer:10mMTris-HCl,pH7.5,and1mMEDTA,
pH 8.0
2. Plasmid template DNA for transcribing RNA oligonu-
cleotide (see Note 2)
3. Restriction enzymes and buffers
4. Proteinase K (5 mg/mL stock solution (100×))
5. H
2
O treated with diethylpyrocarbonate (DEPC)
6. Transcription buffer (5×): Provided with the enzyme or
200 mM Tris-HCl (pH 8.0), 40 mM MgCl
2
, 10 mM sper-
midine, 250 mM NaCl
7. rUTP solution (10 mM)
8. rATP solution (10 mM)
9. rCTP solution (10 mM)
10. rGTP solution ( 10 mM)
11. DNA-dependent RNA polymerase (20 U/μL)
12. RNase-free DNase I (0.5 Kunitz units)
13. DTT (0.1 M)
14. RNasin (40 U/μL).
2.4. RNA
Oligonucleotide–
Cellular RNA
Ligation
1. RNA sample ( TAP-treated and untreated)
2. Ligation buffer (10×): 500 mM Tris-HCl, 100 mM MgCl
2
,
100 mM DTT, 10 mM ATP (pH 7.8 at 25
◦
C) (see Note 3)
3. RNA oligonucleotide (see Note 4)
4. TP (2 mM)
5. T4 RNA ligase (New England BioLabs) (20 U/μL).
2.5. Reverse
Transcription
1. SuperScript II reverse transcriptase (Invitrogen) (200
U/μL). Reverse-transcription buffer (5×) is supplied with
the transcriptase.
2. dNTP solution (containing all four dNTPs, each at 10 mM)
3. Gene-specific antisense primer for 5
end RACE or reverse
complement of NRC3 (new Race Compliment primer) for
3
end RACE (20 ng/μL)
4. RNase H (2 U/μL)
5. DTT (0.1 M)
6. RNasin (40 U/μL)
7. TE buffer: 10 mM Tris-HCl, 1 mM EDTA, pH 8.0.
2.6. PCR
Amplifications
1. Hercules Hot-Start polymerase buffer (10×; Stratagene).
Do not add additional nucleotides if they are already con-
tained in the buffer.
112 Yeku and Frohman
Fig. 8.3. New RACE overview. a Outline of steps involved in the amplification of cDNA 5
ends. See text for detailed
explanation. b Plasmid map and primers used for the example in the text. c In vitro transcription of the T3 plasmid in
(b) produces a 132-nt product. The sequences for NRC1, NRC2 and NRC3 used for the generation of corresponding
primers are underlined. Figure reproduced from citation (21). http://www.nature.com/nprot/journal/v1/n6/fig_tab/
nprot.2006.479_F1.html.
2. Hercules Hot-Start polymerase (Stratagene). Hot-start pro-
tocol is str ongly recommended (see Note 5).
3. User-defined gene-specific oligonucleotide primers GSP1,
GSP2, NRC1, and NRC2 to clone the 5
end and the reverse
compliment of GSP1, GSP2, NRC1, NRC2, and NRC3 to

Rapid Amplification of cDNA Ends (RACE) 113
clone the 3
end (see Note 6 for primer design considerations
and Fig. 8.3 for details of primers NRC1 and NRC2).
2.7. DNA/RNA
Purification and
Electrophoresis
1. Agarose gel (1%) in TAE buffer
2. Ethidium bromide staining bath (0.5 μg/mL ethidium bro-
mide in electrophoresis buffer; prepared from a stock solu-
tion of 10 mg/mL in water)
3. Sodium acetate: 3 M, pH 5.2
4. Phenol/chloroform (1:1 (vol/vol))
5. Chloroform
6. Ethanol
7. Microcon spin filters (Millipore).
3. Methods
If large amounts of RNA are u sed, it is strongly recommended
that the quality control steps be performed. Small samples of the
RNA can be run on an agarose gel to check for degradation.
Aliquots can then be stored at –80
◦
C for future experiments.
The quantities presented in this protocol are the ideal amounts
of RNA to be used. Reactions can be scaled down in the event
that RNA quantities are limited.
3.1.
Dephosphorylation
of Degraded RNA
1. Combine reagents listed in Table 8.1 in a sterile microfuge
tube. Follow the manufacturers guidelines regarding the use
of SAP.
2. The reaction should be incubated for 1 h at 37
◦
C. Dephos-
phorylation of uncapped mRNA prevents degraded frag-
ments from participating in the subsequent ligation step.
Table 8.1
Dephosphorylation of degraded RNA
Component Amount Final
RNA 50 μg50μg
10X phosphatase buffer 5 μL1×
DTT (0.1 M) 0.5 μL1mM
RNasin (40 U/μL) 1.25 μL50U
SAP (1 U/μL) 3.5 μL 3.5 U
H
2
Oto50μL–
