Nielsen H. RNA: Methods and Protocols
Подождите немного. Документ загружается.

124 Meijer and de Moor
poly(A) tail (oligoadenylated RNA) is not necessarily unstable (3).
The length of the poly(A) tail is also thought to be linked to the
translational activity of that mRNA. A long poly(A) tail seems to
coincide with active translation while a short poly(A) tail corre-
lates with translational repression (4).
One of the most challenging parts of studying poly(A) tail
length is that all methods that are dependent on visualisation
of the size of the poly(A) tail by gel electrophoresis tend to
underestimate the length of the poly(A) tail because the signal
is spread out over a large area which weakens the signal. Most
traditional techniques are based on either northern blotting (for
long mRNAs in combination with RNase H cleavage) or PCR.
The northern blotting approach can only be used for r e latively
abundant mRNAs (5). Several PCR-based protocols (6–8)have
improved detection limits but still suffer from an underestimation
of long poly(A) tails. Both northern blotting and the PCR-based
protocols require the preparation of a sample treated with RNase
H in the presence of oligo(dT) to remove the poly(A) tail as a
marker (5). For some systems, such as Xenopus oocytes, it is pos-
sible to inject radiolabelled reporter mRNAs which can then be
analysed by gel electrophoresis (9). The use of radiation and the
possibility to use very short probes limits the underestimation of
the poly(A) tail length but unfortunately cannot be used in many
experimental systems.
When studying poly(A) tail changes on a global scale, mRNAs
need to be separated on the basis of the length of their poly(A)
tail. The resulting fractions can then be used for microarray anal-
ysis. mRNA can be bound to poly(U) agarose and then eluted at
different temperatures (10). While this technique can be used to
separate the RNA into fractions dependent on poly(A) tail length,
it is labour intensive and difficult to get this technique working
reproducibly.
The protocol detailed here can be used for both the analy-
sis of poly(A) tail length, especially for changes in poly(A) tail
length, as well as for the fractionation of mRNA based on the
length of the p oly(A) tail as a preparation of samples for microar-
ray analysis (see Fig. 9.1). Total RNA or crude lysate is mixed
with a radiolabelled and polyadenylated probe (see Fig. 9.2), and
biotinylated oligo(dT). The probe allows for visualisation of the
fractionation and is furthermore used to estimate the poly(A) tail
length in the sample R NA. The poly(A+) mRNA will bind to the
biotinylated oligo(dT), which in turn is bound to paramagnetic
streptavidin beads. The amount of biotinylated oligo(dT) needs
to be optimised for each sample type (see Fig. 9.3). A magnet is
used to separate the bound mRNA from the unbound RNA with-
out a poly(A) tail or a poly(A) shorter than 15 nt. The poly(A+)
mRNA is then eluted from the beads by decreasing the salt con-
centration, which results in elution of mRNAs with increasing
Fractionation of mRNA Based on the Length of the Poly(A) Tail 125
1
2
3
4
5
Unbound RNA
AAAAA
TTTTT
B
AAAAA
Oligoadenylated RNA
AAAAAAAAAA
Polyadenylated RNA
AAAAAAAAAA
5’UTR ORF 3’UTR
AAAAA
AAAAAAAAAA
TTTTT
TTTTT
B
B
AAAAAAAAAA
TTTTT
TTTTT
S
BB
AAAAAAAAAA
TTTTT
TTTTT
S
BB
AAAAAAAAAA
TTTTT
TTTTT
S
BB
TTTTT
TTTTT
S
BB
Fig. 9.1. Schematic representation of the poly(A) fractionation protocol. Total RNA is
mixed with biotinylated oligo(dT) in a GTC buffer (Step 1). The biotin is then allowed to
bind to the streptavidin paramagnetic beads (Step 2). The unbound fraction is collected
(Step 3) and the beads are washed in 0.5× SSC. The oligoadenylated (Step 4)and
polyadenylated RNA (Step 5) are then eluted in two or more steps by decreasing the
SSC concentration in the elution buffer. A radiolabelled and polyadenylated probe is
added to the RNA sample prior to fractionation. This probe is co-fractionated and allows
for analysis of the fractionation and estimation of the poly(A) tail length of the RNA in the
different fractions. UTR = untranslated region, ORF = open reading frame, B = biotin,
S = paramagnetic streptavidin bead.
poly(A) tail length. For the analysis of (changes in) poly(A) tail
length the mRNA is eluted in 6 fractions (see Fig. 9.4). For the
preparation of samples for microarray analysis the mRNA is eluted
in 2 fractions, (oligoadenylated and polyadenylated mRNA) (see
126 Meijer and de Moor
marker
5.0x
7.5x
10x
15x
20x
25x
30x
40x
no E - pap
E - pap dilution
probe
A0
A24
A42
A566
A70
A168
A255
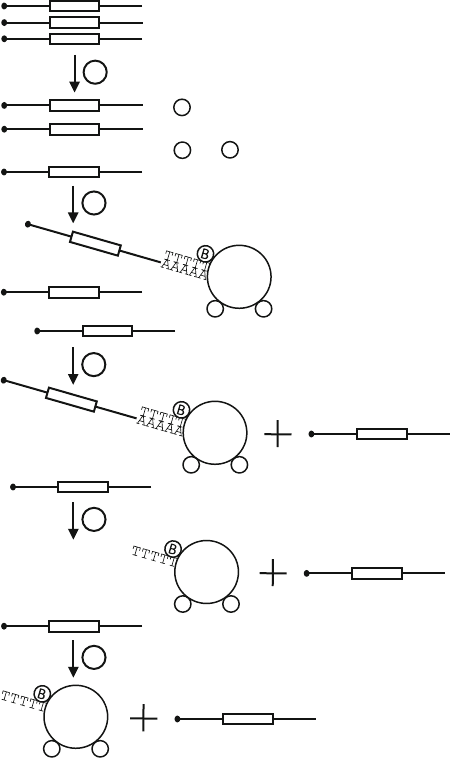
Fig. 9.2. Polyadenylation of radiolabelled probe. A radiolabelled probe was incubated
with several dilutions of E-PAP. 0.5 μL of marker, 0.5 μL of probe and 1 μL of each
polyadenylation reaction was analysed on a 5% acrylamide/urea/TBE gel. The length of
the marker bands is indicated as the number of nucleotides in excess over the unadeny-
lated probe (indicated with A0, actual length 79 nt).
A0
A24
A42
A566
A70
A168
A255
marker
total
unbound
unbound
unbound
unbound
0.075x SSC
0.075x SSC
0.075x SSC
0.075x SSC
water
water
water
water
800 400 200 100
pmole oligo dT
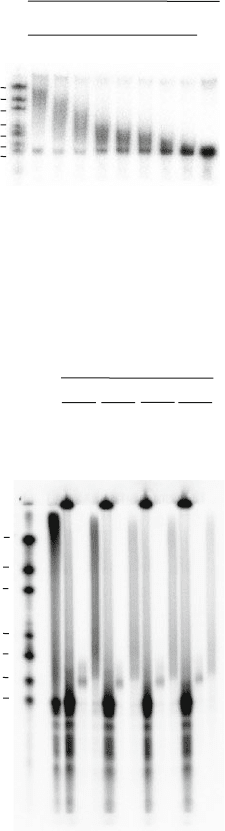
Fig. 9.3. Poly(A) fractionation into two fractions with different amounts of biotinylated
oligo(dT). 80 μg of total RNA from NIH3T3 cells was mixed with polyadenylated radio-
labelled probe and different amounts of biotinylated oligo (dT) and separated using the
poly(A) fractionation protocol. RNA was eluted with 0.075× SSC (oligoadylated RNA)
and water (polyadenylated RNA). This shows that 800 pmole of oligo dT is required for
efficient fractionation of 80 μg of RNA from NIH3T3 cells.
Fig. 9.3). Since poly(A) tail length and translational activity are
thought to be correlated for many mRNAs, poly(A) fractionation
can be used as an alternative for other posttranscriptional profiling
methods. Poly(A) fractionation is a fast and reproducible method
which can be used for a wide variety of samples such as tissue cul-
ture cells, oocytes, embryos, clinical samples and total RNA (3).
Fractionation of mRNA Based on the Length of the Poly(A) Tail 127
marker
A0
A24
A42
A566
A70
A168
A255
total
unbound
0.2x SSC
0.1x SSC
0.075x SSC
0.050x SSC
0.025x SSC
water
elutions
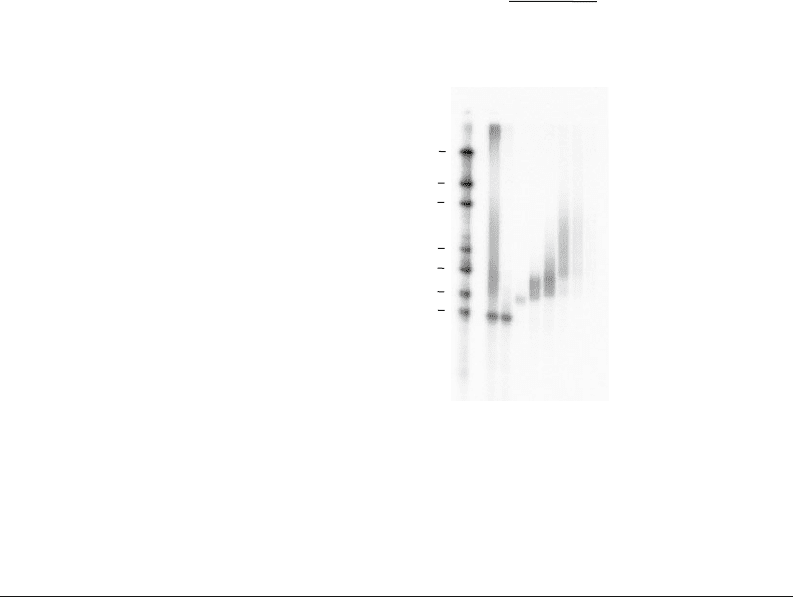
Fig. 9.4. Poly(A) fractionation into 6 f ractions. 80 μg of total RNA from NIH3T3 cells was
mixed with polyadenylated radiolabelled probe and 750 pmole of biotinylated oligo(dT)
and separated using the poly(A) fractionation protocol. The marker can be used to esti-
mate the range of poly(A) tail length in each fraction. Note that the signal in the water
lane is weak due to the lack of probe with a very long poly(A) tail.
2. Materials
2.1. Molecular
Weight Marker
1. Linearised plasmid DNA. The marker is a mixture of radioac-
tively labelled in vitro transcripts of known lengths. In order
to make this, you will need several plasmids which dif-
fer slightly in size when digested with the same restriction
enzyme. Alternatively, you can use one plasmid which can
be digested by several different restriction enzymes (one at a
time).
2. The same materials as for the purification of the template (see
Section 2.2) and for the probe synthesis (see Section 2.3).
2.2. Template
Purification
1. Linearised plasmid DNA (this plasmid will also generate the
lowest band of the molecular weight marker).
2. Water-saturated phenol.
3. Chloroform.
4. 3 M NaAc, pH 5.2.
5. 100% isopropanol.
6. 70% ethanol.
7. RNase-free water.
128 Meijer and de Moor
2.3. Probe Synthesis
1. RNA polymerase (including 5× transcription buffer (TSC)
and 100 mM dithiothreitol (DTT)) (Promega).
2. NTP mix: 5 mM ATP, 5 mM CTP, 0.1 mM UTP,
0.5 mM GTP.
3. 40 U/μL RNasin.
4. 5 mM cap analogue (m
7
GpppG) (Ambion).
5. 1 μg/μL template in R Nase-free water.
6. 800 Ci/mmole, 20 μCi/μL[α-
32
P]UTP.
7. Water-saturated phenol.
8. Chloroform.
9. G-50 Sephadex in water, autoclaved: store at 4
◦
C.
10. 1-mL syringes.
11. Sterilised and silanised glass wool.
2.4. Polyadenylation
1. Poly(A) Tailing Kit (containing 2 U/μL Escherichia coli
poly(A) polymerase (E-PAP), 5× E-PAP buffer, 10 mM
ATP , 25 mM MnCl
2
, nuclease free water) (Ambion).
2. RNase-free TE: 10 mM Tris-HCl, 1 mM ethylenediamine
tetraacetic acid (EDTA) (pH 8.0), autoclaved.
3. Water-saturated phenol.
4. Chloroform.
2.5. Analysis
of Probes
1. Urea.
2. RNase-free 10× TBE: 500 mM Tris-base, 500 mM boric
acid, 1 mM EDTA, pH 8.0; autoclaved.
3. 30% acrylamide/bisacrylamide (37.5:1).
4. 10% ammonium persulfate (APS); fr e shly prepared or
aliquoted and stored at –20
◦
C.
5. N,N,N,N’-tetramethyl-ehylenediamine (TEMED).
6. Gel loading buffer: 95% formamide, 0.025% xylene cyanol,
0.025% bromophenol blue, 18 mM EDTA, 0.025% sodium
dodecyl sulfate (SDS).
2.6. Oligo(dT)-Based
Fractionation of RNA
1. 80 μg of total RNA in RNase-free water.
2. PolyA Tract System 1000, containing paramagnetic
streptavidin beads, guanidinium thiocyanate (GTC)
extraction buffer, dilution buffer, biotinylated oligo(dT),
β-mercaptoethanol (BME), 0.5× SSC and magnetic
separation stand (Promega).
3. Extra 50 p mol/μL biotinylated oligo(dT) (Promega).
4. RNase-free water.

Fractionation of mRNA Based on the Length of the Poly(A) Tail 129
5. 3 M NaAc, pH 5.2.
6. 100% isopropanol.
7. 10 μg/μL yeast tRNA.
8. 70% ethanol.
2.7. Analysis of
Fractionation
1. The same materials as for the analysis of the probes (see
Section 2.5)
3. Methods
(see Note 1)
3.1. Molecular
Weight Marker
Linearise the plasmids as described for the purification of the tem-
plate (see Section 3.2) and then mix them and use for the syn-
thesis of the molecular weight marker as described for the probe
synthesis (see Section 3.3)(see Note 2).
3.2. Template
Purification by
Phenol Extraction
and Ethanol
Precipitation
1. Linearise the p lasmid DNA.
2. Add 0.5 volume of phenol and vortex for 10 s (see
Note 3).
3. Add 0.5 volume of chloroform and vortex for 30 s.
4. Spin at 20,000 ×g for 10 min at room temperature.
5. Transfer the supernatant to a new tube. Add 1 volume of
chloroform and vortex for 30 s.
6. Spin at 20,000 ×g for 10 min at room temperature.
7. Transfer the supernatant to a new tube. Add 0.1 volume of
3 M NaAc, pH 5.2, and vortex.
8. Add 1 volume of isopropanol and vortex.
9. Incubate on ice for 30 min.
10. Spin at 20,000 ×g for 10 min at 4
◦
C.
11. Aspirate the supernatant. Wash the pellet with 2 volumes
of 70% ethanol.
12. Spin at 20,000×g for 5 min at 4
◦
C and remove residual
ethanol.
13. Dry the pellet and dissolve it in 10 μLofH
2
O.
14. Check an aliquot of the template on a 1.5% agarose gel.
15. Quantify the template and dilute it to 1 μg/μL.
3.3. Probe Synthesis
by In Vitro
Transcription
1. Mix:1 μL5×TSC
0.5 μL0.1MDTT
0.5 μL NTP mix
0.5 μL40U/μL RNasin
130 Meijer and de Moor
0.5 μL 5 mM cap analogue
0.5 μL template (1 μg/μL)
1.5 μL 800 Ci/mmole, 20 μCi/μL[α-
32
P]UTP (see
Note 4)
0.25 μL RNA polymerase (see Note 5).
2. Incubate for 1 h at 37
◦
C.
3. In the meantime prepare a G-50 Sephadex column per
sample. Remove the plunger fr om a 1-mL syringe and put
a piece of glass wool in the syringe. Push it down with
the plunger to the bottom of the syringe and then remove
the plunger. Swirl the Sephadex to mix it and pipette it
into the syringe, avoiding air bubbles. Completely fill the
syringe. Place the syringe in a 15-mL Falcon tube. Spin
for 1 min at 1,300×g at 4
◦
C. Discard the flow through.
Keep adding and spinning until the syringe contains 1 mL
of Sephadex. Spin for 5 min at 1,300×g at 4
◦
C. Discard
the flow through and move the syringe to a new 15-mL
Falcon tube. The G-50 Sephadex column is now ready for
the purification of the probe.
4. After the incubation add 45 μLofH
2
O to the probe.
5. Add 25 μL of p henol and vortex for 10 s.
6. Add 25 μL of chloroform and vortex for 30 s .
7. Spin at 20,000 ×g for 10 min at 4
◦
C.
8. Load the supernatant on the G-50 Sephadex column.
9. Spin for 1 min at 1,300×g at 4
◦
C.
10. Transfer the eluate to a 1.5-mL tube.
11. Load 50 μL of RNase-free H
2
O onto the Sephadex col-
umn.
12. Spin for 1 min at 1,300×g at 4
◦
C.
13. Transfer the eluate to the tube with the first eluate. The
combined eluates is the unadenylated probe.
3.4. Polyadenylation
Assemble on ice! (see Note 6)
1. Prepare E-PAP dilutions (7.5×,15×,30× in 1× E-PAP
buffer).
2. For each E-PAP dilution mix:
5 μL5× E-PAP buffer
2.5 μL25mMMnCl2
2.5 μL10mMATP
5 μL diluted E-PAP
10 μL unadenylated probe (see Section 3.3)
3. Incubate for exactly 30 min at 37
◦
C.
4. Immediately put samples on ice.
Fractionation of mRNA Based on the Length of the Poly(A) Tail 131
5. Add 25 μL of TE and mix.
6. Add 25 μL of p henol and vortex for 10 s.
7. Add 25 μL of chloroform and vortex for 30 s.
8. Spin at 20,000 ×g for 10 min at 4
◦
C.
9. Transfer the supernatant to a new tube. The polyadenylated
probe is used without further purification steps or concen-
tration by ethanol precipitation.
3.5. Analysis
of Probes on a 5%
Acrylamide/Urea/TBE
Mini Gel
1. Mix:
6gurea
1.2 mL 10×TBE
2 mL 30% acrylamide/bisacrylamide (37.5:1) (see Note 7)
4mLH
2
O.
2. Microwave briefly to dissolve urea. Do not allow the solu-
tion to get warmer than 60
◦
C.
3. Let the solution cool down to room temperature.
4. Add 24 μL of APS and 24 μL of TEMED (see Note 8).
5. Pour the gel and allow it to set.
6. Pre-run the gel at 100 V for 15 min.
7. Prepare samples:
0.5 μL marker in gel loading buffer
0.5 μL pr obe w/o poly(A) tail in gel loading buffer
1 μL polyadenylated probe (30× diluted E-PAP) in gel
loading buffer
1 μL polyadenylated probe (15× diluted E-PAP) in gel
loading buffer
1 μL polyadenylated probe (7.5× diluted E-PAP) in gel
loading buffer.
8. Denature the samples at 95
◦
Cfor5min.
9. Load the samples and run at 100 V (until bromophenol
blue is almost at the bottom of the gel).
10. Wrap the gel in Saran wrap and expose it to a PhosphoIm-
ager screen or film for 5–30 min.
11. Mix the polyadenylated samples to produce a range from
A0 to A400, including a small amount of probe without
poly(A) tail. This constitutes the probe mix (see Fig. 9.2).
3.6. Oligo(dT)-Based
Fractionation of RNA
All steps ar e at room temperature unless indicated otherwise.
1. Allow the GTC, BME, biotinylated oligo(dT), 0.5×SSC
and H
2
O to r each room temperature.
2. Add 41 μL of BME per mL of GTC (GTC/BME).
3. Add 20.5 μL of BME per mL of dilution buffer
(DIL/BME). Preheat the DIL/BME to 70
◦
C.
132 Meijer and de Moor
4. Make the required SSC dilutions (0.200×, 0.100×,
0.075×, 0.050×, 0.025×).
5. Mix a maximum of 40 μLtotalRNA(max80μg) with
400 μL GTC/BME in a 2-mL tube (see Note 9).
6. Add 5 μL of polyadenylated probe mix (see Section 3.5),
15 μL biotinylated oligo(dT) and 816 μL DIL/BME
(see Note 10,andFig. 9.3).
7. Incubate at 70
◦
Cfor5min.
8. Spin at 12,000 ×g for 10 min at room temperature.
9. In the meantime prepare the paramagnetic streptavidin
beads. Completely resuspend the beads by gently rock-
ing the bottle. Transfer 600 μL of resuspended beads to a
2-mL tube for each sample. Place the tube on the magnetic
stand and slowly move the stand to a horizontal position
until the beads are collected at the tube side. Carefully pour
off the storage buffer by tilting the tube so that the solu-
tion runs over the captured beads. Resuspend the beads in
600 μL0.5×SSC and capture the beads again using the
stand. Repeat this wash step twice. Resuspend the beads in
600 μL0.5×SSC.
10. After the spin add the supernatant to the beads. Allow the
biotinylated oligo(dT) to bind to the beads by rotation at
room temperature for 15 min.
11. After the biotinylated oligo(dT) has bound to the beads
capture the beads using the magnetic stand. Transfer the
supernatant to a new tube and keep on ice (unbound frac-
tion).
12. Wash the beads three times as described above (See
Step 9). Rotate for at least 5 min between each wash.
13. Resuspend the beads in 400 μL 0.200× SSC. Rotate for at
least 5 min at room temperature. Capture the beads using
the magnetic stand. Transfer the eluate (400 μL) to a new
tube. Be careful not to disturb the beads when doing so.
Keep the 400 μL in a tube on ice (eluate 1) (see Note 11).
14. Repeat the elution (Step 13) with 400 μL 0.100× SSC,
0.075× SSC, 0.050× SSC, 0.025× SSC and H
2
O (eluates
2–6) (see Note 11).
15. Spin all collected fractions (unbound and eluates 1–6) at
20,000×g for 5 min at 4
◦
C to remove any transferred
beads.
16. Transfer 2× 650 μL of the unbound supernatant to two
new tubes. Add 650 μL of isopropanol to each. Mix and
precipitate overnight at –20
◦
C.
Fractionation of mRNA Based on the Length of the Poly(A) Tail 133
17. Transfer 360 μL of each eluate supernatant to a new tube.
Add 2 μL yeast tRNA, 36 μL 3 M NaAc pH 5.2 and
360 μL isopropanol. Mix and precipitate overnight at –
20
◦
C.
18. Spin all samples at 20,000×g for 30 min at 4
◦
C.
19. Remove the supernatant and wash with 800 μL 70%
ethanol.
20. Spin at 20,000 ×g for 10 min at 4
◦
C.
21. Completely remove the supernatant with a drawn out glass
pipette and allow the pellets to dry.
22. Resuspend the pellets from the eluates in 10 μLofH
2
O.
Resuspend the pellets from the unbound fraction in 5 μL
of H
2
O per tube and pool the two samples.
3.7. Analysis of the
Fractionation on a
5% Acrylamide/
UREA/TBE Maxi Gel
1. Mix per gel:
30 g urea
6mL10×TBE
10 mL 30% acrylamide/bisacrylamide (37.5:1)
20 mL H
2
O.
2. Microwave briefly to dissolve urea. Do not allow the solu-
tion to get warmer than 60
◦
C.
3. Let the solution cool down to room temperature.
4. Add 120 μL of APS and 120 μL of TEMED.
5. Pour the gel and allow to set.
6. Pre-run the gel at 200 V for 15 min.
7. Prepare samples:
1 μL of molecular weight marker 20× diluted in gel load-
ing buffer.
0.9 μL of probe mix in gel loading buffer.
1 μL of unbound 5× diluted in gel loading buffer.
1 μL for each eluate in gel loading buffer (see Note 12).
8. Denature the samples at 95
◦
Cfor5min.
9. Load the samples and run at 200 V (until the dark blue
colour of the dye indicator is almost at the bottom of the
gel).
10. Wrap the gel in Saran wrap and expose it to a PhosphoIm-
ager screen or film for 24 h at –20
◦
C. Allow the cassette to
reach room temperature before opening (see Fig. 9.4).
3.8. Further Analysis
The remainder of the samples can be used for subsequent analysis
by RT-qPCR and/or northern blotting to show in which fractions
endogenous and/or reporter RNAs are (see Note 13).
