Nielsen H. RNA: Methods and Protocols
Подождите немного. Документ загружается.

84 George and Tenenbaum
References
1. Griffiths-Jones, S., et al. (2005) Rfam:
annotating non-coding RNAs in complete
genomes. Nucleic Acids Res 33, suppl_1,
D121–D124.
2. Mignone, F., et al. ( 2005) UTRdb and UTR-
site: a collection of sequences and regulatory
motifs of the untranslated regions of eukary-
otic mRNAs. Nucleic Acids Res 33 Suppl 1,
D141–D146.
3. Huang, H., et al. (2006) RegRNA: an inte-
grated web server for identifying regulatory
RNA motifs and elements. Nucleic Acids Res
34, Web Server issue, W429–W434.
4. Jacobs, G. H., et al. (2006) Transterm–
extended search facilities and improved inte-
gration with other databases. Nucleic Acids
Res 34, Database issue, D37–D40.
5. Abreu-Goodger, C., et al. (2004) Conserved
regulatory motifs in bacteria: riboswitches
and beyond. TIGS 20, 475–479.
6. Abreu-Goodger, C., Merino, E. (2005)
RibEx: a web server for locating riboswitches
and other conserved bacterial regulatory ele-
ments. Nucleic Acids Res 33, Web Server
issue, W690–W692.
7. Pedersen, J. S., et al. (2006) Identification
and classification of conserved RNA sec-
ondary structures in the human genome.
PLoS Comput Biol 2, e33.
8. Torarinsson, E., et al. (2008) Compara-
tive genomics beyond sequence-based align-
ments: RNA structures in the ENCODE
regions. Genome Res 18, 242–251.
9. Washietl, S., Hofacker, I. L., Stadler, P.
F. (2005) Fast and reliable prediction of
noncoding RNAs. Proc Natl Acad Sci 102,
2454–2459.
10. Berman, H. M., et al. (1992) The nucleic
acid database. A comprehensive relational
database of three-dimensional structures of
nucleic acids. Biophys J 63, 751–759.
11. Griffiths-Jones, S., et al. (2006) miRBase:
microRNA sequences, targets and gene
nomenclature. Nucleic Acids Res 34, suppl_1,
D140–D144.
12. Bindewald, E., et al. (2008) RNAJunction: a
database of RNA junctions and kissing loops
for three-dimensional structural analysis and
nanodesign. Nucleic Acids Res 36, Database
issue, D392–D397.
13. Lestrade, L., Weber, M. J. (2006) snoRNA-
LBME-db, a compr ehensive database of
human H/ACA and C/D box snoR-
NAs. Nucleic Acids Res 34, Database issue,
D158–D162.
14. Xie, J., et al. (2007) Sno/scaRNAbase:
a curated database for small nucleolar
RNAs and cajal body-specific RNAs.
Nucleic Acids Res 35, Database issue,
D183–D187.
15. Rocheleau, L., Pelchat, M. (2006) The Sub-
viral RNA Database: a toolbox for viroids,
the hepatitis delta virus and satellite RNAs
research. BMC Microbiol 6, 24.
16. Zhou, Y., et al. (2008) GISSD: group
I intron sequence and structure database.
Nucleic Acids Res 36, Database issue,
D31–D37.
17. Cannone, J. J., et al. (2002) The comparative
RNA web (CRW) site: an online database of
comparative sequence and structure informa-
tion for ribosomal, intron, and other RNAs.
BMC Bioinformatics 3, 2.
18. Eddy, S. R. (2006) Computational analysis
of RNAs. Cold Spring Harb Symp Quant Biol
71, 117–128.
19. Klein, R. J., Eddy, S. R. (2003) RSEARCH:
finding homologs of single structured RNA
sequences. BMC Bioinformatics 4, 44.
20. Pesole, G., Liuni, S. (1999) Internet
resources for the functional analysis of 5
and 3
untranslated regions of eukaryotic
mRNAs. TIGS 15, 378.
21. Dsouza, M., L arsen, N., Overbeek, R.
(1997) Sear ching for patterns in genomic
data. Trends Genet 13, 597–498.
22. Reeder, J., Reeder, J., Giegerich, R.
(2007) Locomotif: from graphical motif
description to RNA motif search. Bioin-
formatics 23, i392–i400. data. TIGS, 13,
497–498.
23. Sakakibara, Y. (2003) Pair hidden Markov
models on tree structures. Bioinformatics 19
Suppl 1, i232–i240.
24. Sakakibara, Y., et al. (2007) Stem kernels
for RNA sequence analyses. J Bioinformatics
Comput Biol 5, 1103–1122.
25. Matsui, H., Sato, K., Sakakibara, Y. (2004)
Pair stochastic t ree adjoining grammars
for aligning and predicting pseudoknot
RNA structures, in Proceedings/IEEE Com-
putational Systems Bioinformatics Conference,
CSB. IEEE Computational Systems Bioinfor-
matics Conference, pp. 290–299.
26. Hofacker, I. L. (2003) Vienna RNA sec-
ondary structure server. Nucleic Acids Res 31,
3429–3431.
27. Hofacker, I. L. (2004) RNA secondary struc-
ture analysis using the V ienna RNA package.
Curr Protoc Bioinformatics (Editoral Board,
Andreas D. Baxevanis et al., Chapter 12,
Unit 12.2).
28. Le, S. Y., Zhang, K., Maizel, J. V. (1995) A
method for predicting common structures of
Web-Based RNA Resources 85
homologous RNAs. Comput Biomed Res Int
J 28, 53–66.
29. Macke, T. J., et al. (2001) RNAMotif,
an RNA secondary structure definition and
search algorithm. Nucleic Acids Res 29,
4724–4735.
30. Gautheret, D., Lambert, A. (2001) Direct
RNA motif definition and identification from
multiple sequence alignments using sec-
ondary structure profiles. J Mol Biol 313,
1003–1011.
31. Lambert, A., et al. (2005) Computing expec-
tation values for RNA motifs using dis-
crete convolutions. BMC Bioinformatics 6,
118.
32. Liu, J., et al. (2005) A method for aligning
RNA secondary structures and its application
to RNA motif detection. BMC Bioinformat-
ics 6, 89.
33. Xue, C., Liu, G. (2007) RScan: fast search-
ing structural similarities for structured
RNAs in large databases. BMC Genomics 8,
257.
34. Busch, A., Backofen, R. (2006) INFO-RNA
– a fast approach to inverse RNA folding.
Bioinformatic 22, 1823–1831.
35. Bafna, V., Zhang, S. (2004) FastR: fast
database search tool for non-coding RNA,
in Proceedings/IEEE Computational Systems
Bioinformatics Conference, CSB. IEEE Com-
putational Systems Bioinformatics Conference,
pp. 52–61.
36. Zhang, S., et al. (2005) Searching
genomes for noncoding RNA using FastR.
IEEE/ACM Trans Comput Biol B ioinformat-
ics 2, 366–379.
37. Thébault, P., et al. (2006) Searching RNA
motifs and their intermolecular contacts
with constraint networks. Bioinformatics 22,
2074–2080.
38. Veksler-Lublinsky, I., et al. (2007) A
structure-based flexible search method for
motifs in RNA. J Comput Biol J Comput Mol
Cell Biol 14, 908–926.
39. Chang, T., et al. (2006) RNAMST: efficient
and flexible approach for identifying RNA
structural homologs. Nucleic Acids Res 34,
Web Server issue, W423–W428.
40. diBernardo, D, Down, T. and Hubbard,
T (2003) ddbRNA: detection of conserved
secondary structures in multiple alignments.
Bioinformatics 19, 1606–1611.
41. Hofacker, I. L. (2007) RNA consensus struc-
ture prediction with RNAalifold. Methods Mol
Biol 395, 527–544.
42. Rivas, E., Eddy, S. R. (2001) Noncod-
ing RNA gene detection using compara-
tive sequence analysis. BMC Bioinformatics
2, 8.
43. Kiryu, H., Kin, T., Asai, K. (2007) Robust
prediction of consensus secondary structures
using averaged base pairing pr obability matri-
ces. Bioinformatics 23, 434–441.
44. Coventry, A., Kleitman, D. J., Berger, B.
(2004) MSARI: multiple sequence align-
ments for statistical detection of RNA sec-
ondary structure. Proc Natl Acad Sci 101,
12102–12107.
45. Knudsen, B., Hein, J. (2003) Pfold: RNA
secondary structure prediction using stochas-
tic context-free grammars. Nucleic Acids Res
31, 3423–3428.
46. Ruan, J., Stormo, G. D., Zhang, W. (2004)
An iterated loop matching approach to the
prediction of RNA secondary structures with
pseudoknots. Bioinformatics 20, 58–66.
47. Knight, R., Birmingham, A., Yarus, M.
(2004) BayesFold: rational 2 degrees folds
that combine thermodynamic, covaria-
tion, and chemical data for aligned RNA
sequences. RNA 10, 1323–1336.
48. Bindewald, E., Shapiro, B. A. (2006)
RNA secondary structure prediction from
sequence alignments using a network of k-
nearest neighbor classifiers. RNA 12, 342–
352.
49. W ilm, A., Linnenbrink, K., Steger, G. (2008)
ConStruct: Improved construction of RNA
consensus structures. BMC Bioinformatics
9,
219.
50. Meyer, I. M., Miklós, I. (2007) Simul-
Fold: simultaneously inferring RNA struc-
tures including pseudoknots, alignments, and
trees using a Bayesian MCMC framework.
PLoS Comput Biol 3, e149.
51. Dalli, D., et al. (2006) STRAL: progressive
alignment of non-coding RNA using base
pairing probability vectors in quadratic time.
Bioinformatics 22, 1593–1599.
52. W ilm, A., Higgins, D. G., Notredame, C.
(2008) R-Coffee: a method for multiple
alignment of non-coding RNA. Nucleic Acids
Res 36, e52.
53. Moretti, S., et al. (2008) R-Coffee: a web
server for accurately aligning noncoding
RNA sequences. Nucleic Acids Res 36, Web
Server issue, W10–W13.
54. Steffen, P., et al. (2006) RNAshapes: an
integrated RNA analysis package based on
abstract shapes. Bioinformatics 22, 500–503.
55. Hamada, M., et al. (2006) Mining fre-
quent stem patterns from unaligned RNA
sequences. Bioinformatics 22, 2480–2487.
56. Tabei, Y., et al. (2008) A fast structural
multiple alignment method for long RNA
sequences. BMC Bioinformatics 9, 33.
57. Kin, T., Tsuda, K., Asai, K. (2002) Marginal-
ized kernels for RNA sequence data analysis.
86 George and Tenenbaum
Genome Inform Int Conf Genome Inform 13,
112–122.
58. Le, S., Maizel, J. V., Zhang, K. (2004)
An algorithm for detecting homologues of
known structured RNAs in genomes, in Pro-
ceedings/IEEE Computational Systems Bioin-
formatics Conference, CSB. IEEE Computa-
tional Systems Bioinformatics Conference, pp.
300–310.
59. Touzet, H. (2007) Comparative analysis of
RNA genes: the caRNAc software. Methods
Mol Biol 395, 465–474.
60. Ji, Y., Xu, X., Stormo, G. D. (2004) A graph
theoretical approach for predicting common
RNA secondary structure motifs including
pseudoknots in unaligned sequences. Bioin-
formatics 20, 1591–1602.
61. Hu, Y. (2003) GPRM: a genetic program-
ming approach to finding common RNA sec-
ondary structur e elements. Nucleic Acids Res
31, 3446–3449.
62. Yao, Z., Weinberg, Z., Ruzzo, W. L. (2006)
CMfinder–a covariance model based RNA
motif finding algorithm. Bioinformatics 22,
445–452.
63. Pavesi, G., et al. (2004) RNAProfile: an algo-
rithm for finding conserved secondary struc-
ture motifs in unaligned RNA sequences.
Nucleic Acids Res 32, 3258–3269.
64. Xu, X., Ji, Y., Stormo, G. D. (2007) RNA
Sampler: a new sampling based algorithm for
common RNA secondary structure predic-
tion and structural alignment. Bioinformatics
23, 1883–1891.
65. Horesh, Y., et al. (2007) RNAspa: a shortest
path approach for comparative prediction of
the secondary str ucture of ncRNA molecules.
BMC Bioinformatics 8, 366.
66. Katoh, K., Toh, H. (2008) Improved accu-
racy of multiple ncRNA alignment by
incorporating structural information into a
MAFFT-based framework. BMC Bioinfor-
matics 9, 212.
67. Do, C. B., Foo, C., Batzoglou, S. (2008) A
max-margin model for efficient simultaneous
alignment and folding of RNA sequences.
Bioinformatics 24, i68–i76.
68. Holmes, I. (2005) Accelerated probabilistic
inference of RNA structure evolution. BMC
Bioinformatics 6, 73.
69. Lindgreen, S., Gardner, P. P., Krogh, A.
(2007) MASTR: multiple alignment and
structure prediction of non-coding RNAs
using simulated annealing. Bioinformatics 23,
3304–3311.
70. Kiryu, H., Tabei, Y., et al. (2007) Murlet: a
practical multiple alignment tool for struc-
tural RNA sequences. Bioinformatics 23,
1588–1598.
71. Hofacker, I. L., Bernhar t , S. H. F., Stadler,
P. F. (2004) Alignment of RNA base pair-
ing probability matrices. Bioinformatics 20,
2222–2227.
72. W ill, S., et al. (2007) Inferring noncod-
ing RNA families and classes by means
of genome-scale structure-based clustering.
PLoS Comput Biol 3, e65.
73. Siebert, S., Backofen, R. (2005) MARNA:
multiple alignment and consensus str uc-
ture prediction of RNAs based on sequence
structure comparisons. Bioinformatics 21,
3352–3359.
74. Anwar, M., Nguyen, T., Turcotte, M. (2006)
Identification of consensus RNA secondary
structures using suffix arrays. BMC Bioinfor-
matics 7, 244.
75. Bauer, M., Klau, G. W., Reinert, K. (2007)
Accurate multiple sequence-structure align-
ment of RNA sequences using combi-
natorial optimization. BMC Bioinformatics
8, 271.
76. Doyle, F., et al. (2008) Bioinformatic tools
for studying post-transcriptional gene regu-
lation: the UAlbany TUTR collection and
other informatic resources. Methods Mol Biol
419, 39–52.
77. Gardner, P., Giegerich, R. (2004) A com-
prehensive comparison of comparative RNA
structure prediction approaches. BMC Bioin-
formatics 5, 140.

Chapter 7
Northern Blotting Analysis
Knud Josefsen and Henrik Nielsen
Abstract
Northern blotting analysis is a classical method for analysis of the size and steady-state level of a specific
RNA in a complex sample. In short, the RNA is size-fractionated by gel electrophoresis and transferred
by blotting onto a membrane to which the RNA is covalently bound. Then, the membrane is analysed
by hybridization to one or more specific probes that are labelled for subsequent detection. Northern
blotting is relatively simple to perform, inexpensive, and not plagued by artefacts. Recent developments
of hybridization membranes and buffers have r esulted in increased sensitivity closing the gap to the more
laborious nuclease protection experiments.
Key words: Gel electrophoresis, northern blotting, probe, hybridization analysis, mRNA.
1. Introduction
Northern blotting analysis is a method for obtaining information
on the size and abundance of a specific RNA in a complex mix-
ture. In short, the RNA sample (e.g. whole cell R NA or a fraction
hereof) is size-fractionated by gel electrophoresis. Then, the RNA
is transferred onto a membrane (“blotted”) and analysed by bind-
ing (“hybridization”) of one or more labelled probes specific for
the RNA in question. Northern blotting (1, 2) is a further devel-
opment of a similar technique for DNA analysis, Southern blot-
ting, developed by Ed Southern (3). Thus, its name, like western
blotting, was introduced as a joke. Because northern blotting,
unlike Southern blotting, does not refer to a person’s name, it is
spelled in lower case.
Northern blotting analysis gives information on the length of
the RNA molecule and the possible existence of length variants
H. Nielsen (ed.), RNA, Methods in Molecular Biology 703,
DOI 10.1007/978-1-59745-248-9_7, © Springer Science+Business Media, LLC 2011
87
88 Josefsen and Nielsen
because the RNA is electrophoresed under denaturing conditions
in parallel with a molecular weight marker (an RNA ladder). RNA
molecules that are branched or circular have aberrant mobility’s
and their analysis requires specialized procedures (4). Northern
blotting is f requently used simply to demonstrate the presence of
a specific RNA in a sample, but the method also allows for quan-
titative measurements. It is the steady-state level of the RNA, that
is the sum of its production and removal, that is being measured.
If the transcriptional activity is of interest, this is measured by e.g.
nuclear run-on experiments. Decay of the RNA is measured inde-
pendently by measurement of the transcript level at various time-
points following addition of a transcription initiation inhibitor.
When used as a quantitative method, northern blotting is usually
used to compare the RNA level in different experimental situ-
ations rather than for determination of absolute amounts. This
is because hybridization to filter-bound RNA is suboptimal and
strongly dependent on experimental parameters, such as degree of
covalent attachment to the filter. Addition of known amounts of
an inter n al standard of different length but with similar sequence
made by in vitro transcription could be used to estimate absolute
transcript levels, but this is rarely seen.
An outline of a northern blotting analysis is shown in Fig. 7.1.
Only the most common variations of the technique are covered
by this chapter. For a more comprehensive description, see (5–7).
The input RNA can be of any kind, but mostly it is whole cell
RNA (“total RNA”) prepared by the acid phenol/guanidinium
thiocyanate extraction method (8) or by purification on an affin-
ity matrix as in many commercial kits for preparation of RNA. If
an mRNA is being analysed and the sensitivity of the analysis is
an issue, polyA+ RNA (mostly mRNA) is isolated by oligo(dT)
chromatography (9) prior to gel electrophoretic fractionation.
Cytoplasmic, polyadenylated RNA constitute approximately 3% of
whole cell RNA in human cells. By isolating this fraction, more of
the relevant RNA can be loaded on the gel and consequently less
abundant RNAs can be detected. Another application that calls
for fractionation is analysis of the small RNAs. Simple fraction-
ation protocols based on RNA binding columns can be applied
to size-fractionate whole cell RNA and thus used to increase the
capacity of gels, e.g. for analysis of cellular micr oRNAs.
Large RNAs (mRNA size) are size-fractionated on agarose
gels. The useful range of agarose-concentration is 0.8% (for very
large RNAs) to 1.4% (w/v). Outside this range, the gels either
become difficult to handle or less efficient in blotting. Small
RNAs are size-fractionated on polyacrylamide gels in the range
4–20% acrylamide. The size-fractionation is performed at dena-
turing conditions. RNA molecules fold into complex structures
that involve the formation of double-stranded segments by base
pairing. Thus, size-fractionation at native conditions will depend
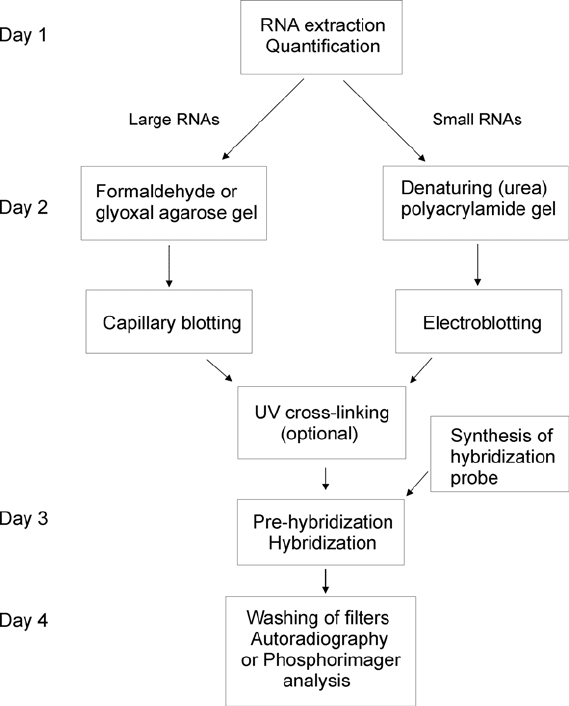
Northern Blotting Analysis 89
Fig. 7.1. Outline of a northern blotting analysis. The individual steps, except the extrac-
tion of RNA, are discussed in the text. A standard experiment takes 3–4 days. The
overnight blotting step by upward capillary transfer can be shortened to few hours by
application of alternatives discussed in the main text. The overnight hybridization step
can similarly be performed in hours using “fast” hybridization solution alternatives.
not only on the length of the RNA chain but also on the fold-
ing. The two main methods for denaturing agarose gels are based
on formamide/formaldehyde (10) and glyoxal (11), respectively.
Formaldehyde is used both in the loading buffer (together with
formamide) and in the gel. Formamide denatures the RNA and
formaldehyde maintains the denatured state by re acting cova-
lently with the amine groups of adenine, guanine, and cytosine
bases. Since these bases are directly involved in formation of the
A-U, G-U, and G-C base pairs in RNA, formation of secondary
structure during the gel run is prevented. The reaction is reversed
by treatment of the filter after blotting to allow the RNA to base
pair with the probe. Glyoxal is not used in the gel but only in
pre-treatment of the RNA sample. Glyoxal denatures RNA by
90 Josefsen and Nielsen
reacting covalently with guanine bases with the effect of inhibit-
ing base pair formation similarly to formaldehyde. The gly-
oxal reaction is also reversed by treatment of the filter prior to
hybridization analysis. Glyoxal gels tend to produce sharper bands
than formaldehyde gels but care must be taken to prevent pH-
dependent reversal of the glyoxalation during the gel run. In
northern blotting analysis of small RNAs, the sample is denatured
by heating in a loading buffer containing formamide or urea and
the gels are made with 50% (approximately 8 M) urea (12).
One of the most common uses of northern blotting analysis
is comparison of RNA levels in different experimental situations
represented by different lanes on the gel. Thus, equal loading
of RNA samples is critical. When whole cell RNA is used, equal
amounts (up to 20 μg) based on spectr ophotometry is applied to
the gel. This is robust because cytoplasmic ribosomal RNA make
up 70–80% of total RNA and together with other stable RNA are
unlikely to be affected by the experimental variable. When enrich-
ment by isolation of polyA+ RNA or by other means is used, the
RNA of interest can be enriched to different degrees in the sam-
ples that are compared. In this case, the RNA levels are normally
expressed in relation to an RNA that is detected in parallel by a
different probe and known to be unaffected by the experimen-
tal variable. Popular examples of standards for comparing mRNA
levels are cytoskeletal mRNAs (actin, tubulin), cyclophilin, and
the mRNA coding for the glycolytic enzyme glucose-6-phosphate
dehydrogenase (GAPDH). However, there are numerous exam-
ples of variations in the expression levels of these and the choice
of standard should be considered carefully for each type of exper-
iment. It has been suggested that ribosomal RNA would be a
convenient general standard based on the arguments presented
above. This runs into technical problems in northern blotting
analysis because the ribosomal RNAs are so prominently present
that they saturate the filter at the position of the ribosomal RNA
bands making quantitative hybridization impossible.
Northern blotting analysis of mRNA is very sensitive to even
slight degradation of the sample. For this reason, it is important
to visually inspect the RNA after gel electrophoresis. RNA is con-
veniently stained by ethidium bromide similarly to DNA albeit
less efficiently because binding is by ionic interaction rather than
intercalation. One approach is to include ethidium bromide in the
loading buffer for formaldehyde gels. This allows inspection dur-
ing the gel run. This is not an option with glyoxal gels because
ethidium bromide reacts with glyoxal and interferes with its abil-
ity to denature the RNA. Alternatively, the gel is stained after
the run. Inclusion of ethidium bromide in the gel is not rec-
ommended because it migrates towards the cathode creating a
front of stain in the gel. An important point in staining of whole
cell RNA is that the quality of the RNA can be assessed from
Northern Blotting Analysis 91
inspection of the ribosomal RNA bands. The large (LSU) and
small (SSU) subunit ribosomal RNAs are found in 1:1 ratio in
cells. Thus, the differences in size (5,035 nt for LSU (28S) and
1,871 nt for SSU (18S) in humans) which is 2.7:1 should be
reflected in the intensities of the two bands in ethidium bromide
staining. If the ratio is less, this indicates that the RNA is partially
degraded. Binding of ethidium to the RNA may reduce transfer
and hybridization efficiency. If this is a problem, a test lane of the
RNA should be run in parallel and stained after the gel run.
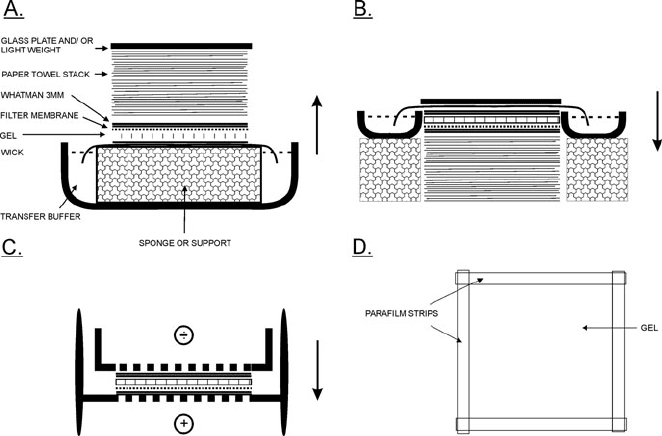
There are essentially two approaches to the blotting step of
northern blotting analysis, capillary blotting and electroblotting.
The traditional passive capillary transfer method (similar to the
classical Southern blotting method (3)) is beautiful in its sim-
plicity and can be performed with materials that are generally at
hand in the laboratory. In essence, a stack of paper towels or sim-
ilar absorbent material is used to draw a high salt transfer buffer
from a reservoir, through the gel and an RNA binding membrane
placed on top of the gel. The flow of buffer carries the RNA out of
the gel so that it is trapped on the membrane. The setup for such
an upward capillary transfer is shown in Fig. 7.2a. Transfer takes
several hours and is usually performed over night. A faster and
equally simple setup is the downward capillary transfer depicted
Fig. 7.2. Three different configura tions for the gel transfer (blotting) step in northern blotting analysis. a Standard upward
capillar y transfer. This is used with agarose gels and takes several hours (3–6) to overnight to go to completion. b
Downward capillary transfer. This is used with agarose gels and is completed in 1 h but can be left for longer times. c
Electroblotting. This is used with polyacrylamide gels and (rarely) with agarose gels. The time depends on the type of gel
but is usually complete in 1 h. d In all three transfer configurations is the gel lined with overlapping strips of parafilm to
avoid short circuiting.
92 Josefsen and Nielsen
in Fig. 7.2b (13). Transfer in this setup takes around 1 h but
can be left for longer periods of time without problems. Other
ways to speed up the transfer is to use a vacuum below the gel
or to apply a pr essure above the gel. Specialized equipments for
these configurations are commercially available. Capillary transfer
is the method of choice for agarose gels, whereas electroblotting
(14) is used for polyacrylamide gels. In this method, the gel and
the membrane are placed in a special cassette placed in an elec-
trophoresis cell (see Fig. 7.2c). An electrical field is applied per-
pendicular to the cassette and the RNA is transferred onto the
membrane. This appr oach is also used for western blotting of
proteins and a variety of electroblotting apparatus are available.
One particular useful variety is the semi-dry type in which gel and
membrane are sandwiched between Whatman 3MM paper wet-
ted with transfer buffer. Electroblotting can be performed in less
than an hour in some cases.
The choice of filter membrane is critical in nor thern blotting
analysis. There are several different options, but for most pur-
poses, positively charged nylon membranes are optimal. The pos-
itive charge is due to amino groups on the membrane surface and
has the dual effect of increasing the affinity for nucleic acid and
facilitating the immobilization of the RNA on the membrane. For
the very same reasons, nylon membranes tend to produce more
background due to unspecific binding of the probe unless care is
taken to avoid this. Nylon membranes are characterized by high
tensile strength and high binding capacity (4–500 μg/cm
2
). They
are produced with different pore sizes (0.22 and 0.45 μm) and
some are optimized, e.g. for repeated use or for binding of small
RNAs. It is important to notice the manufacturer’s recommenda-
tions on handling of the membrane. For convenience, it is advis-
able to use membranes that are charged on both sides.
If the gel was stained with ethidium bromide, successful trans-
fer can be confirmed by inspection of the filter in UV light (or
even in daylight). It is also possible to inspect the gel to check
that transfer was complete and not biased towards transfer of
small RNAs. Alternatively, filter-bound RNA can be stained at
later stages by methylene blue (15). RNAs transferred to posi-
tively charged nylon membranes at alkaline conditions become
covalently bound to the filter. In all other situations, it is impor-
tant to ensure that the sample RNA is immobilized on the filter
in order to prevent loss during subsequent hybridization steps.
It is advisable to follow the recommendations of the manufac-
turer of the membrane, but some general rules apply. The most
common method is to use a UV light source to immobilize the
RNA (16). UV light activates pyrimidine bases (thymine and
uracil) to form covalent bonds with the surface amines of pos-
itively charged membranes (17). Wet nylon membranes gener-
ally require a dose of 1.6 kJ/m
2
, whereas air-dried membranes
Northern Blotting Analysis 93
require only 0.16 kJ/m
2
. This is most conveniently delivered
by specialized equipment with a calibrated UV light source (e.g.
Stratalinker from Stratagene). These instruments generally have
an “auto-crosslink” setting at a dose optimized for damp mem-
branes. Other UV sources can be used, but this may take con-
siderable optimization and care should be taken to avoid exten-
sive radiation at short (254 nm) wavelengths that will damage
the RNA. An alternative way to immobilize RNA on nylon mem-
branes is to incubate the filter at 65
◦
C for 1–2 h. The exact princi-
ple behind fixation is not known in this case. Some manufacturers
claim that the ionic binding to the membrane is sufficient and that
there is no need for further steps.
Membranes with immobilized RNA can be stored for
extended periods of time and can be used as a record of RNA
experiments for later analysis. The next step in the procedure
is hybridization analysis. This involves application of a probe to
the membrane bound material at conditions that favours spe-
cific binding to complementary target molecules on the mem-
brane. The probe is labelled to allow subsequent detection of its
complex with the target. The most common approach is radioac-
tive labelling (preferably
32
P), but non-isotopic alternatives exist.
These probes are generally much less sensitive than
32
P-labelled
probes. The pr obes can be DNA or RNA that can be labelled in
various ways. The most popular type of probes has been double-
stranded plasmids or PCR-products labelled with the random-
priming labelling method (18), but asymmetric (single-stranded)
PCR-products and RNA probes made by in vitro transcription
are more sensitive due to the absence of a competing strand in
the probe. Oligonucleotide probes are much less sensitive and
are used when discrimination between target molecules of closely
related sequences is the issue.
A large variety of hybridization buffers or systems are avail-
able and can be used with success for membrane hybridizations.
Hybridization theory does not apply to nucleic acids immobilized
on a membrane and the success is mainly empirically based. How-
ever, some rules of thumb exist. The critical value is the melting
temperature (T
m
), defined as the temperature at which 50% of all
hybrids are dissociated into single strands. The maximum rate of
hybridization is typically found around 20
◦
C below the melting
temperature. For long probes, the T
m
for DNA:RNA hybrids is
T
m
= 79.8
◦
C + 18.5 log[Na
+
] + 58.4 (%G+C) + 11.8 (%G+C)
2
– 0.5 (% formamide) – (820/L) (19). (%G+C) is the percentage
of guanine+cytosine content expressed as a mole fraction, [Na
+
]
is the molar sodium concentration, and L is the effective length
of the probe taking part in base pairing with the target. The
constants are slightly different for DNA:DNA and RNA:RNA
hybrids. Formamide is used to reduce the hybridization temper-
ature, which is particularly important when using RNA probes
