Nielsen H. RNA: Methods and Protocols
Подождите немного. Документ загружается.


54 Torarinsson
with some more concrete examples to help us find information
relevant to the structure of the transcription unit. I r ecommend
that you read this chapter while using a computer to follow the
instructions. In selected sections I have added questions (in the
Notes section) to make this chapter more interactive and to help
you understand the potential of genome browsers.
2. Getting Started
The three major genome browsers at UCSC, Ensembl and NCBI
all provide two main entry points to the browsers. These are with
a known sequence or querying for known coor dinates or some
search term. In this chapter we will focus on the case when we
know which gene we are interested in. If you want to enter the
browsers with an unknown sequence, instead of a known gene,
do not worry, the browser navigation described below is exactly
the same, the only difference is that you use BLAT (UCSC) or
BLAST (Ensembl and NCBI) to compare your sequence to their
database and enter browser via the results, and not by accessing
it directly with a known gene. Beware, although major updates
on the browsers’ appearances are rare, some things might have
changed since this was written.
2.1. UCSC
1. Point your browser to http://genome.ucsc.edu.
2. Choose either “Genomes” or “Genome Browser” in the top
left corner.
3. Here you can choose the clade, the genome, and which
assembly. In the fields named “position or search term” you
can enter different kinds of information. These include the
following:
– Gene names → BRCA1
– Specific region → chr7:1–10,000 or simply chr7
– Keywords → kinase, receptor, specific disease
– IDs → NP, NM, OMIM and more
4. To demonstrate we will use the Human assembly from
March 2006 and search for DTNBP1 (see Fig. 5.1). When
you search for this you get a list of data matching your search
term. In our case I chose the third listing under UCSC genes
Fig. 5.1. How you search for DTNBP1 in the UCSC genome browser.
Genome Browsers 55
“dystrobrevin binding protein 1 isoform a.” By following
this link you will reach the heart of the browser.
2.1.1. Genome Browser
To better understand how this browser works it is good to know
the basic organization of the underlying data. Everything in the
browser is organized along the genomic sequence backbone.
The data exist in so-called “tracks,” which are kept in MySQL
databases. For example, there is a track called “UCSC Genes”
and the corresponding database for this track holds information
like the name and the ID of this gene, which chromosome it
belongs to and start and stop positions (also start positions for
the UTR and exons, etc.). The positions are all relative to the
genomic backbone, so if we are studying a region on chromo-
some 7 between position 10 and 10,000, the genome browser
will check the “UCSC Genes” track and plot a gene on the image
if it finds an overlap. The tracks are grouped together so that each
group contains similar type of information, i.e., the “Genes and
Gene Prediction Tracks.” Continuing with our DTNBP1 exam-
ple we are now at the heart of the genome browser.
5. If you have never used your current computer to access the
UCSC genome browser it will display the default tracks; oth-
erwise, if you used it before, it will remember if you removed
or added tracks and show these tracks again. If you are inter-
ested in the default tracks, simply click the “default tracks”
button. To begin with all this information can be over-
whelming so we start by removing all the tracks by selecting
the “Hide all” button, located just below the image.
6. Now the image is almost empty, only displaying where we
are on chromosome 6. Now let us add the “UCSC Genes”
track. The “UCSC Genes” track is located under the “Genes
and Gene Prediction Tracks.” By clicking on the pulldown
menu you will usually have five options:
– “Hide”: completely removes a track from your image.
– “Dense”: all items become collapsed into a single line –
fuses all the rows of data into one.
– “Squish”: each item is on a separate line, but at 50% of its
regular height.
– “Pack”: each item is separate, but efficiently stacked like
sardines. However, they are full height, which makes it
different from squish.
– “Full”: each item, e.g., gene, is on a separate line.
By selecting the link above the pulldown menus you can read
the information about the track, how it was generated, and so on.
Sometimes you may further specify how the track should be dis-
played. In the “UCSC Genes” case you can, for example, change
which ID it displays and read that this track is based on Ref-
56 Torarinsson
Seq, UniProt, CCDS, and Comparative Genomics. Let us choose
“full” for the “Known Genes” track. Then we update the image
by clicking on the “Refresh” button, either just below the image
or at the bottom of the page.
7. The image now displays a few versions of the DTNBP1 gene,
with different colors (see Note 2). Our selected isoform is
the one with dark blue background in its name (DTNBP1).
The full-size boxes indicate exons, the half size-boxes indi-
cate UTRs, and the arrows indicate the direction of the tran-
scription.
8. To get mor e information about the gene you click on one
of the genes (see Note 3), (see Note Q-1). This takes you
to a new page with many information and links to other
databases with further details about this gene. On the top
of this page there are links to all the information within this
page, and just below ther e are links to external databases and
other resources within UCSC like the “Proteome Browser”
and “VisiGene.”
9. Finally, it is worth mentioning that when you ar e on the page
with the browser image, there are a couple of useful links on
the top in the blue horizontal bar. Selecting “DNA” will
able you to get the DNA sequence for the region where the
browser is located; furthermore, there are several options to
manipulate the DNA output, like repeats in lower case and
coloring of some features. The “PDF/PS” link gives you a
PDF and a PS-formatted fi le of the image, which is useful
for publications or presentations.
Although we only used two tracks and one genome in this
little example, the beauty of the genome browser is that the pro-
cedure is exactly the same for all the tracks and genomes. The
procedure is always the same but the information available varies
between tracks and genomes. So if you can follow this small exam-
ple, you should be able to study every track and genome in the
browser. The best way to learn to navigate the browser is by exper-
imenting on your own.
2.2. Ensembl
1. Point your browser to http://www.ensembl.org/index.
html.
2. We stay true to our species and select Homo sapiens, assem-
bly GRCh37 (the link to the right next to the “Michelan-
gelo” icon).
3. Here we can search with some search term similar to UCSC.
Here we search for DTNBP1 again. Below “By Species”
click on “Homo sapiens” and then “Gene” to go to the
search results (you can also enter “By Feature type” her e ,
it does not matter).
Genome Browsers 57
4. This gives us two matches, either the Havana or the Ensembl
protein coding gene. We choose Ensembl since it contains
more information. There are two different links, a long one
with the name and a shorter one named “Region in detail.”
The long link will take you directly to gene report for this
gene, whereas the “Region in detail” link will take you to the
Ensemble equivalent of the UCSC genome browser. Let us
start by selecting the “Region in detail.” Like UCSC every-
thing in the viewer is organized along the genomic sequence
backbone in different tracks.
5. This V iew displays three image boxes. The top image titled
“Chromosome 6” shows chromosome 6 with a red box sur-
rounding the region where we are. The next image zooms
in on chromosome 6, again with a red box surrounding
the region where we are. The information in this view
includes “Contigs,” Ensembl/Havana genes, non-coding
RNA (ncRNA) genes, and ncRNA pseudogenes. Here, the
red box surrounds our DTNBP1 gene and we can see that
there is a gene named JARID2 upstream of our gene. Fur-
thermore, the ncRNA gene U6 is upstream of our gene.
Here, we can click on every gene to obtain further infor-
mation about each gene, but before you do that, study the
bottom image.
6. The bottom image is where we can hide and show all the
tracks available at Ensembl. To add or remove tracks, follow
the “Configure this page” link in the left-side navigation.
You select the group of tracks on the left and click on the
box in front of the track you are interested to select if and
how it should be displayed. Finally, click on the “Save and
Close” icon in the top right corner of the popup window,
this will update the image (see Note 4). If you have a look
at the “Ensembl/Havana gene” track you see that exons are
indicated with filled boxes and UTRs with non-filled boxes.
7. In the bottom view, if you click on one of the transcripts
in the “Ensembl/Havana genes” track like the “DTBP1-
001” you will see a popup box. In this box you can choose
between accessing the gene, transcript, or peptide informa-
tion page. Choose the gene (“Gene:ENSG00000047579”).
This will take you to the gene report for that gene.
8. This page displays the usual information at the top like the
ID and a description. Below that, there are some data con-
cerning the transcripts. In the left menu you can find links
to features that are often very relevant in understanding
the gene structure and potential regulation of this gene (see
Note Q-2). These will be discussed in more details in the
next section.

58 Torarinsson
3. Comparative
Genomics
UCSC and Ensembl are useful in different ways, when studying
the conservation of a given gene in different organisms. UCSC is
very good to quickly locate highly conserved regions, for exam-
ple, high conservation upstream, downstream, or in the UTR
of a given gene, indicating a possible regulatory role of that
region. With UCSC you can find links, from the gene informa-
tion page, to a few orthologues and view them separately in the
browser. Ensembl on the other hand has many more orthologue
predictions, with emphasis on predictions, and it is possible to
view them simultaneously. This makes things like comparing exon
structure and genomic context much easier with Ensembl. Fur-
thermore , it is easy to retrieve pairwise or multiple alignments.
Let us work with an example to illustrate these different strengths.
3.1. UCSC
1. Like in our earlier example, we go to http://genome.
ucsc.edu and choose “Genome Browser” in the top left
corner.
2. In this example we will work with homeobox C8. Select
Human, assembly March 2006, and either search for hoxc8
(and choose the first match in “UCSC Genes”) or go directly
to the location “chr12:52,689,157-52,692,812.”
3. To ease the visual inspection of the tracks, start by clicking
on “hide all” tracks just below the image. Now select “pack”
in the pulldown menus for the “UCSC Genes,” “Conser-
vation” and “28-way Most Cons” (see Note 5) tracks (in
the “Comparative Genomics” group). Click on “Refresh”
to apply your changes.
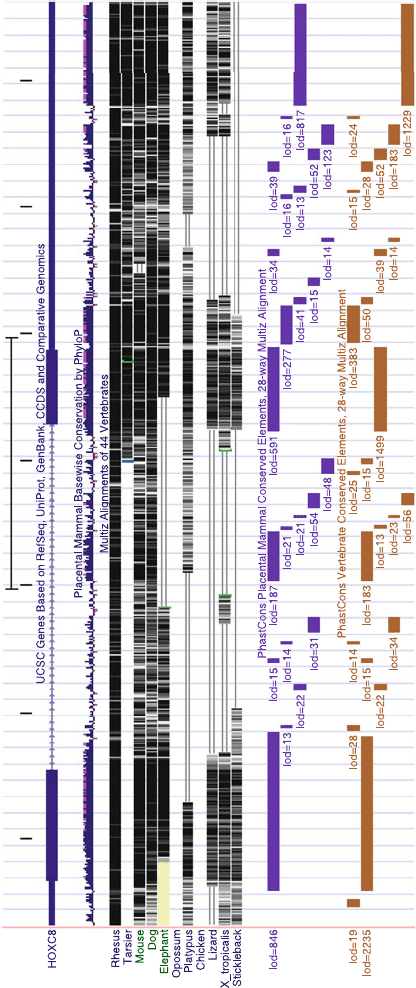
4. The image (see Fig. 5.2) now displays the HOXC8 gene.
Below the “UCSC Genes” track you see the “Conservation”
track. The histogram indicates the level of conservation and
below you can see where the conserved regions lie in the
respective organisms. Finally, at the bottom you see the “28-
way Most Cons” track. It is often interesting to study the
gene and the conservation simultaneously, like for this gene
we can see that the 3
UTR is extremely well conserved. It
is often good to be aware of simple things like this when
studying the transcription and regulation of this gene (see
Note Q-3).
3.2. Ensembl
1. Point your browser to http://www.ensembl.org/index.
html.
2. Select the human genome and then search for HOXC8.
Click on “Homo sapiens” and the “Gene.” Go to the
gene report page for the Ensembl gene (the Ensembl ID is
Genome Browsers 59
Scale
chr12:
1 kb
52689500 52690000 52690500 52691000 52691500 52692000 52692500
Vertebrate Multiz Alignment & Conservation (44 Species)
PhastCons Conserved Elements, 28-way Vertebrate Multiz Alignment
Fig. 5.2. A UCSC genome browser image displaying three tracks, the “UCSC Genes” track, the “Conservation” track, and the “28-way Most Cons” track.
60 Torarinsson
ENSG00000037965). Do NOT click on “Region in detail”
click on the long link with the gene name; this will take you
directly to the gene report page.
3. The gene report for HOXC8 reveals, amongst other things,
that there is only one known transcript, several putative
orthologues, and several putative paralogues in human
(Orthologues and Paralogues links are in the left-side
menu).
4. From the link “Genomic alignments” you can view this
gene in genomic alignments to other species. Select the
“11 eutherian mammals EPO” from the “Select an align-
ment” pulldown. Right click on “Go to a graphical
view” and open it in a new window/tab. This win-
dow makes it quite easy to compare genomic contexts
in several species simultaneously. Now we are at zoom
level two (the bar on the right surrounded by “+” and
“−” icons is at position 2) and only see HOXC8; let
us change to zoom level five by clicking on bar num-
ber five (corresponds to region 54354719–54454718) (see
Fig. 5.3).
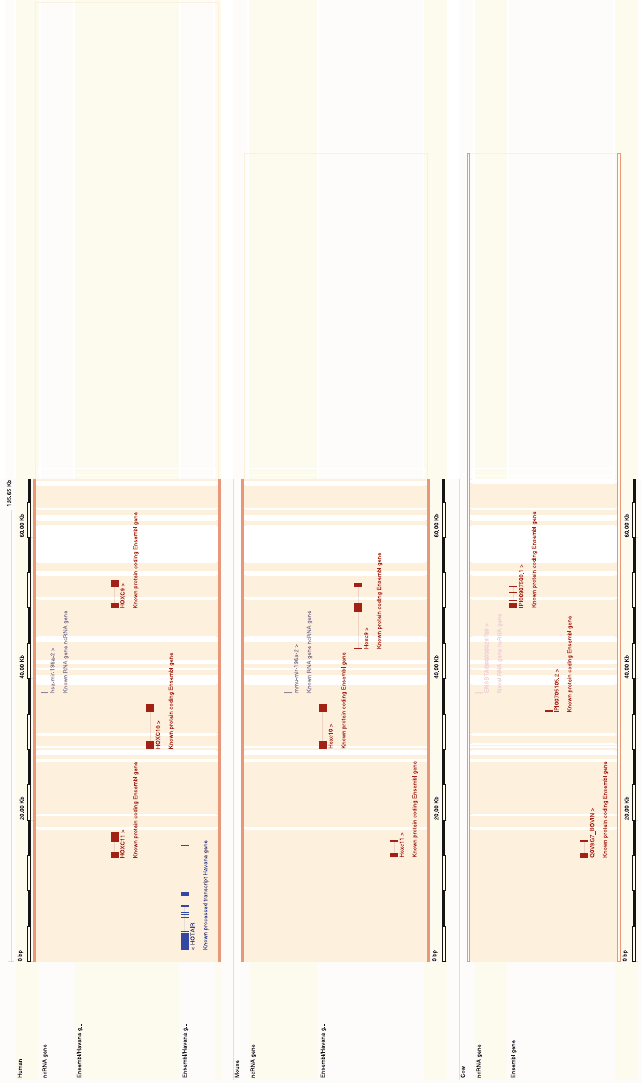
5. Studying the genomic context of a given gene in sev-
eral organisms can often be very useful. For example,
when studying how the gene might be regulated, but
also to do things like annotate genes. One could say that
it is quite likely that the “Novel RNA genes” ENSB-
TAG00000029788 and ENSECAG00000026361 in cow
and horse, respectively, is the micro RNA miR-196, consid-
ering the annotation in the other mammals. So here we have
a relatively easy way of using well-annotated organisms, to
help annotate other less annotated organisms.
6. Now go back to the gene report for human HOXC8; if you
do not have it open, just go back or click on the HOXC8
gene in the human box. In the Orthologues view you can
do four things: (i) click on the first link and view the gene
report for the orthologue, (ii) click on “Multi-species view”
where you can view the orthologue, together with your
gene, in a similar way to what we just did in Step 5, (iii)
click on “Align” to obtain the alignment between your gene
and your orthologue. Via “Configure this page” you can
choose between DNA or peptide, several output formats,
and species, and (iv) view the gene tree.
7. Still in the Orthologue view, we can click on “View sequence
alignments of these homologues.” As the name implies, this
will show all the pairwise orthologues and paralogues align-
ments (the same as clicking on “Align” for every ortho-
logue).
8. Finally in the transcript view, in accessed through the
“Transcript: HOXC8-201” link at the top (next to “Gene:
Genome Browsers 61
Fig. 5.3. A simplified image of the region surrounding the HOXC8 gene in humans. This image only shows the Ensembl/Ha vana genes and ncRNAs for human, mouse and cow (i.e.,
in “Configure this page” I removed some default tracks and species).

62 Torarinsson
HOXC8”) ther e is more interesting data to be obtained.
These include the following links:
– “Gene Ontology”: where you can see which GO terms
(see Note 6) have been mapped to this gene, and by fol-
lowing the links there you can further information con-
cerning the GO term.
– “Domains & features”: where you see which domains the
gene has and view all the genes with the same domain.
– “Population comparison”: where you can see variations in
this transcript (i.e., to Watson and Venter).
4. Expression
and Regulation
Again, when studying expression and regulation, the strengths of
UCSC and Ensembl are different. UCSC, with its simple way of
viewing many tracks simultaneously, makes it very e asy to com-
pare your gene with various expression and regulation tracks. To
some extent this is also possible in Ensembl, though it is mor e dif-
ficult and time consuming. What Ensembl has is a nice view of the
regulatory factors from the cisRED database, predicted miRNA
target sites from miranda analysis, and regulatory features from
the Ensembl Regulatory Build, to mention a few. Again, with
custom tracks, this is also possible in UCSC but more difficult.
Here is a simple example.
4.1. UCSC
1. We continue studying the HOXC8 gene. If you do not have
it open fr om Section 3.1, repeat Steps 1–3.
2. Select “configure” under the browser image. Scroll down to
the “Expression” and “Regulation” groups, click on “show
all” for both groups and then "submit" at the top of the
page.
3. Here you can compare our gene with several expression
tracks from GNF, Yale and Affymetrix. Regulatory tracks
include, for example, CpG islands, conserved transcription
factor binding sites, regulatory elements from the ORe-
gAnno database, and a track displaying ESPERR regulatory
potential scores computed from alignments of seven organ-
isms (the darker the color, the higher regulatory potential)
(see Note Q-4).
4.2. Ensembl
1. We continue studying the HOXC8 gene. If you do not have
it open fr om Section 3.2, repeat Steps 1 and 2.

Genome Browsers 63
2. In the left menu, select “Regulation.” This page contains
a graphical display and a listing with relevant links of reg-
ulatory features from the cisRED database and regulatory
features from the Ensembl Regulatory Build, among others
(see Note Q-5).
5. Other
I have just covered a fraction of the functionality at UCSC and
Ensembl. There are several other interesting features in both
browsers.
5.1. UCSC
Other interesting features at UCSC include the VisiGene browser
and the ENCODE tracks. The VisiGene browser is a virtual
microscope for viewing in situ images. These images show where
a gene is used in an organism, sometimes down to cellular res-
olution. With VisiGene users can retrieve images that meet spe-
cific search criteria, then interactively zoom and scroll across the
collection. A link to the VisiGene browser is available from the
UCSC browser home page.
If your gene of interest happens to be located in the
ENCODE regions, there are many ENCODE specific tracks avail-
able. This r eveals all the ENCODE tracks, which can be viewed
like any other track we have looked at so far. The ENCODE tracks
include groups like “Transcription,” “Chromatin Immunoprecip-
itation,” “Chromatin Structure,” and additional “Comparative
Genomics and Variation” tracks.
5.2. Ensembl
No matter if you are looking at a gene report or the “Region in
detail” many of the most interesting features at Ensembl are often
located in the left menu. For example, when viewing a gene report
you can view a multiple alignment of the genomic sequence with
several organisms, or get a nice graphical phylogenetic tree (the
“Gene Tree (image)” link) or variations (the “Variation table” and
“Variation image” links). The “Variation table” site has a listing
over the variations, where they are, which alleles are involved and
if they are synonymous or non-synonymous when located in a
coding region.
6. Notes
1. Much more detailed information and good online tutorials
are available for the browsers. OpenHelix (http://www.
openhelix.com/downloads/ucsc/ucsc_home.shtml)
