Moss Tom. DNA-protein interactions: principles and protocols
Подождите немного. Документ загружается.

Cleavage of DNA by Histone–Fe(II) EDTA 285
6. Several assays for the correct binding of linker histones to DNA have been per-
formed (17,18). One of the easiest involves a brief digestion with micrococcal
nuclease in the chromatosome stop assay (13).
3.6.2. Site-Directed Hydroxyl Radical Mapping
of Linker Histone-DNA Interaction
1. Scale up the binding reaction to include 40,000–50,000 cpm of labeled reconsti-
tuted nucleosomes and add enough modified mutant linker histone to form
H1–nucleosome complexes.
2. Add glycerol to 0.5% final concentration (see Note 9).
3. Add sodium ascorbate to a final concentration of 1 mM.
4. Add H
2
O
2
to a final concentration of 0.0075%.
5. Incubate for 30 min at room temperature in the dark.
6. After 30 min, add 1/10 vol of 50% glycerol and 10 mM EDTA solution.
7. Load samples immediately onto a running (90 V) preparative 0.7% agarose and
0.5X TBE gel.
8. Separate the samples so that the H1–nucleosome complexes are well resolved
from tetramer and free DNA bands.
9. Next, wrap the gel tightly with plastic wrap so that the gel cannot move within
the plastic. Lay fluorescent markers onto various portions of the gel for align-
ment purposes (can be obtained from Stratagene) or accurately mark the position
of the gel on the film.
10. Expose the wet gel for several hours at 4°C.
11. Next, develop the autoradiograph and overlay onto the wet gel, lining up the
fluorescent markers.
12. Cut and remove the agarose containing the H1–nucleosome complexes or bands
of interest and place them into Series 8000 Microcentrifuge Filtration Devices.
13. Freeze the filtration tubes containing the agarose plugs on dry ice for 15 min.
14. Spin down the agarose in a microcentrifuge at maximum speed for 30 min at
room temperature. The fluid from the agarose matrix will be collected in the
2-mL centrifuge tube surrounding the filtration device.
15. Gently remove the agarose plug from the bottom of the filtration device and place
into a clean Eppendorf tube. Save the centrifugation devices for use later.
16. Using a microcentrifuge pestle, crush the agarose pellet and add 500 µL of 10 mM
Tris-HCl, pH 8.0, and 0.1% SDS and continue to crush the agarose.
17. After the agarose is crushed into tiny pieces, place all samples at 4°C
overnight.
18. Place the crushed agarose into the same centrifugation device and pellet.
Spin down the agarose in a microcentrifuge at maximum speed for 30 min at
room temperature.
19. Combine identical samples from both spins and precipitate the DNA.
20. Dissolve the DNA in 15 µL of TE buffer.
21. Heat the samples to 90°C for 2 min to denature.

286 Chafin and Hayes
3.7. Sequencing Gel Analysis of H1
°
C-EPD Cleavage
3.7.1. Sequencing Gel Electrophoresis
1. Add equal numbers of counts from each sample, including the G specific reac-
tion, to clean eppendorf tubes.
2. Place the samples into a Speedvac concentrator and dry to completeness.
3. Dissolve the samples in 4 µL of formamide loading buffer.
4. Place samples directly onto ice to prevent renaturation.
5. Separate samples on a 6% polyacrylamide and 8 M urea sequencing gel running
at constant 2000 V.
3.7.2 Example of Site-Directed Cleavage
of Nucleosomal DNA by EPD
An example of a linker histone site-directed DNA cleavage reaction is pre-
sented in Fig. 2. A schematic of the 5S mononucleosome is shown in the left
panel. The thick black line represents the 5S ribosomal DNA fragment that
contains the transcriptional coding sequence for this gene (gray arrow). The 5S
ribosomal gene fragment was used because it contains a nucleosomal position-
ing sequence that precisely wraps the DNA around the core histones and pro-
vides a homogeneous population of nucleosomes. Furthermore, because one
major translational position predominates within this population of nucleo-
somes, the precise orientation of the DNA as it wraps around the core histones
is known. This enables us to determine to base-pair resolution, the sites of
cleavage by EPD with respect to the nucleosome structure. A singly end-
labeled 5S DNA fragment was incorporated into nucleosomes via the salt
dialysis procedure detailed earlier. Labeled mononucleosomes were bound by
an EPD-modified linker histone containing a single cysteine substitution for
the lysine residue at position 59, referred to as K59C-EPD. After allowing
hydroxyl radical cleavage for 30 min, the protein–DNA complexes were sepa-
rated on a 0.7% agarose gel (Fig. 2A) and the labeled DNA fragments corre-
sponding to the H1–nucleosome complexes were purified. These purified
DNAs were then analyzed on a 6% sequencing gel (Fig. 2B, right panel).
Fig. 2 (opposite page). (A) Various DNAs from control or hydroxyl radical cleav-
age reactions were separated on a 0.7% agarose gel. The wet gel was exposed to auto-
radiographic film for 3–4 h according to Subheading 3.6.2. Lanes 1 and 2 contain free
DNA (FD) and bulk nucleosomes (Nuc.), respectively, not exposed to hydroxyl
radical cleavage. Lanes 3–5 contain nucleosomes or H1–nucleosome complexes
(H1–Nuc.) subjected to hydroxyl radical cleavage. Nucleosomes–K59C (lane 3),
nucleosomes with unmodified K59C (lane 4) or nucleosomes with K59C–EPD (lane 5).
(B) A linear schematic of the 5S nucleosome is shown (left). The DNA (black line)
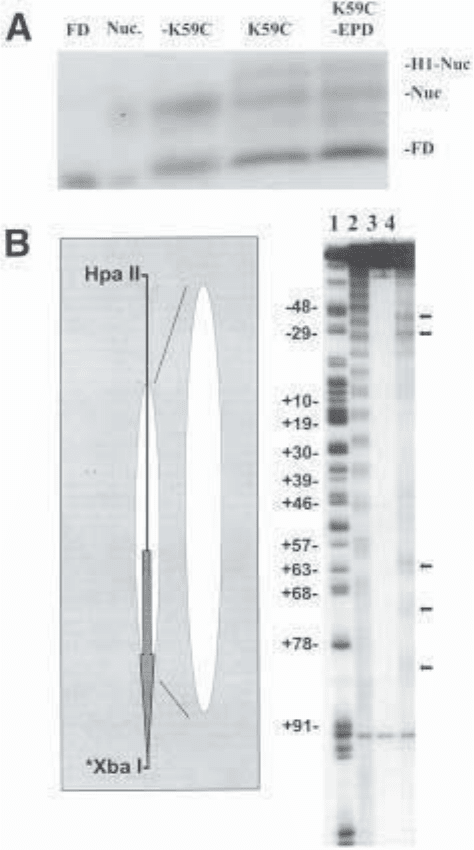
Cleavage of DNA by Histone–Fe(II) EDTA 287
was restricted with the restriction enzymes shown and radiolabeled (
*
) at the XbaI site.
The position of the 5S nucleosome (white oval) is shown with respect to the start site
of transcription (gray arrow) and with respect to the size of DNAs in the sequencing
gel (larger white oval). Labeled DNAs from hydroxyl radical cleavage reactions were
analyzed on a 6% sequencing gel (right). Lane 1: Maxim–Gilbert G-specific reaction;
lane 2: hydroxyl radical cleavage in the absence of histone H1; lanes 3 and 4: hydroxyl
radical cleavage with K59C (lane 3) or K59C–EPD (lane 4). Specific cleavages are
shown to the right of the gel (black arrows).
288 Chafin and Hayes
The gel reveals that the reconstituted mononucleosomes used for this
experiment give a characteristic 10-bp protection when footprinted by general
cleavage with hydroxyl radical in the absence of linker histone (15). This indi-
cates that the 5S DNA has been properly assembled with the histone octamer
into a nucleosome (Fig. 2B, lane 2). The linker histone-directed cleavage
experiments are shown in lanes 3 and 4. No cleavages are observed when the
reaction is carried out in the presence of unmodified K59C (Fig. 2B, lane 3). In
contrast, when the cleavage reagents are added in the presence of K59C-EPD
bound nucleosomes, two sets of cleavages are evident (Fig. 2B, lane 4). The
cleavages at +62, +72, and +82 correspond to the end of the nucleosome where
the DNA exits. This result is consistent with previous data suggesting that
the linker histone binds the nucleosome at the periphery, tucked inside a super-
helical gyre of DNA (12,19). A second set of cleavages occurs at –29 and –39.
These cleavages occur on the DNA strand directly underneath that of the +62/
+72/+82 cleavages as the DNA makes one full superhelical turn around the
histone octamer. It is possible that amino acid 59 makes close contacts with
both strands of the DNA, consistent with the strong cleavages seen at each site.
It is also possible that hydroxyl radicals have diffused away from the –29/–39-
cleavage site and cleave the DNA in other areas. Inconsistent with this, glyc-
erol, a very good hydroxyl radical scavenger, does not seem to have an effect
on the cleavages obtained with K59C-EPD at concentrations known to elimi-
nate hydroxyl radical cleavage (Chafin and Hayes, unpublished results).
4. Notes
1. A complication of the in vitro reconstitution procedure is that purified histone
proteins are often obtained in two fractions, H2A/H2B and H3/H4 (22). Thus, in
addition to total histone mass, the ratio between these two substituents must be
empirically adjusted to yield maximum octamer–DNA complexes (13).
2. Many ligation procedures are available from primary literature or commercial
sources. Ligation of two DNA fragments occurs more rapidly at room tempera-
ture or 37°C if the base-pair overlap is sufficiently stable.
3. Many DNA mini-prep procedures are described in detail in ref. (20). The DNA isolated
for the techniques described here were from the boiling DNA mini-prep procedure (20).
4. Before proceeding, it is recommended that a small amount of the culture be
checked for overexpression of the protein of interest. This can be done by remov-
ing 1 mL of the culture before and after induction by IPTG.
5. Sonication techniques tend to increase the temperature of the sample quickly,
which could induce proteolysis of the proteins. The sample must therefore be
cooled before and during sonication. Allow several min between sonication runs
to keep the sample as cold as possible.
6. Histones H2A or H2B do not bind to Bio-Rex 70 when purified individually.
However, we have found that when allowed to heterodimerize, they bind to the
Cleavage of DNA by Histone–Fe(II) EDTA 289
column and elute off consistently in 1 M NaCl (13). This characteristic could be
the result of the fact that histone H2A and H2B are completely unfolded when
separated from each other (21).
7. Storing labeled DNA in a concentrated form is not advised, as autodegradation of the
DNA takes place. DNA can be stored for several weeks at approx 5000 cpm/µL.
8. Several methods can be used for the incorporation of linker histones into recon-
stituted mononucleosomes. The method described here involves direct addition
of linker histones to mononucleosis in 50 mM NaCl. Linker histones are folded in
low-salt solutions in the presence of DNA (23). Indeed, we find that linker his-
tones can be directly mixed to nucleosomes in either 5- or 50-mM NaCl solutions
and these proteins then bind in a physiologically relevant manner (17).
9. Glycerol is a good scavenger for hydroxyl radicals and will generally inhibit
hydroxyl-radical-based cleavage if added at a final concentration over 0.5% and
therefore should be avoided. However, adding small concentrations of glycerol
will allow hydroxyl radical cleavage to occur if the EPD moiety is in close prox-
imity to the DNA backbone but not cleavage from sites farther away.
References
1. Wolffe, A. P. (1995) Chromatin: Structure and Function. Academic, London.
2. van Holde K. E. (1989) Chromatin. Springer-Verlag, New York.
3. Thoma, F., Koller, T., and Klug, A. (1979) Involvement of histone H1 in the
organization of the nucleosome and the salt-dependent superstructures of chro-
matin. J. Cell Biol. 83, 403–427.
4. Carruthers, L. M., Bednar, J., Woodcock, C. F. L., and Hansen, J. C. (1998)
Linker histones stabilize the intrinsic salt-dependent folding of nucleosomal
arrays: mechanistic ramifications for higher-order folding. Biochemistry 37,
14,776–14,787.
5. Crane-Robinson, C. (1997) Where is the globular domain of linker histone located
on the nucleosome? Trends Biochem. Sci. 22, 75–77.
6. Zhou, Y.-B., Gerchman, S. E., Ramakrishnan, V., Travers, Andrew, and
Muyldermans, S. (1998) Position and orientation of the globular domain of linker
histone H5 on the nucleosome. Nature 395, 402–405.
7. Ebright, Y. W., Chen, Y., Pendergrast, P. S., and Ebright, R. H. (1992)
Incorportation of an EDTA–metal complex at a rationally selected site within a
protein: application to EDTA–iron DNA affinity cleaving with catabolite gene
activator protein (CAP) and Cro. Biochemistry 31, 10,664–10,670.
8. Ermacora, M. R., Delfino, J. M., Cuenoud, B., Schepartz, A., and Fox, R. O.
(1992) Conformation-dependent cleavage of staphlyococcal nuclease with a dis-
ulfide-linked iron chelate. Proc. Natl. Acad. Sci. USA 89, 6383–6387.
9. Neelin, J. M., Neelin, E. M., Lindsay, D. W., Palyga, J., Nichols, C. R., and Cheng,
K. M. (1995) The occurrence of a mutant dimerizable histone H5 in Japanese
quail erythrocytes. Genome 38, 982–990.
10. Chen, Y. and Ebright, R. H. (1993) Phenyl-azide-mediated photocrosslinking
analysis of Cro–DNA interaction. J. Mol. Biol. 230, 453–460.
290 Chafin and Hayes
11. Lee, K.-M. and Hayes, J. J. (1997) The N-terminal Tail of Histone H2A Binds to
Two Distinct Sites Within the Nucleosome Core. Proc. Natl. Acad. Sci. USA 94,
8959–8964.
12. Hayes, J. J. (1996) Site-directed cleavage of DNA by a linker histone–Fe(II)EDTA
conjugate: localization of a globular domain binding site within a nucleosome.
Biochemistry 35, 11,931–11,937.
13. Hayes, J. J. and Lee, K.-M. (1997) In vitro reconstitution and analysis of
mononucleosomes containing defined DNAs and proteins. Methods 12, 2–9.
14. Rhodes, D. (1985) Structural analysis of a triple complex between the histone
octamer, a Xenopus gene for 5S RNA and transcription factor IIIA. EMBO J.
4(13A), 3473–3482.
15. Hayes, J. J., Tullius, T. D., and Wolffe, A. P. (1990) The structure of DNA in a
nucleosome. Proc. Natl. Acad. Sci. USA 87, 7405–7409.
16. Simpson, R. T. (1991) Nucleosome positioning: occurrence, mechanisms and
functional consequences. Prog. Nucleic Acids Res. Mol. Biol. 40, 143–184.
17. Hayes, J. J. and Wolffe, A. P. (1993) Preferential and asymmetric interaction of
linker histones with 5S DNA in the nucleosome. Proc. Natl. Acad. Sci. USA 90,
6415–6419.
18. Allan, J., Hartman, P. G., Crane-Robinson, C., and Aviles, F. X. (1980) The struc-
ture of histone H1 and its location in chromatin. Nature 288, 675–679.
19. Pruss, D., Bartholomew, B., Persinger, J., Hayes, J. J., Arents, G., Moudrianakis,
E. N., et al. (1996) A new model for the nucleosome: a binding site for linker
histone inside the DNA gyre. Science 274, 614–617.
20. Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989) Molecular Cloning: A
Laboratory Manual, 2nd ed., Cold Spring Harbor Laboratory, Cold Spring Har-
bor, NY.
21. Karantza, V., Baxevanis, A. D., Freire, E., and Moudrianakis, E. N. (1995) Ther-
modynamic studies of the core histones: Ionic strength and pH dependence of
H2A–H2B dimer stability. Biochemistry 34, 5988–5996.
22. Simon, R. H. and Felsenfeld, G. (1979) A new procedure for purifing histone
pairs H2A + H2B and H3 + H4 from chromatin using hydroxyapatite. Nucleic
Acids Res. 6, 689–696.
23. Clark, D. J. and Thomas, J. O. (1986) Salt-dependent co-operative interaction of
histone H1 with linear DNA. J. Mol. Biol. 187, 569–580.

Nitration of Tyrosine Residues 291
291
From:
Methods in Molecular Biology, vol. 148: DNA–Protein Interactions: Principles and Protocols, 2nd ed.
Edited by: T. Moss © Humana Press Inc., Totowa, NJ
20
Nitration of Tyrosine Residues
in Protein–Nucleic Acid Complexes
Simon E. Plyte
1. Introduction
Chemical modification is a powerful tool for investigating the accessibility
and function of specific amino acids within folded proteins. It has provided
significant information regarding the role of different amino acids at the bind-
ing sites of numerous enzymes and DNA-binding proteins. The identification
of such residues by chemical modification has then often be used to plan sub-
sequent site-directed mutagenesis experiments. These data complement those
from crystallographic and nuclear magnetic resonance (NMR) studies in deter-
mining the residues located at the active site; thus, one needs to consider all
these techniques when elucidating protein structure and function. For example,
chemical modification of leukotriene A4 hydrolase, 3-hydroxyisobutyrate
dehydrogenase, and lactate dehydrogenase (1–3) have contributed significantly
to the understanding of active-site mechanisms in these proteins and in
elucidating the mechanisms of DNA binding in the Fd and Pf1 gene 5 pro-
teins (4–5).
Reagents exist to modify cysteine, methionine, histidine, lysine, arginine,
tyrosine and carboxyl groups selectively. However, in this chapter, we are only
concerned with the selective modification of tyrosine residues (for reagents
and conditions for the modification of the other amino acids, see ref. 6). The
side chain of tyrosine can react with several compounds, the most commonly
used being N-acetylimidizole and tetranitromethane (TNM). N-acetylimidizole
will O-acetylate tyrosine residues in solution (7), and this reagent has been
used to modify numerous proteins including the Fd gene 5 protein (4). How-
ever, this reagent can also N-acetylate primary amines, and in the study on the
Fd gene 5 protein (4) in addition to acetylation of three tyrosine residues, all
292 Plyte
five lysine residues were found to be modified. Tetranitromethane is a reagent
highly specific for tyrosine residues and reacts under mild conditions to form
the substitution product 3-nitrotyrosine (8). The modified tyrosine has a char-
acteristic adsorption maximum at 428 nm, and this can be used to quantitate
the number of tyrosine residues modified (8). However, under harsher condi-
tions, there have been some reports of modification of sulfhydryl groups and
limited cases of reaction with histidine and tryptophan (9).
1.1. Strategies
1.1.1. Tyrosine Accessibility
The general strategy employed in chemical modification experiments is to
determine the accessibility of the target residues within the native protein and
the extent of protection offered by the bound substrate. Peptide mapping of the
labeled protein then allows the roles of the individual residues to be assessed.
First, the free protein is nitrated and then digested into fragments by proteoly-
sis. These peptides are then separated to enable identification of the modified
residue(s). The nucleoprotein complex is also nitrated and the modified resi-
dues identified in a similar way. From a comparison of these data, the extent of
protection at each site can be established.
For peptide mapping, a protease should be chosen that, on digestion of the
target protein, will place each tyrosine in a separate peptide. However, this is
not essential if the modified residues are identified by N-terminal sequencing.
It is possible that tyrosine modification may affect the efficiency of α-chymot-
rypsin digestion and this enzyme should be avoided if possible. The peptides
can be separated by reverse-phase high-performance liquid chromatography
(HPLC), and those containing tyrosine purified for further analysis. The
tyrosine-containing peptide can be easily identified directly after HPLC purifi-
cation by the characteristic fluorescence emission maximum of 3-nitrotyrosine
at 305 nm (when excited at 278 nm). A particular tyrosine residue can then be
identified by N-terminal sequence analysis.
The identification of nitrated tyrosine residues in the free protein provides
information concerning the solvent accessibility of these residues in the pro-
tein and indicates which residues are likely to be buried within the protein.
DNA protection studies will indicate which of these residues may be involved
in protein–DNA interactions. However, the protection from nitration by bound
DNA is only an indication of a functional role for a particular residue. The
bound DNA may confer protection to a residue several angstroms away or may
induce protein oligomerization (cooperative binding) which protects the
tyrosine by protein–protein interactions. Consequently, functional studies need
to be performed to further determine the role of the protected residue(s). The
Nitration of Tyrosine Residues 293
situation is analogous to the two types of analysis frequently used in the
investigation of the DNA bases involved in complexes: “footprinting” and
“interference” techniques. The data obtained from chemical modification and
protection studies can then be used to design site-directed mutagenesis experi-
ments to look at the function of an individual residue by observing the effects
of its replacement with other amino acids.
1.1.2. Functional Studies
A protocol for functional studies will not be described in this chapter, but
some general considerations will be mentioned here. One should nitrate the
free protein and determine whether the modified protein still binds to DNA.
This information should indicate whether the residues protected in the nucle-
oprotein complex are implicated in DNA binding. However, with proteins that
bind cooperatively to DNA, a reduction in DNA-binding affinity may result
from a disruption of protein–protein interactions rather than from protein–DNA
interactions. A possible way to resolve this ambiguity is to bind the native and
modified proteins to short oligonucleotides where the cooperativity factor is
negligible. Modification of residues involved in protein–protein interactions
should not significantly affect the intrinsic binding of the modified protein to
DNA, when compared to the native protein.
Tyrosine residues can interact with DNA either by hydrophobic interactions
via stacking with DNA bases or by hydrogen-bonding with the nucleotide
through the phenolic OH group (10). Nitrotyrosine has a pK
a
of 8.0, which may
disrupt H-bonding as well as base stacking interactions. However, the addition
of sodium dithionate reduces 3-nitrotyrosine to 3-amino tyrosine (which has a
pK
a
similar to that of native tyrosine) and may restore H-bonding interactions
(11). Reduction with this reagent may provide further information concerning
the nature of the tyrosine–nucleic acid interaction.
1.1.3. Rates of Modification
Nitration of a protein will initially report on the accessibility of specific
tyrosine residues in the presence and absence of DNA. However, if the modi-
fied tyrosine residues can be analyzed individually, one can look at the nitra-
tion rates of the tyrosines and determine the degree of accessibility of each
residue. This is achieved by removing aliquots of protein (at various time inter-
vals) from a nitration experiment and determining the percentage nitration of
each tyrosine for a given time-point. This can be done by quantitating the
nitrated and unnitrated products after digestion, either by measuring the peak
areas (recorded at 214 nm), taken directly from the HPLC profile (5), or by
amino acid analysis of the purified peptides.
294 Plyte
2. Reagents
All chemicals should be of AnalaR grade or higher and dissolved in double-
distilled water. For HPLC analysis, trifluoroacetic acid (TFA), water and
acetonitrile should be of HPLC grade. Buffers for HPLC should be filtered
(0.2 µm) and degassed before use.
1. Tetranitromethane (TNM) stock solution: a 300 mM stock solution of TNM in
ethanol. Store in the dark at 4°C. Note that TNM can cause irritation to the skin
and lungs, and the solution should be made up in the fume hood. Additionally,
TNM can be explosive in the presence of organic solvents such as toluene.
2. Nitration buffer: 150 mM NaCl and 10 mM Tris, pH 8.0.
3. Desalting column: Disposable “10DG” Econo columns (Bio-Rad, Richmond,
CA) are preferred.
4. µBondapak C
18
HPLC column (Waters Associates, Milford, MA) or a similar
reverse-phase column.
5. Trypsin (TPCK treated).
6. Standard sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-
PAGE) equipment with DC power supply capable of 150 V.
7. SDS–polyacrylamide gel stock solutions:
Solution A: 152 g acrylamide and 4 g bis-acrylamide. Make up to 500 mL.
Solution B: 2 g SDS and 30 g Tris base, pH 8.8. Make up to 500 mL.
Solution C: 2 g SDS and 30 g Tris base, pH 6.8. Make up to 500 mL.
When making up these solutions, they should all be degassed and filtered using a
Buchner filter funnel. They should be stored in lightproof bottles; they will keep
for many months.
8. 10% ammonium persulfate (APS): dissolve 0.1 mg in 1 mL of dH
2
O.
9. 15% SDS–polyacrylamide gel: Mix together 8.0 mL of solution A, 4.0 mL of
solution B, and 3.9 mL of dH
2
O. Add 150 µL of 10% APS and 20 µL of
N,N,N',N'-tetramethylethylene diamine (TEMED). Mix well and then pour
between the plates. Immediately place a layer of dH
2
O (or butanol) on top of
the gel to create a smooth interface with the stacking gel. When the resolving
gel has set, pour off the water and prepare the stacking gel. This is done by
adding 750 mL of solution A and 1.25 mL of solution C to 3.0 mL of dH
2
O.
Finally, add 40 µL of APS and 10 µL of TEMED, pour on the stacking gel and
insert the comb. To avoid the gel sticking to the comb, remove the comb as
soon as the gel has set.
10. 10X SDS running buffer: 10 g SDS, 33.4 g Tris base, and 144 g glycine made up
to 1 L.
11. High methanol protein stain: Technical-grade methanol 500 mL, 100 mL glacial
acetic acid and 0.3 g PAGE 83 stain (Coomassie blue), made up to 1 L.
12. Destain solution: 100 mL methanol and 100 mL glacial acetic acid, made up
to 1 L.
13. 2X SDS-PAGE loading buffer: 4% (w/v) SDS, 60 mM Tris-HCl, pH 6.8, 20%
glycerol, 0.04% (w/v) bromophenol blue, and 1% (v/v) β-mercaptoethanol.
