Moss Tom. DNA-protein interactions: principles and protocols
Подождите немного. Документ загружается.

306 Taylor and Webb
dilute to 1 mL with 50 mMTBS. Load the samples into dialysis cassettes and
dialyze overnight against 2 L of 50 mM TBS using at least 3 changes.
6. After extensive dialysis, remove all the samples from the cassettes. For free-
protein samples, continue from step 7 onward. Samples from the DNA–protein
complex time-course need to be processed to remove the DNA as follows. Equili-
brate a small-volume (1 mL or less) high-performance anion exchange (see Note 7)
HPLC column (analytical TSK-GEL, DEAE-NPR [4.6 × 35 mm] or an equiva-
lent) in 50 mM TBS at a flow rate of 1 mL/min. Set the UV detector to 280 nm.
Apply each sample from the time-course to the column and collect any
flowthrough. Elute the protein (if bound) and the DNA by application of an
increasing NaCl gradient from 0.05 M to 1.0 M over 50 column volumes. Collect
the protein-containing fractions and proceed.
7. Determine the molar concentration of the samples from each time-course
(including the “zero”) using the absorbance at 280 nm. Add 10 µL of each sample
to 1 mL of liquid scintillant, mix well, then determine the amount of incorporated
radioactivity at each time-point by liquid scintillation counting.
8. Calculate the specific activity (in nCi/nmol) of the labeled protein (σ
protein
) at
each time-point of the reaction using Eq. 3. Then, using the value for the effec-
tive specific activity of the [
3
H] formaldehyde determined in Subheading 3.2.
calculate the number of lysine residues modified at each point in the time-course
using Eq. 4 (see Note 8):
σ
protein
= (dpm)/(No. nmoles counted × 2220) (3)
No. modified lysines = σ
(protein)
= 2σ
([3H] formaldehyde)
(4)
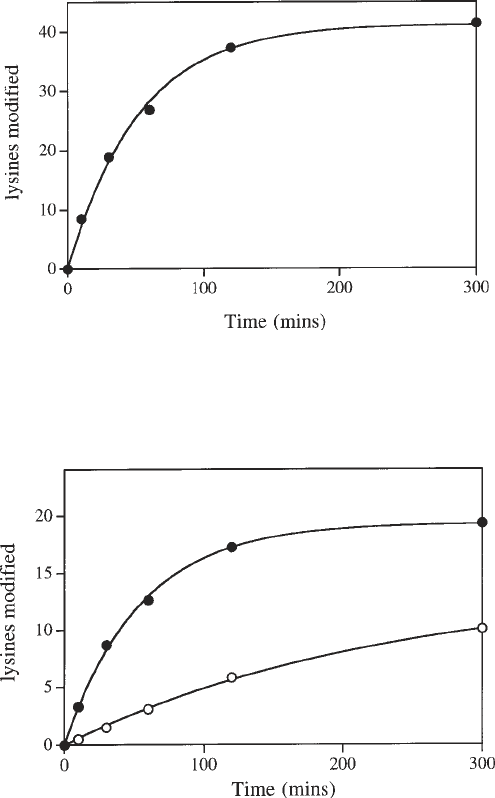
9. Plot the number of lysine residues modified against time and fit the data to a
single exponential process using Eq. 5 (Fig. 2). In most cases, the data should fit
well to this model (see Note 9) and the total number of modifiable lysine residues
is then given by the limit value (L). Fitting the data in this manner also allows a
rate constant (k) for the incorporation of radiolabel to be derived. Both of these
parameters can be affected by DNA binding.
No. modified lysines = L(1–e
–kt
) (5)
10. Compare the fitted curves of the time-course for the reaction of free protein and
for the DNA–protein complex (Fig. 3). Formation of the DNA–protein complex
may well reduce L, indicating the presence of a population of strongly protected
lysines. At the same time, differences in k are likely to arise from an overall
lowering of the rate of modification because of decreased accessibility of lysine
residues in the presence of DNA.
3.4. Pulse Chase Labeling of Proteins
If the fraction of lysine residues protected by DNA is large, as determined in
Subheading 3.3., then a pulse-labeling procedure carried out on the free pro-
tein will reveal which lysine residues are surface accessible and likely to be
Modification of Lysine by Reductive Methylation 307
involved in DNA binding. If only a small number of lysine residues are
protected by DNA, then a modification to the procedure should be undertaken
(see Note 10).
Fig. 2. Time-course of reductive methylation of the type I DNA methyltransferase
M.EcoR124I. The curve is the best fit of the data to a single exponential (L = 41, k =
0.019/min).
Fig. 3. The effect of DNA binding on reductive methylation of the DNA recogni-
tion subunit (HsdS) from M.EcoR124I. The upper curve is a best fit to the data from a
time-course for the modification reaction of free protein (L = 19, k = 0.019/min). The
lower curve is the best fit to the data from a time-course for modification of the protein
in the DNA–protein complex (L = 14, k = 0.004/min).
308 Taylor and Webb
1. Prepare 1 mL of DNA-binding protein at a concentration of 1–2 mg/mL in 50 mM
TBS by either dialysis or buffer exchange.
2. Add sodium cyanoborohydride to a 30-fold molar excess over the total lysine
content of the protein and then initiate the chemical modification reaction by
the addition of [
3
H] formaldehyde at a 10-fold molar excess. Incubate at 24°C,
the length of time will depend on the results from the experiments in Sub-
heading 3.3. Aim to modify for a length of time when the reaction is about 50%
complete (see Note 11). This will probably be between 10 and 60 min.
3. At the end of the pulse, quench the reaction by the addition of 50 mM glycine and dialyze
overnight against 1 L of 50 mM TBS. Change the dialysis buffer at least three times.
4. Equilibrate a semipreparative C
3
reverse-phase column (9.4 × 250 mm) in 5%
acetonitrile and 0.05% (v/v) TFA at a flow rate of 3 mL/min and set the UV
detector to 225 nm. Remove the protein from the dialysis cassette, add urea to a
final concentration of 8 M and DTT to 50 mM. Incubate the sample briefly at
room temperature, acidify by the addition of 100 mM acetic acid, and apply the
protein to the column. Elute with a 5–65% gradient of acetonitrile and 0.05% (v/v)
TFA over 60 min. Collect the protein containing fractions and lyophilize them.
5. Redissolve the modified protein in 1 mL of 8 M urea and 10 mM HEPES (pH 7.5)
and determine the protein concentration from the absorbance at 280 nm. At this
point, the extent of label incorporation should be determined as in Subheading
3.3., steps 7 and 8.
6. To “chase” the reaction with unlabeled reagent, add a 30-fold molar excess of
sodium cyanoborohydride over the total lysine content followed by a 10-fold
excess of unlabeled formaldehyde. Incubate for 3 h at 24°C, then add a second
aliquot of these reagents and continue the reaction for a further 3 h.
7. Add DTT to a final concentration of 50 mM, incubate briefly at room tempera-
ture then acidify with 100 mM acetic acid. Purify the fully modified protein by
reverse-phase chromatography as in step 4. Lyophilize the fractions containing
protein and store in aliquots of approx 2 nmol at –20°C in a box containing silica gel.
8. Redissolve a 2 nmol aliquot of modified protein in 100 µL of 0.9% formic acid
(see Note 12). Ensure that the sample is fully dissolved then dilute to 500 µL with
dH
2
O and titrate to pH 8 by the addition of 35 µL of 2 M Tris base.
9. Dissolve the contents of a vial of sequencing grade trypsin (see Note 13) in 1 mM
HCl to give a concentration of 1 mg/mL. Add the trypsin to the protein to give an
enzyme to a substrate ratio of 1:10 (w/w) and incubate at 37°C for approx 18 h.
To increase the efficiency of cleavage add the trypsin in three aliquots at roughly
4-h intervals. Terminate the digest by the addition of 1 mM Pefabloc and store at
–20°C until required.
10. Equilibrate an analytical C
3
reverse-phase HPLC column (4.6 × 250 mm) in 2%
acetonitrile and 0.05% (v/v) TFA at a flow rate of 1 mL/min and set the UV
absorbance detector to 214 nm. Adjust the tryptic digests to 8 M urea and 50 mM
DTT and incubate briefly at room temperature. Acidify the mixture by the addi-
tion of 100 mM acetic acid and apply to the column. Elute the peptides with an
increasing gradient of acetonitrile collecting 250 µL fractions. For a complex
Modification of Lysine by Reductive Methylation 309
mixture of peptides, the following gradient works well: 2–35% in 45 min fol-
lowed by 35–60% in 20 min. It may be necessary to alter this for the particular
protein under investigation.
11. Remove 25 µL from each fraction, add 1 mL of liquid scintillant, and determine
the level of radioactivity by liquid scintillation counting. Pool the fractions
across each peak and lyophilize them. Store at –20°C in a box containing silica
gel until required.
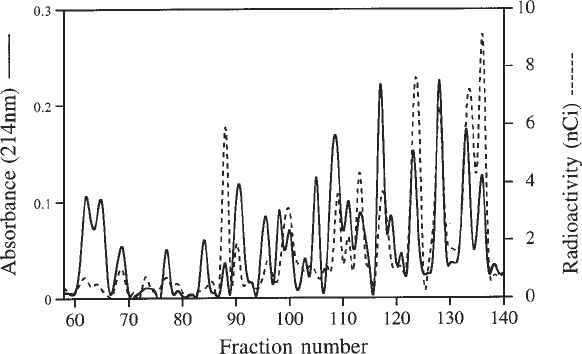
12. Construct an overlaid chromatogram as in Fig. 4 and use this to select peaks with
an apparently high specific activity (see Note 14). These peaks require further
fractionation by C
18
reverse-phase chromatography (see Note 15) before ulti-
mately submitting them for N-terminal amino acid sequencing.
13. Equilibrate an analytical C
18
reverse-phase column in 2% acetonitrile and 0.05%
TFA at a flow rate of 1 mL/min and set the UV detector to 214 nm. Redissolve
each selected peak in 500 µL of 8 M urea and 100 mM acetic acid and apply to the
column. Elute the bound peptides with a linear gradient of acetonitrile (2–50% in
70 min) and collect 250-µL fractions.
14. Remove 25 µL from fractions across each peak and determine the incorporated
radioactivity as in step 11. Lyophilize the remainder and store at –20°C in a box
containing silica gel until required.
15. Analyze each purified peptide using automated N-terminal amino acid sequenc-
ing. The objective is to determine the number of pmoles of each amino acid
released at each cycle of the sequencing reaction and also to determine the amount
of radioactivity associated with the residue. A suggested method for doing this is
described in Note 16.
Fig. 4. Separation of reductively methylated peptides from pulse-labeled HsdS by
C
4
reverse-phase chromatography. The absorbance at 214 nm and the amount of
radioactivity (nCi) are plotted for each fraction.
310 Taylor and Webb
3.5. Data Analysis
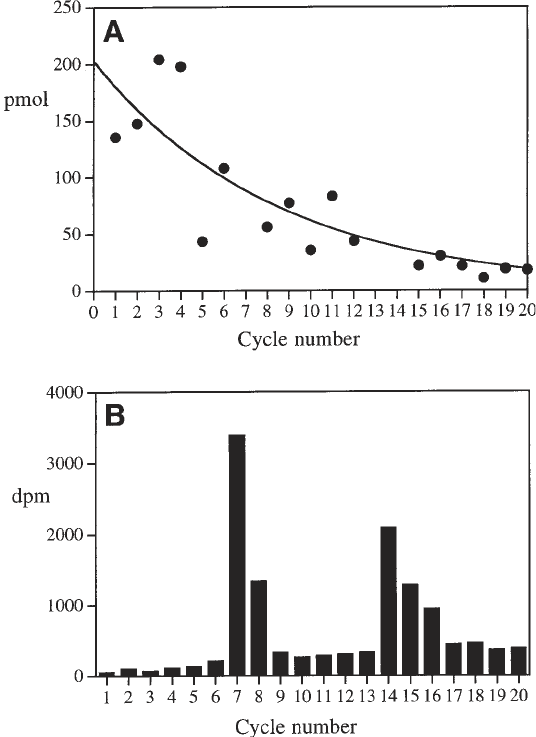
The data from the N-terminal sequencing are used to determine the specific
activity of each modified lysine as follows. Plot the number of pmoles of each
residue released at each cycle pmole(n) versus the cycle number (n); then, fit
these data to Eq. 5 (see Fig. 5A). E is the efficiency of the sequencing process
(usually around 90%) and pmole(0) is the amount of starting material.
pmole(n) = pmole(0) × E
n
(5)
As PTH-dimethyl-lysine is not a standard amino acid, the number of pmoles
of modified lysine is not determined directly from integration of the HPLC
trace. Instead, the value can be calculated by interpolation of the fitted curve
for the cycle at which the dimethyl-lysine was released. Plot the amount of
radioactivity released at each cycle versus the cycle number (Fig. 5B). Signifi-
cant quantities of radioactivity should only be present at a cycle where a modi-
fied lysine is present. Combine the data from the two plots and use Eq. 1 to
calculate the specific activity (σ
lysx
); then, determine the fractional modifica-
tion (σ
lysx
/σ
α-MSH
) for each dimethyl-lysine in a peptide. The value of this ratio
is proportional to the accessibility of the residue during the pulse part of the
chemical modification reaction. Values close to unity indicate a high degree of
accessibility, whereas values close to zero indicate a residue that is inacces-
sible to chemical modification.
These data can be used to build up a picture of the protein surface and
identify clusters of lysines that are potential surfaces for DNA binding.
Residues identified by these methods are then targets for site-directed mutagen-
esis experiments.
4. Notes
1. The α-MSH peptide contains only a single lysine and has a blocked N-terminus.
The presence of the two aromatic residues allow its concentration to be deter-
mined accurately from its UV absorbance at 280 nm (ε
280
= 7000/M/cm). We
have extensively characterized the reductive methylation reaction with this pep-
tide using NMR and mass spectroscopy (10). Under the reaction conditions used
in Subheading 3.2., >95% of the product is dimethylated and the remainder
monomethylated. In principle, any peptide substrate with a known number of
free amino groups and for which the concentration can be measured accurately
could be used to determine the effective specific activity. However, if a different
peptide substrate is used, it is advisable to extensively characterize the reaction
in the same way. On a routine basis, if access can be gained to a mass spectrom-
eter, it may be worthwhile checking the completeness of the reaction in this way.
2. [
3
H] Formaldehyde from NEN is supplied as a 0.3 M aqueous solution in snap-
off glass vials. Make sure the whole contents of the vial are at the bottom and
then leave on ice for 10 min before breaking the seal. After opening, transfer the
Modification of Lysine by Reductive Methylation 311
contents to a screw-cap microfuge tube and store at 4°C. If possible, use all of the
reagent within 1–2 d of opening.
3. The efficiency of the reductive methylation of proteins is greatly reduced by the
presence of amines in the solution because of competitive inhibition. Thus, com-
Fig. 5. Identification of radiolabeled lysine residues in the tryptic peptide D190-
N220 from the reductively methylated pulse-labeled HsdS subunit from M.EcoR124I.
(A) The pmole yield at each cycle of the Edman degradation sequencing reaction.
Fitting of the data to Eq. 5 allows the yield of dimethyl-lysine to be determined by
interpolation. (B) A histogram showing the amount of radioactivity released at each
cycle of the same sequencing reaction. The combination of radioactivity and picomole
released at each cycle allows estimation of the specific activity of individual residues.
312 Taylor and Webb
monly used buffers such as Tris–HCl and triethanolamine have to be avoided.
HEPES and phosphate are good alternatives. A further problem is the potential
for the formaldehyde–lysine adducts to undergo side reactions leading to
unwanted protein crosslinking. To prevent this, the sodium cyanoborohydride
should be added to the protein solution prior to addition of the formaldehyde.
4. The stoichiometry of the reductive methylation reaction dictates that two mol-
ecules of formaldehyde are required to complete the modification of a lysine
residue to ε-N,N-dimethyl-lysine (see Fig. 1). Because of this, the effective spe-
cific activity of the [
3
H] formaldehyde is half the value determined for the fully
dimethylated peptide.
5. For dialysis of small volumes, (0.5–2 mL) Slide-A-Lyser cassettes (Pierce) are
extremely useful. We find that a 1-mL sample volume is easy to inject and recover
from the cassette without large losses of sample and without large dilution. If
necessary, a smaller volume could be used.
6. The amount and the exact ratio of the reagents used for the reaction are somewhat
empirical. The major concern is the prevention of side reactions resulting from
reactive formaldehyde–lysine adducts. For a detailed account, see ref. 6. Briefly,
the concentration of formaldehyde needs to be at an excess over the number of
lysine residues to drive the reaction to completion, but not so high as to favor
protein crosslinking. The other requirement is that the sodium cyano-
borohydride be in excess over the formaldehyde to ensure efficient reduction of
the Schiff bases.
7. To separate the DNA from protein, DEAE or Q ion-exchange columns are the
method of choice. DNA oligonucleotides will bind very strongly to these matri-
ces and the protein either can be recovered from the flowthrough or will elute
earlier in a NaCl gradient. An alternative is to use heparin–Sepharose or, for
basic proteins, a cation-exchange resin. If the chromatographic separation of the
DNA from the protein is problematic, treat each sample with DNase I (FPLCpure,
Pharmacia) before application to the column.
8. The N-terminus of the protein can also be reductively methylated. If the protein
is relatively small or the total number of modified residues is low, then it is worth-
while to consider this when calculating the extent of modification.
9. If the time-course is extended to 5 h incubation, the reaction should be complete
and the data will usually fit well to a single exponential process. Occasionally,
this is not the case—for instance, if the protein contains several distinct popula-
tions of lysines with different kinetics. In this case, a more complex model will
be needed to deconvolve the various classes of reacting species.
10. The pulse-labeling method involves treating the protein with a short pulse of
labeled formaldehyde followed by a “cold” chase. This will identify all the sur-
face lysines and provide information about their accessibility. If a large propor-
tion of the total number of modifiable lysine residues are protected by DNA, then
a pulse chase procedure of this kind will identify residues likely to be involved in
DNA binding. However, if only a small proportion of lysines are protected by
DNA, then an initial “cold” labeling should be performed on the DNA–protein
Modification of Lysine by Reductive Methylation 313
complex, followed by separation of the protein from the DNA before the pulse
with labeled reagents is applied.
11. This is a compromise between getting enough label into the protein to allow the
sites of modification to be easily determined and providing for differential label-
ing at individual sites so that information about the accessibility of each lysine
can be obtained.
12. There may be some difficulty in redissolving the lyophilized protein in the aque-
ous buffers required for tryptic digestion (e.g., 20 mM Tris-HCl, pH 8.0). The
formic acid strategy described works well in some cases but is not guaranteed.
An alternative is to dissolve the sample in 100 mM Tris-HCl and 8 M urea pH 8.0
and then to add an equal volume of trypsin in 1 mM HCl such that the final urea
concentration is 4 M and the trypsin concentration 1:10 (w/w).
13. The objective of the proteolytic digest is to produce peptides of an optimal length
(5–30 amino acids) for quantitative analysis by automated Edman degradation. A
tryptic digest of a reductively methylated protein will produce an arginine spe-
cific digest, as ε-N,N-dimethyl-lysine residues are not substrates for tryptic cleav-
age (11). Such a digest will yield some peptides that are suitable for N-terminal
sequencing but will probably not cover the entire protein. In order to produce
further peptides of suitable length, other proteases and chemical cleavage reagents
should be investigated. The usefulness of these agents can vary substantially. In
general, the best enzymatic alternatives are chymotrypsin and V8 protease.
Cyanogen bromide is the best alternative for chemical cleavage.
14. The main criterion for selection of peaks is an apparent high specific activity,
indicating that surface accessible lysines are present in peptides eluted within the
peak. Additional information about the location of buried lysines within the
protein can be gained by sequencing peptides which show very low or appar-
ently no label incorporation. Although this could yield valuable data, one
should be aware that some of these “cold” peaks are likely to be peptides derived
from trypsin.
15. After an initial separation of a complex mixture of reductively methylated pep-
tides, by C
3
reverse-phase chromatography, a further fractionation of peptides
using either C
8
or C
18
is highly recommended. Often a peak with an apparently
high specific activity taken from an initial C
3
separation will resolve into mul-
tiple components on C
8
or C
18
, only some of which are labeled. Avoid loading
peptides eluted from a C
3
column at high acetonitrile concentrations (>50%) onto
columns with longer alkyl-chain-bonded phases as the interaction between the
sample and the bonded phase may be too strong for efficient recovery. For these
larger, more hydrophobic peptides, it may be better to reapply to a C
3
column and
then elute with a different gradient to the one used initially. Alternatively, redigest
with a different enzyme and separate the products on a C
18
column.
16. A simple and effective way to quantify the degree of incorporated label at indi-
vidual lysine residues involves splitting the peptide sample during the automated
Edman degradation sequencing reaction; most sequenators are equipped with this
facility. After extraction of each 2-analino–5-thiazolinone-derivatized amino
314 Taylor and Webb
acid, split the sample and divert 50% to a fraction collector. Convert the remain-
der to the phenylthiohydantoin derivative and identify the residue by on-line
HPLC analysis in the usual way. Calculate the number of picomoles of the newly
released residue by integration of the HPLC peak. Determine the amount of
radioactivity associated with the residue by liquid scintillation counting of the
material that was diverted to the fraction collector. This method may require
modification, depending on the configuration of the available sequenator.
References
1. Lundblad, R. L. and Noyes, C. M. (1984) Chemical Reagents for Protein Modifi-
cation. CRC, Boca Raton, FL.
2. Hunter, M. J. and Ludwig, M. L. (1962) J. Am. Chem. Soc. 84, 3491–3497.
3. Means, G. E. and Feeney, R. E. (1968) Reductive alkylation of amino groups in
proteins. Biochemistry 7, 2192–2201.
4. Means, G. E. and Feeney, R. E. (1995) Reductive alkylation of proteins. Anal.
Biochem. 224, 1–16.
5. Jentoft, N. and Dearborn, D. G. (1983) Protein labelling by reductive alkylation.
Methods Enzymol. 91, 570–579.
6. Jentoft, N. and Dearborn, D. G. (1979) Labelling of proteins by reductive methy-
lation using sodium cyanoborohydride. J. Biol. Chem. 254, 4359–4365.
7. Zhang, M., Thulin, E., and Vogel, H. J. (1994) Reductive methylation and pKa
determination of the lysine side chains in calbindin D9k. J. Protein Chem. 13,
527–535.
8. Lambert, S. F. and Thomas, J. O. (1986) Lysine-containing DNA-binding regions
on the surface of the histone octamer in the nucleosome core particle. Eur. J.
Biochem. 160, 191–201.
9. Thomas, J. O. and Wilson, C. M. (1986) Selective radiolabelling and identifica-
tion of a strong nucleosome binding site on the globular domain of histone H5.
EMBO J. 5, 3531–3537.
10. Taylor, I. A., Webb, M., and Kneale, G. G. (1996) Surface labeling of the type I
methyltransferase M.EcoR124I reveals lysine residues critical for DNA binding.
J. Mol. Biol. 258, 62–73.
11. Poncz, L. and Dearborn, D. G. (1983) The resistance to tryptic hydrolysis of pep-
tide bonds adjacent to N-epsilon, N-dimethyllysyl residues. J. Biol. Chem. 258,
1844–1850.

Limited Proteolysis of Protein–Nucleic Acid 315
315
From:
Methods in Molecular Biology, vol. 148: DNA–Protein Interactions: Principles and Protocols, 2nd ed.
Edited by: T. Moss © Humana Press Inc., Totowa, NJ
22
Limited Proteolysis
of Protein–Nucleic Acid Complexes
Simon E. Plyte and G. Geoff Kneale
1. Introduction
Limited Proteolysis is a useful structural probe for investigating the globu-
lar nature of proteins by preferentially digesting the more accessible regions
often found between domains. Generally, proteases require a small region of
polypeptide chain possessing conformational flexibility for accommodation in
the active site (1). The regions of a protein possessing conformational flexibil-
ity are often found between tightly folded domains and are, therefore, prefer-
ential sites for proteolysis. In practice, limited proteolysis is achieved by
dilution of the enzyme sufficiently so that it will only digest the most acces-
sible regions leaving the domains intact. Digestion of protein–nucleic acid is
often advantageous in that the DNA may provide steric protection of the DNA-
binding domain not afforded by the free protein. The generation of domains by
limited proteolysis relies directly on the tertiary structure of the protein under
investigation and provides much firmer evidence for their existence than that
provided by sequence homology.
An increasing number of nucleic-acid-binding proteins are known in which
regions of their polypeptide chain are folded separately into compact globular
domains, each possessing a distinctive function. For example, digestion of the
A1 heterogeneous nuclear ribonucleoprotein (A1 hnRNP) with Staphylococ-
cus aureus V8 protease produces two discrete domains, both capable of bind-
ing single-stranded nucleic acids (2,3). Similarly, digestion of the Pf1 gene 5
nucleoprotein complex results in the production of a 12-kDa domain that retains
much of the single-stranded DNA-binding ability of the intact protein (4). Fur-
ther, using limited proteolysis, a cryptic DNA-binding domain was revealed in
the COOH terminus of yeast TFIIIB70 and a core ssDNA-binding domain was
