Moss Tom. DNA-protein interactions: principles and protocols
Подождите немного. Документ загружается.

316 Plyte and Kneale
generated, possessing increased binding affinity, in human replication protein A
(5,6). In addition to its use for the analysis of domain structure, limited proteolytic
fragments from Escherichia coli DNA gyrase B, for example, permitted the suc-
cessful crystallization and structure determination of one of its domains (7).
1.1. Strategy
The strategy adopted for the limited proteolysis of nucleoprotein complexes
can be considered in four parts: optimization of the proteolysis, characteriza-
tion of the proteolysed complex, purification of the DNA-binding domains,
and sequence characterization of the fragment(s).
1.1.1. Proteolysis of Nucleoprotein Complex
The nucleoprotein complex should be digested with various proteases to
establish which conditions are optimal for generating a protease-resistant
domain. We routinely vary two parameters (enzyme/substrate ratio and time of
digestion) when determining the best conditions for limited proteolysis.
However, other parameters, such as temperature, ionic strength, and pH may
also be varied. To determine the appropriate enzyme/substrate ratio for a par-
ticular protease, the nucleoprotein complex is digested at several enzyme/sub-
strate ratios, removing samples at regular time intervals for sodium dodecyl
sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) analysis. The appear-
ance of a discrete domain, resistant to further degradation (even if only tran-
siently), is evidence for the existence of a domain, although not necessarily
one that binds DNA. Choice of protease is often critical (see Table 1). Ini-
tially, it is best to try a relatively nonspecific enzyme (e.g., papain) because
this decreases the likelihood of activity being dependent on primary sequence
rather than tertiary structure.
1.1.2. Preliminary Characterization of DNA-Binding Properties
of the Proteolyzed Nucleoprotein Complex
An initial indication of DNA binding can be found during the proteolysis
experiment by removing two aliquots for gel analysis that can be run on poly-
acrylamide or agarose gels appropriate for the size of complex in the presence
and absence of the denaturant SDS. A retardation in the mobility of the DNA
(seen under ultraviolet [UV] light) in the absence of SDS implies that the frag-
ment is still associated with DNA and constitutes a DNA-binding domain.
However, this does not prove that the proteolyzed fragment is a discrete DNA-
binding domain; it is possible that the nucleoprotein complex has only been
“nicked” by the protease and maintains its native tertiary structure by noncova-
lent interactions. Therefore, it is necessary to purify the domain and fully char-
acterize its DNA-binding properties.
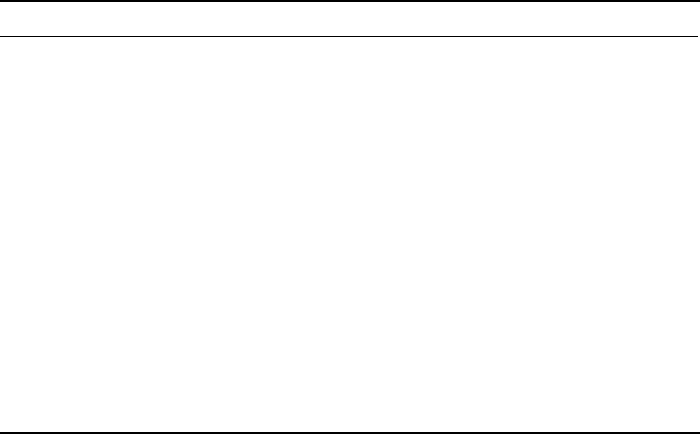
Limited Proteolysis of Protein–Nucleic Acid 317
1.1.3. Purification of the DNA-Binding Domain
Purification of the fragment can make use of the fact that it will still be
associated with DNA. Ultracentrifugation of the proteolyzed nucleoprotein
complex (if large fragments of DNA are used) concentrates the domain and
removes residual protease and small proteolytic fragments. The proteolyzed nucle-
oprotein complex can then be dissociated and the domain further purified if
necessary. Alternatively, the DNA-binding fragment can be purified by affinity
chromatography on DNA agarose. Several techniques are available to deter-
mine whether the purified domain binds DNA (discussed in several chapters)
and include gel retardation assay, a variety of footprinting techniques, fluores-
cence spectroscopy, and circular dichroism.
1.1.4. Determination of the Amino Acid Sequence of the Domain
N-Terminal sequencing and amino acid analysis of the purified DNA-bind-
ing domain should be sufficient to establish the sequence of the domain, if the
Table 1
Useful Enzymes for Limited Proteolysis
Enzyme Substrate specificity Inhibitors
α-Chymotrypsin Preferentially cuts C-terminally Aprotinin, PMSF, DFP,
to aromatic amino acids TPCK, cymostatin
Elastase Cuts C-terminally to aliphatic PMSF, DFP
noncharged amino acids
(e.g., Ala, Val, Leu, Ile, Gly, Ser)
Endoproteinase Arg-C Cuts C-terminally to arginine residues DFP, TLCK
Endoproteinase Lys-C Cuts C-terminally to lysine residues Aprotinin, DFP
Papain Nonspecific protease but shows some PMSF, TPCK, TLCK,
preference for bonds involving leupeptin, heavy
Arg, Lys Gln, His, Gly, and Tyr metal ions
Pepsin Nonspecific protease Pepstatin
Subtilisin Nonspecific protease DFP, PMSF
Trypsin Cuts C-terminally to lysine DFP, PMSF, TLCK
and arginine residues
Endoproteinase Glu-C Cuts C-terminally to glutamic acid DFP
(V8 protease) and or aspartic acid residues
a
Abbreviations used: DFP, diisopropyl fluorophosphate (extremely toxic!); PMSF, phenyl-
methyl sufonyl fluoride; TPCK, N-tosyl-l-phenylalanine chloromethyl ketone; TLCK, Nα-p-
tosyl-l-lysine chloromethly ketone.
a
Will cut C-terminally to glutamic acid residues in ammonium bicarbonate, pH 8.0, or ammo-
nium acetate, pH 4.0; will cut C-terminally to glutamic and aspartic acid residues in phosphate
buffer, pH 7.8.
318 Plyte and Kneale
native amino acid sequence is known. Alternatively, N-terminal sequencing
and mass spectroscopy should enable unambiguous identification of the
domain. If certain proteases have been used to generate the domain (e.g.,
trypsin, α-chymotrypsin, endoproteinase Arg-C, and so forth), the C-terminal
amino acid may also be known. If there are still ambiguities, carboxypeptidase
digestion of the fragment can also be used to help identify the C-terminal resi-
dues, although this is not always reliable. If this still does not yield an unam-
biguous result, one must resort to amino acid sequencing of the entire fragment.
2. Materials
1. Spectra-Por dialysis membrane washed thoroughly in double-distilled water.
2. All proteases should be of the highest grade available and treated for contaminat-
ing protease activity, if necessary. A list of useful enzymes and their inhibitors is
given in Table 1.
3. Buffers should be AnalR grade or higher and made up in double-distilled water.
4. SDS–polyacrylamide gel stock solutions:
Solution A: 152 g acrylamide and 4 g bis-acrylamide. Make up to 500 mL.
Solution B: 2 g SDS and 30 g Tris base, pH 8.8. Make up to 500 mL.
Solution C: 2 g SDS, 30 g Tris base, pH 6.8. Make up to 500 mL.
When making up these solutions, they should all be degassed and filtered using a Buchner
filter funnel. They should be stored in lightproof bottles and will keep for many months.
6. 10% ammonium persulfate (APS): dissolve 0.1 mg in 1 mL of dH
2
O.
7. 15% SDS–polyacrylamide gel: Mix together 8.0 mL of solution A, 4.0 mL of
solution B, and 3.9 mL of dH
2
O. Add 150 µL of 10% APS and 20 µL of N,N,N',N'-
tetramethylethylene diamine (TEMED). Mix well and then pour between the
plates. Immediately place a layer of dH
2
0 (or butanol) on top of the gel to create
a smooth interface with the stacking gel. When the resolving gel has set, pour
off the water and prepare the stacking gel. This is done by adding 750 µL of
solution A and 1.25 mL of solution C to 3.0 mL of dH
2
O. Finally, add 40 µL
of APS and 10 µL of TEMED, pour on the stacking gel, and insert the comb.
Remove the comb as soon as the gel has set to avoid the gel sticking to the comb.
8. 10X SDS running buffer: 10 g SDS, 33.4 g Tris base, and 144 g glycine made up to 1 L.
9. High-methanol protein stain: technical-grade methanol 500 mL, 100 mL glacial
acetic acid and 0.3 g PAGE 83 stain (Coomassie blue), made up to 1 L.
10. Destain solution: 100 mL methanol and 100 mL glacial acetic acid, made up to 1 L.
11. 2X SDS-PAGE loading buffer: 4% (w/v) SDS, 60 mM Tris-HCl, pH 6.8, 20%
glycerol, 0.04% (w/v) bromophenol blue, and 1% (v/v) β-mercaptoethanol.
12. 6X agarose gel loading buffer: 0.25% (w/v) bromophenol blue, 0.25% (w/v)
xylene cyanol, and 30% glycerol.
13. 6X agarose gel loading buffer plus SDS: as in item 12 plus 12% SDS (w/v).
14. TE buffer: 10 mM Tris-HCl, pH 7.5, and 1 mM EDTA.
15. 5 M NaCl or MgCl
2
(or other concentrated salt solutions for dissociation of the
nucleoprotein complex [e.g., NaSCN]).
Limited Proteolysis of Protein–Nucleic Acid 319
3. Methods
The method given here covers the first three objectives outlined in Sub-
heading 1.1. Experimental details for the determination of the amino acid
sequence of the fragment can be found in any standard text on protein
chemistry. The following protocol was used for the generation of an 11-kDa
DNA-binding domain from the Pf1 gene 5 protein (8). This protein binds
cooperatively to ssDNA to produce a nucleoprotein complex of several million
Daltons. Different nucleoprotein complexes will require different conditions
of digestion and purification, but the basic principles remain the same.
3.1. Limited Proteolysis
1. Dialyze the nucleoprotein complex into the appropriate digestion buffer (see the
manufacturer’s recommendations for the buffer, temperature of reaction, and
inhibitor). We routinely digest the nucleoprotein complex at approx 1 mg/mL,
but the concentration is not too critical.
2. Prepare 40 tubes containing 5 mL of 2X SDS loading buffer plus 1 µL of the
appropriate protease inhibitor. Leave on ice.
3. Pipet 55 µL of the nucleoprotein complex (55 mg) into each of four tubes labeled
1:100, 1:200, 1:500, 1:1000, respectively. Place on ice until needed.
4. Dissolve the protease in digestion buffer to a concentration that will give an
enzyme/substrate ratio of 1:100 (w/w) when 1 µL of the protease is added to
50 µL of nucleoprotein complex (i.e., 0.5 mg/mL).
5. Prepare three dilutions of the protease. In this case, the protease is diluted 1:2,
1:5, and 1:10 with digestion buffer that will result in a final enzyme substrate
ratio of 1:200, 1:500, and 1:1000 (w/w).
6. Remove 5 µL of the nucleoprotein complex from each of the four tubes and add
to 1 of the 40 tubes containing 2X loading buffer (plus inhibitor) and place on
ice. This is the time = 0 tube and should be labeled accordingly.
7. Add 1 µL of the protease to the appropriate nucleoprotein solution (e.g., pro-
tease diluted 1:5 to the nucleoprotein solution marked 1:500) and incubate at
the specified temperature.
8. Remove 5 µL samples every 15 min and add to 2X loading buffer (in the appro-
priately marked tube) and then place on ice.
9. At the end of the experiment, boil the samples and run an SDS polyacrylamide
gel. The presence of a degraded fragment(s), resistant to further proteolysis, is
evidence for a discrete domain (see Note 1).
10. Adjustment of the enzyme/substrate ratios, time course, and choice of enzymes is
often necessary. The optimum conditions must be found by trial and error.
3.2. Purification of the DNA-Binding Domain
1. Digest a large quantity (several milligrams) of the nucleoprotein complex under
the optimized conditions determined in Subheading 3.1. to produce the DNA-
binding domain (see Note 2). Add the appropriate inhibitor and run a sample on
SDS-PAGE to check the digestion.
320 Plyte and Kneale
2. For very large nucleoprotein complexes, the proteolyzed complex can be puri-
fied away from the protease and small proteolytic fragments by ultracentrifuga-
tion. Spin the nucleoprotein complex at 229,000g (Beckman 70.1 Ti rotor)
for 3 h at 4°C (see Note 3). Carefully wash the centrifuge tube with 4 mL of TE
buffer, discard the washings, and resuspend the nucleoprotein complex in 2 mL
of TE buffer on ice. Another ultracentrifugation step can be performed to remove
all traces of the protease. For smaller nucleoprotein complexes, the DNA can
be immobilized on a large resin (e.g., DNA cellulose) prior to interaction with
the DNA-binding protein. Low-speed centrifugation can then be used to purify
the DNA-associated domain. Sometimes, limited proteolysis can generate sev-
eral fragments that bind DNA. These may arise from the same region of the pro-
tein; if so, this can be overcome by allowing the proteolysis to proceed further or
by increasing the amount of protease.
3. Dissociate the proteolyzed nucleoprotein complex by the addition of salt to the
appropriate concentration (see Note 4). The DNA can then be removed by ultra-
centrifugation (if sufficiently large) or nuclease digestion. If the DNA was
originally bound on a solid support, then it can be removed by low-speed centri-
fugation (see Note 5).
4. Remove the high-salt buffer by desalting or dialysis. If the sample contains sev-
eral different domains or a residual undigested protein, it will be necessary to
purify the domains to homogeneity. Various chromatographic techniques are
available to further purify the domains, including chromatofocusing, ion
exchange, affinity, and gel filtration chromatography. These techniques permit
recovery of the domain in a native state for further biochemical analysis. Alterna-
tively, if the fragment is only to be used for sequence analysis, the mixture can be
applied to a C
3
reverse-phase high-performance liquid chromatography column
and separated in an acetonitrile gradient.
5. If the sequence of the native protein is known, then the sequence of the DNA-
binding domain can be established by N-terminal sequencing and amino acid
analysis. Additionally, the mass of the fragment (determined by mass spectros-
copy) should help locate the sequence of the DNA-binding domain.
4. Notes
1. Often during the experiment, a protease-resistant fragment is only transiently
formed during complete digestion of the protein. If this occurs, vary some of the
parameters (enzyme dilution, temperature, etc.) to try and prolong the lifetime of
the fragment.
2. Scaling up of the digestion is not generally a problem and we routinely digest
several milligrams (>10) of nucleoprotein complex if necessary.
3. The speed and duration of centrifugation will vary depending on the size of the
nucleoprotein complex. For smaller complexes, ultracentrifugation may not be
appropriate.
4. In many cases, a NaCl concentration between 1 M and 2 M is sufficient to disso-
ciate the nucleoprotein complex. However, some nucleoprotein complexes
Limited Proteolysis of Protein–Nucleic Acid 321
remain associated above 2 M NaCl and require 1 M MgCl
2
or 1 M NaSCN for
dissociation (8). The appropriate salt concentration can be determined by SDS-
PAGE analysis of the pellet and supernatant after ultracentrifugation at different
ionic strengths.
5. The DNA can also be removed by DNase digestion followed by gel fitration (i.e.,
desalting column) or by extensive dialysis against TE buffer.
References
1. Vita, C., Dalzoppo, D., and Fontana, A (1987) Limited proteolysis of globular
proteins: molecular aspects deduced from studies on thermolysin, in Macromo-
lecular Biorecognition (Chaiken, I., Chaiancone, E., Fontana, A., and Veri, P.,
eds.), Humana Press, Clifton, NJ.
2. Merril, B., Stone, K., Cobianchi, F., Wilson, S., and Williams, K. (1988)
Phenylalanines that are conserved among several RNA-binding proteins form part
of a nucleic acid binding pocket in the heterogeneous nuclear ribonucleoprotein.
J. Biol. Chem. 263, 3307–3313.
3. Bandziulis, R., Swanson, M., and Dreyfuss, G. (1989) RNA binding proteins as
developmental regulators. Genes Dev. 4, 431–437.
4. Plyte, S.E. and Kneale, G.G. (1993) Characterization of the DNA-binding domain
of the Pf1 gene 5 protein. Biochemistry 32, 3623–3628
5. Huet, J., Conesa, C., Carles, C., and Sentenac, A. (1997) A cryptic DNA-binding
domain at th COOH terminus of TFIIIB70 affects formation, stability and func-
tion of perinitiation complexes. J. Biol. Chem. 272, 18,341–18,349.
6. Bochareva, E., Frappier, L., Edwards, A., and Bochareva, A. (1998) The RPA32
subunit of human replication protein A contains a single stranded DNA-binding
domain. J. Biol. Chem., 273, 3932–3936.
7. Wigley, D., Davies, G., Dodson, E., Maxwell, A., and Dodson, G. (1991) Crystal
structure of an N-terminal fragment of the DNA gyrase B protein. Nature 351,
624–629.
8. Kneale, G.G. (1983) Dissociation of the Pf1 nucleoprotein assembly complex and
characterization of the DNA-binding protein. Biochem. Biophys. Acta 739, 216–224.

DNA–Protein Photocrosslinking 323
323
From:
Methods in Molecular Biology, vol. 148: DNA–Protein Interactions: Principles and Protocols, 2nd ed.
Edited by: T. Moss © Humana Press Inc., Totowa, NJ
23
Ultraviolet Crosslinking of DNA–Protein
Complexes via 8-Azidoadenine
Rainer Meffert, Klaus Dose, Gabriele Rathgeber,
and Hans-Jochen Schäfer
1. Introduction
In biological systems, photoreactive derivatives have been widely applied
to study specific interactions of receptor molecules with their ligands by
photoaffinity labeling (1–3). While the receptors are generally proteins (e.g.,
enzymes, immunoglobulins, or hormone receptors), the ligands differ widely
in their molecular structure (e.g., sugars, amino acids, nucleotides, or oligo-
mers of these compounds).
The advantage of photoaffinity labeling compared with affinity labeling, or
chemical modification with group-specific reagents is that photoactivatable
nonreactive precursors can be activated at will by irradiation (Fig. 1). These
reagents do not bind covalently to the protein unless activated. On irradiation
of the precursors, highly reactive intermediates are formed that react indis-
criminately with all surrounding groups. Therefore, after activation, a
photoaffinity label, interacting at the specific binding site, can label all the
different amino acid residues of the binding area. Today, aromatic azido com-
pounds are mostly used as photoactivatable ligand analogs. They form highly
reactive nitrenes upon irradiation because of the electron sextet in the outer
electron shell of these intermediates (Fig. 2).
In addition to the azido derivatives, photoreactive precursors forming
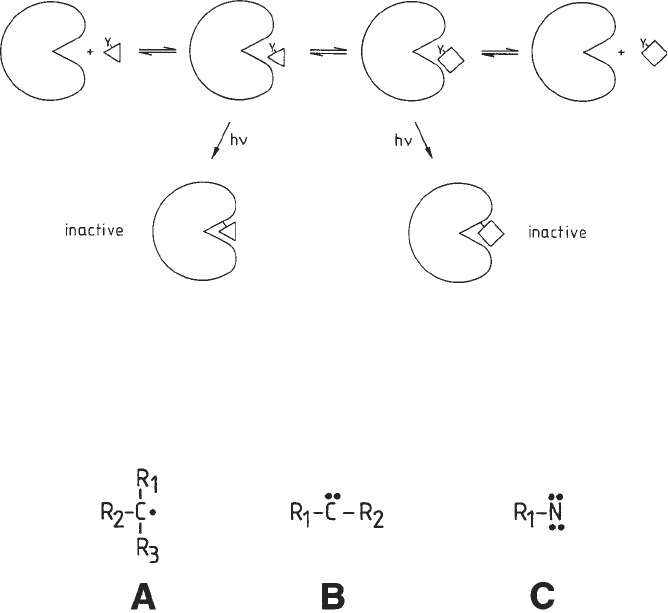
radicals or carbenes on irradiation can be used as photoaffinity labels. All of
these intermediates (nitrenes, e.g.) vigorously try to complete an electron octet
(Fig. 3).
To produce covalent crosslinks between proteins and DNA, various meth-
ods have been applied (4–11): ultraviolet (UV) irradiation, γ-irradiation, chemi-
324 Meffert et al.
Fig. 1. Photoaffinity labeling of receptor proteins (e.g., enzymes) by photo-
activatable ligand analogs (e.g., substrate analog/product analog). In the dark (upper
line), the biological interactions of the protein with the ligand analog can be studied.
On irradiation (lower line), the protein (enzyme) is labeled and inactivated by the
substrate analog/product analog.
Fig. 2. Highly reactive photogenerated intermediates: radical (A), carbene (B), and
nitrene (C).
cal methods, and even vacuum or extreme dryness. Besides these methods,
photoaffinity labeling and photoaffinity crosslinking are helpful tools for the
study of specific interactions between proteins and deoxyribonucleic acids. To
date, many successful attempts have been made to photocrosslink proteins
to nucleic acids using different photoactivatable deoxynucleotides. 5-bromo-,
5-iodo-, 5-azido-, and 5-[N-(p-azidobenzoyl)-3-aminoallyl]-2'-deoxyuridine-
5'-monophosphate (12–18), 4-thio-2'-deoxythymidine-5'-monophosphate (19),
and 8-azido-2'-deoxyadenosine-5'-monophosphate (20,21) have been incorpo-
rated into deoxyribonucleic acids to bind DNA covalently to adjacent proteins
(for a review see ref. 22).
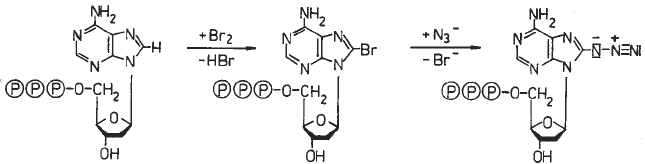
Here, we describe the synthesis of 8-azido-dATP (8-N
3
dATP), its incorpo-
ration into DNA by nick translation, and the procedure to photocrosslink azido-
modified DNA to proteins (20,21).
DNA–Protein Photocrosslinking 325
2. Materials
2.1. Synthesis of 8-N
3
dATP
1. dATP (disodium salt, Boehringer Mannheim, Mannheim, Germany).
2. Potassium acetate buffer: 1 M, pH 3.9.
3. Bromine.
4. Sodium disulfite (Na
2
S
2
O
5
).
5. Ethanol.
6. DEAE–Sephadex A-25.
7. Triethylammonium bicarbonate buffer: 0.7 M, pH 7.3.
8. Dimethylformamide.
9. Hydrazoic acid: 1 M in benzene.
10. Triethylamine.
2.2. Characterization of 8-N
3
dATP
1. Silica gel plates F
254
(Merck, Darmstadt, Germany).
2. Cellulose plates F (Merck).
3. Isobutyric acid/water/ammonia (66:33:1 v/v).
4. n-Butanol/water/acetic acid (5:3:2 v/v).
2.3. Preparation of Azido-Modified DNA
1. DNA (e.g., pBR 322 or pWH 106).
2. Deoxyribonucleotides (dATP, dGTP, dCTP, dTTP, [α-
32
P]-dCTP).
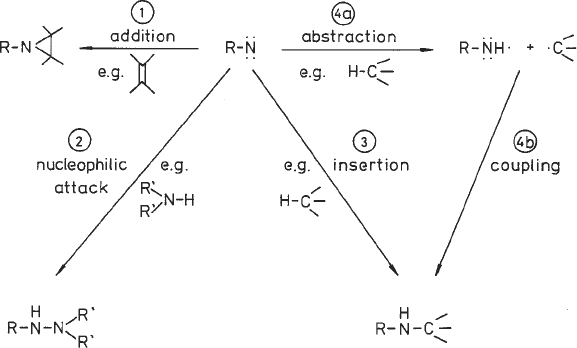
Fig. 3. Reactions of nitrenes. Cycloaddition to multiple bonds forming three-mem-
bered cyclic imines (1), addition to nucleophiles (2), direct insertion into C-H bonds
yielding secondary amines (3), and hydrogen atom abstraction followed by coupling
of the formed radicals to a secondary amine (4a, 4b).
326 Meffert et al.
3. DNase I (Escherichia coli, 2000 U/mg, Boehringer Mannheim) in 0.15 M NaCl
and 50% glycerol.
4. 50 mM Tris-HCl, pH 7.2.
5. Magnesium sulfate (MgSO
4
).
6. Bovine serum albumin.
7. DNA polymerase I (E. coli, Boehringer Mannheim, No. 104493, purchased con-
taining definite amounts of DNase I).
8. Ethylenediaminetetraacetic acid disodium salt (EDTA).
9. Sephadex A-25.
2.4. Photocrosslinking
An ultraviolet lamp (e.g., Mineralight handlamp UVSL 25 at position “long
wave”) emitting UV light at wavelengths of 300 nm and longer.
3. Methods
3.1. Synthesis of 8-N
3
dATP
The synthesis of 8-N
3
dATP (Fig. 4) is performed principally by analogy to
the synthesis of 8-N
3
ATP (23) (see Note 1). In the first step, bromine exchanges
the hydrogen at position 8 of the adenine ring. Then, the bromine is substituted
by the azido group.
1. Dissolve 0.2 mmol (117.8 mg) of dATP in 1.6 mL of potassium acetate buffer
(1 M, pH 3.9) and add 0.29 mmol (15 µL) of bromine. Keep the reaction mixture
in the dark at room temperature for 6 h (the absorption maximum shifts from 256 nm
to 262 nm; see Note 2).
2. Reduce excessive bromine by addition of traces of (approx 5 mg) Na
2
S
2
O
5
until the
reaction mixture looks colorless or pale yellow. Pour the reaction mixture into 20 mL
of cold ethanol (–20°C) and allow to stand for at least 30 min at –20°C in the dark.
3. Collect the precipitated deoxynucleotide by centrifugation and redissolve the resi-
due in 0.5 mL of double-distilled water. Further purification is achieved by ion-
exchange chromatography over DEAE–Sephadex A-25 column (50 × 2 cm) with
a linear gradient of 1000 mL each of water and triethylammonium bicarbonate
(0.7 M, pH 7.3).
Fig. 4. Synthesis of 8-N
3
dATP.
