Lewin Benjamin (ed.) Genes IX
Подождите немного. Документ загружается.

tw vNo
q]!M
au^zul alol
Jo
uor.lPllossv
slolluo]
dr.uPll aql
0l'8I
.luau6P.lJ
lxeu
oql
ozrsaqlu^s
ol
dool olqdulel
papuells-elEurs
eql ur.lauud
P
qllM selell
-ossPar
uaql
puP
luau6PrJ
qezPl0
uP
Jo
puo
oql
lP
salPtlosstp
puPlls
6u166e1 aq1 sazrsaqlu{s
1eq1
aseraLufilod aq1'filsnonurluol
sa^oul
puPlls
6urpeel aq1 sazLseqlufs
1eq1
aseratuf1od aq1'due1r
6utptts e
[q
VUC
ol uo
plaq
sr
qrrqMJo qrpa'slrunqns
rLy{1e1er
asetaufi1od
VN6
oml
o1
pallouuol
sr
l.loJ
uorlelrldar aq1 6urlearl osprrleq
eH-L
,..
i
+
L
iiSit-{.:lr*
.JSPIUIJd
Jqt 8ur)eldsrp roJ alqlsuodsJr
sI
III
JSerJru
-A1od
yNq
Llruareddy
'seseq
uJJunoy
ot
tq8ta
ot
palrurl
sl
VNU
Sururrd
eqt
Jo
qtSuel rqJ
'pJSpJIer
sr aserurrd eql
'slseqlu.ds
rarutrd
8ut
-,lrollod'al1s
ater,rdordde
uB
le
JSpruIJd
geuo
Jql
qlrM
slJPrelul
1I
puP
5lroJ uoIlPJIIdar
aqt
sladord
leqt
eserrlJq
eqt sl
1I
:geu(
;o
sargadord
lenp
aqt
.dq
paprno,rd
sI
>lroJ uotlertldar
aqt
pue
3ur
-rurrd
uaa^,vr.lJq
uorlJJuuor aql
'suorqdJJ
)tJl
uI
;sluaru8er;
DIezp>lO
;o
srsaqtuds
SurtBttrut
roJ
salrs Jqt
SurzruSorar ro; alqrsuodsar
sI
leql4
'xJIClIuO)
Surlerrldar
Jql IUoJJ
Sutletrosstp
tnoqll.rt
'apz{.r
qrea
uI
txJu
aql ot
duep
fl
auo
ruorl
sraJSuerl aseraruLlod
pupJls
3u63e1
eqJ
'luJru
-3er;
qeze>16
lxeu
eql JlpIlIuI
ol
JJpPol
druep
aqt
,(q
petlnJf,eJ
ueql sI
duep
$
,r,rau
y
'doo1
aqt Sursealar
'pataldruol
st
luaur8eJI
q)ea
Jo
srsaqlur(s
eq1 ueqM elerf,ossrp
Laqr
teqr
sMoqs
r1''t:,
E:ii'i{ri,i
'druep
f,
aql
pue
aseurtrd
Jql
JoJ
ldarxa
'(VNq
eW
qrlu pJlPIJossP ureuar
Aaql
'sr
teqt)
Llantssarord uolttun;
snleredde
uolleJ
-qdar
aqt
;o
sluauoduroJ eq]
Jo
11y
ipataldurol
st
tuaru8er;
DIeze>lO Jql
uJqM suaddeq
leq14
'xJu
Jql ut8aq
ol
LPear
st
pue
luaruSeJJ
Juo salalduor
ase.raru.dlod
3ur3
-3e1
aqt JroJJq
lueu8er;
ulezelg Juo
lseel
le
Jo
qfuel
Jql
JoJ
puJtxJ
tsnru
aleldruel
pJpuerls
-apurs
JqJ'pueJts
,lrJU eql
3uzrsaqlu.,{.s
apqn
druep
fl
aqr
q8norqt aleldural
papue:1s-apurs
aqt s11nd
xaldruor
a:or
pueJts
8ur33e1
Jql
'luaru
-8er;
qezelo
ue
Jo
uolteltpl
rrtJv
'seJue^pe
lurod
Surpulmun
Jqt
se ra3;e1
sauoraq
dool
srql
pue
'srsaqtuu(s
puprts
3ur33e1
ro; ateldrual
aql
sapuord
tpql
vNq1
papuerls-a18urs
1o
dool
e sJlpJrJ
pupJls Surpeal
Jqt
Jo
srsaqluz(5
'.{lrrrrssarord
sll
asperJul
ol
'sl
leql
'JJo
Suqpy
rxoJJ aserJluLlod
puerls
3upea1
eql
luJA
-ard
o1 sI
puoJes JqJ
'ploJuJl
trq aror JSeJJru
-z(1od
y1qq
r{q
tuaruarrou
Jo
eler aql
Surseanur
Lq srsaqtur(s
VNo
;o
paads Jql aseaJJul
ol
sr euo
'sIJJJJJ
omt
seq
>lull slql'saror
rtl,{1eler
eql
pue
xalduror
aserurrd-aseJllel{
Jq1
ueJmlaq
uorl)euuoJ
lJaJIp
e sJqsllqelse
slqJ
'JJpPol
druep
aqt;o
(s)ilunqns
r,
eql
spetuoJ
geu(
'uoB)eJIp
,,pJeMro!,,
Jqr
uI
VN(
aqr Suop
saleJolsuerl
pue
'1urod
Surpurmun
aqt satpeJJ
gPu(I
'VN(
aql ul
dool
e Surtean
snql
,,'q8norqt
pa[nd,, sI
puerls
3ur33e1
aqt
roJ alelduat
eql
lpueJls
Sutpeal
aql roJ
aleldruat
aqr 3uo1e,{lsnonuttuoJ
sJAoIu
aruLzuaoloq
Jr{J
'VNq1
Jo
pueJls a1e1dura1
qrea
qum
paterJosse sr ero):ril1eler
r/'Et"*E
]$filt3i
ur
peteJlsnll se
'xaldruor
aru,{zua
eql
Jo
sluau
-odruor
IpnpIAIpuI
Jql
Jo
sruJal
uI
III
JSPJJTU
-d1od
y51q
roJ
Irpotu
slql
eqlrrsap
uer
rM
'druep
Surpls
e
qtlm
petelJosse
sI
qJIqM
Jo
q)ee
'slrunqns
rpIle
-ler
aserarudlod
vN(
oml
ol
pJlJauuoJ
sI
esPJ
-llrq
eqJ
'pu€ns
3ur83e1
aql
roJ
aleldruat
aqt
uo
uorDalrp
,€-,g
Jql uI
sel€lolsueJl
teql-8utr
)rJaruexaq
e sIrrJoJ
.d11ertd,{1
qJIqM-JSP)ITJTI
e
.dq
palean
sI
>lJoJ uotlerqdar
aq1
'aserqdal
e
q)ns
Jo
uorlerado
Jql JoJ
Iapou
ruaua8
P sM€Jp
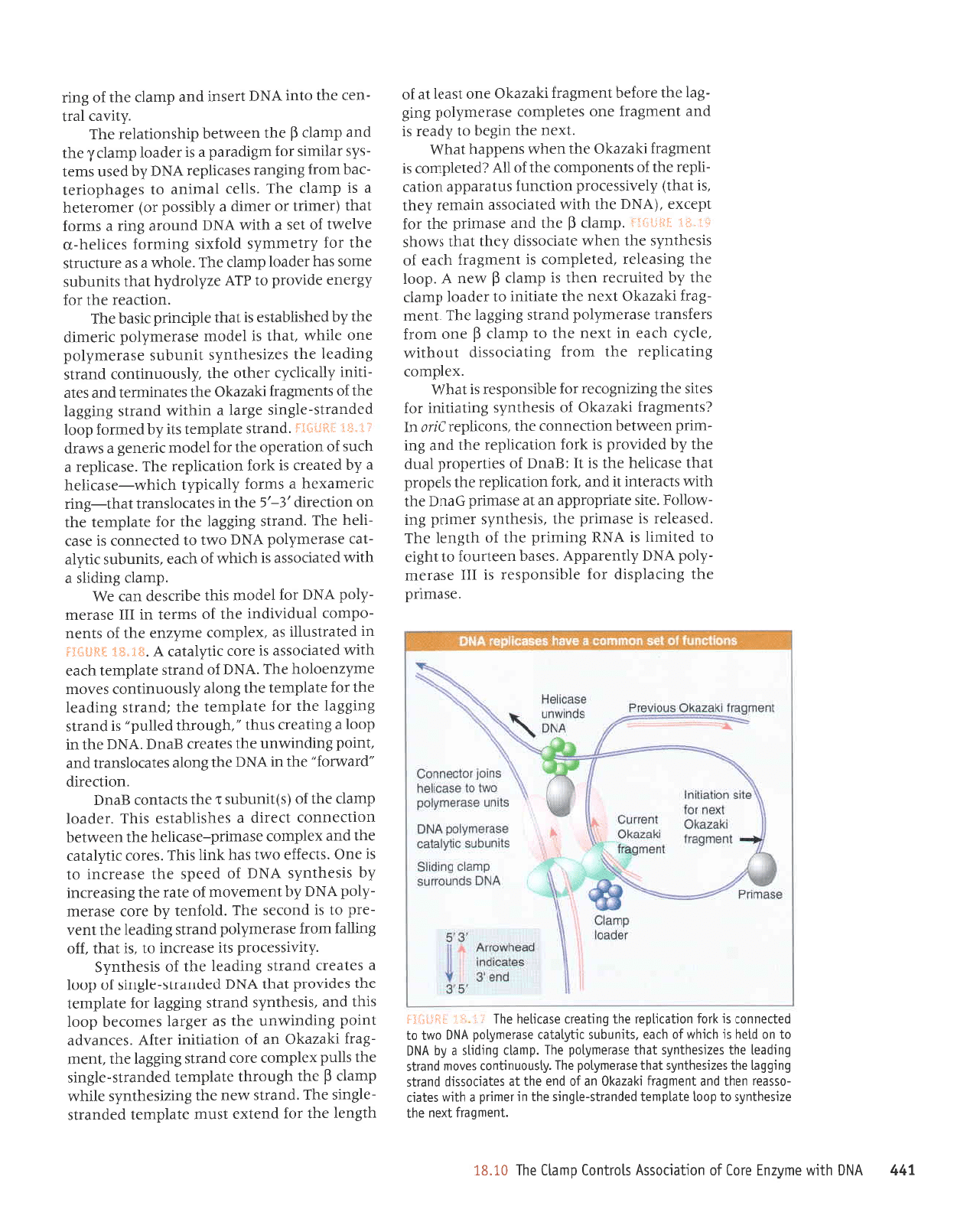
i.t'fJi. tHfi*IjJ
'puerls
aleldrual
str
Lqpautro;
dool
papuerls-a13uts a8rel
e uIqlIM
puerls
3u63e1
eql
Jo
stuaru8er;
qeze>lg
eq1
seleuruual
pue
sele
-1trur
,(1ergrfu
.raqro
aql
,{lsnonulluoJ
pueJls
Surpeal
aql
sezlsJqluLs
ltunqns
aseraruLlod
Juo alrqM
'leq1
sl
Iepour
aseraru,{.1od
f,IJJruIp
aqt
Lq
paqqlqelsr sl
leql
aldourrd
rrseq
aq1
'uoIl)PeJ
Jql JOJ
d8raua
apnord
ot
dJV
az,{.1orpLq
leqt
suunqns
auos
spq
Jepeol
dtuep
aql'JIoqM
e se ernlJnrls
eql
roJ
.drtaruru.ds
ploJXIS
Sutruro;
sJ)IIJq-n
JAIeMI
Jo
les
e
qq,lr
VNo
punoJe
Sutr
e sruro;
leql
(raurrrl
to
Jaulp
e
,(lqrssod
ro) raruoralaq
e sr
druelr
JqJ
'sllJ)
Ierulue
ol
sa8eqdotral
-Jeq
uoJJ
Sut8uer
sasertldar
y51q
z(q
pasn sual
-s,{.s
regrurs
roy
ur8rpered
e sI JJpPol
druepl"
aqr
pue
druelr
f,
aqr
uaa,r,uaq
dtqsuoqelJ]
eqJ
'AlIApJ
IeJl
-uJJ
aql
olul
VNC
uJsul
pu€
druell
aqt;o
3uu
FtSUftf
i8.18
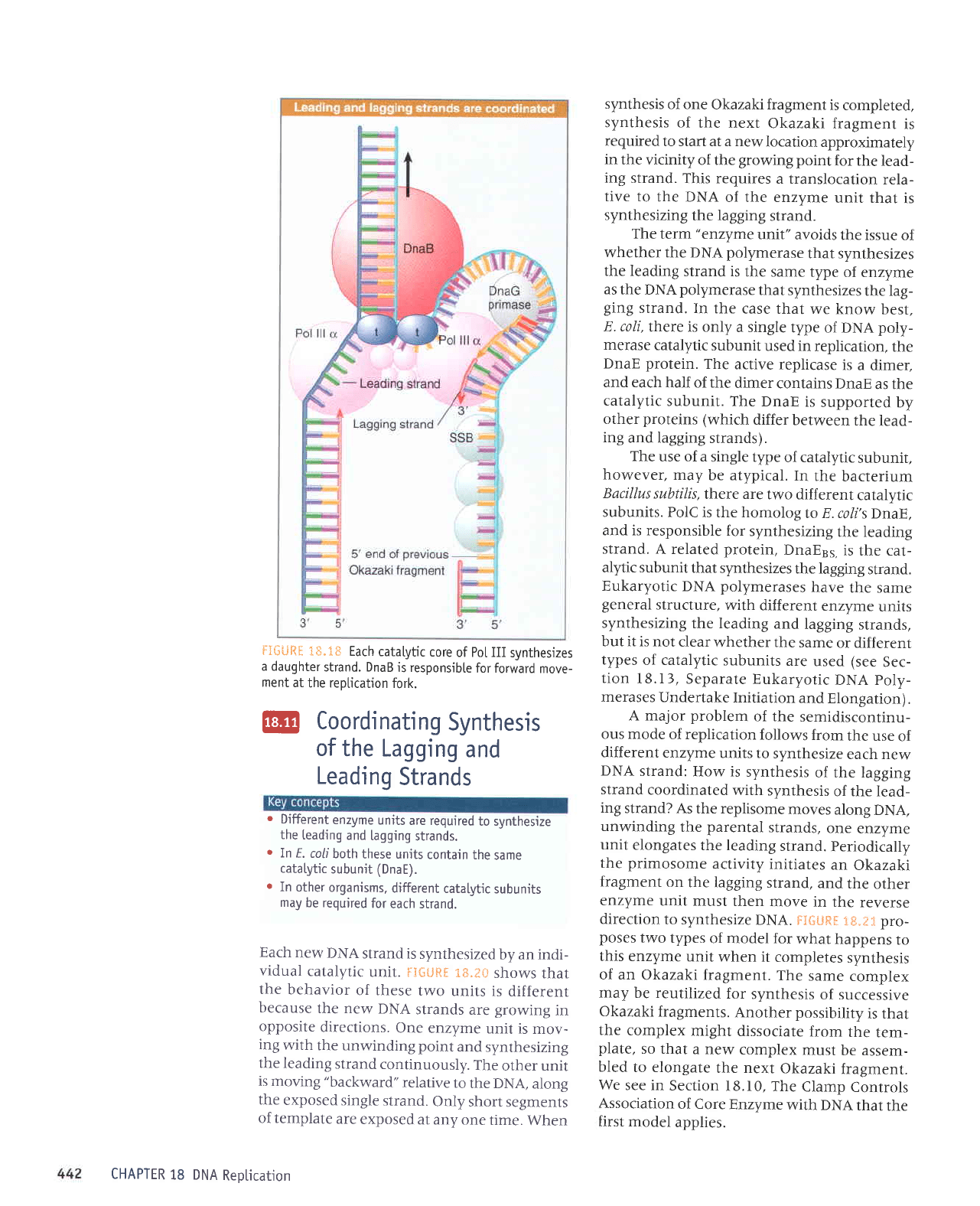
Each
catalytic
core
of Po[ III
synthesizes
a daughter
strand. DnaB
is responsibte
for forward
move-
ment
at
the reptication
fork.
synthesis
of one
Okazaki fragment
is
completed,
synthesis of
the next
Okazaki
fragment
is
required
to start at a new
location
approximately
in
the vicinity
of the
growing
point
for the
lead-
ing
strand.
This requires
a translocation
rela-
tive to
the DNA
of the enzyme
unit that
is
synthesizing
the lagging
strand.
The
term
"e\zyme
unit"
avoids
the issue
of
whether the DNA
polymerase
that synthesizes
the leading
strand is
the
same type
of enzyme
as
the DNA
polymerase
that synthesizes
the lag-
ging
strand.
In the
case that we
know
best,
E. coli,
there is only
a single
type
of DNA
poly-
merase catalytic
subunit
used in
replication.
the
DnaE
protein.
The
active
replicase
is
a dimer,
and each
half of the
dimer
contains
DnaE
as the
catalytic
subunit. The
DnaE
is
supported
by
other
proteins (which
differ
between
the
lead-
ing
and lagging
strands).
The
use of a
single type
of catalytic
subunit,
however,
may
be atypical.
In
the
bacterium
Bacillus
subtills,
there are
two different
catalytic
subunits. PolC
is the homolog
Io E. coli's
DnaE,
and is responsible
for
synthesizing
the leading
strand.
A related
protein,
DnaEss.
is
the cat-
alltic subunit
that
synthesizes
the lagging
strand.
Eukaryotic
DNA
polymerases
have
the same
general
structure,
with
different
enzyme
units
synthesizing
the leading
and
lagging
strands,
but it is not
clear
whether the
same
or different
types
of catalytic
subunits
are
used
(see
Sec-
tion
18.13,
separate
Eukaryotic
DNA
poly-
merases
Undertake
Initiation
and
Elongation).
A major
problem
of the
semidiscontinu-
ous mode
of replication
follows
from
the use
of
different
enzyme
units to
synthesize
each new
DNA
strand:
How is
synthesis
of the lagging
strand
coordinated
with
synthesis
of the lead-
ing
strand?
As the replisome
moves
along
DNA,
unwinding
the
parental
strands,
one
enzyme
unit
elongates
the leading
strand.
Periodically
the
primosome
activity
initiates
an
Okazaki
fragment
on
the lagging
strand,
and
the
other
enzyme
unit must
then
move
in the
reverse
direction
to
synthesize
DNA. FIG{JRf,
?S.*i. pro-
poses
two
types
of model for
what happens
to
this enzyme
unit
when it
completes
synthesis
of an
Okazaki fragment.
The
same
complex
may
be reutilized
for
synthesis
of successive
Okazaki
fragments.
Another possibility
is that
the
complex
might
dissociate
from
the
tem-
plate,
so that
a new
complex
must
be assem-
bled
to elongate
the next
Okazaki
fragment.
We
see in
Section
18.I0,
The
Clamp
Controls
Association
of Core
Enzyme
with
DNA
that
the
first
model
applies.
@
Coordinating
Synthesis
of
the Lagging
and
Leading
Strands
r
Different
enzyme
units
are required
to synthesize
the
[eading
and
lagging
strands.
t
ln
E. coLi
both
these units
contain
the
same
catatytic
subunit
(DnaE).
o
In
other
organisms,
different
catatytic
subunits
may
be required
for
each strand.
Each
new
DNA
strand is
synthesized
by an
indi-
vidual
catalytic
unit. fIG#Rf
ts.I#
shows
thar
the
behavior
of these
two
units is
different
because
the new
DNA
strands
are
growing
in
opposite
directions.
One
enzyme
unit is
mov-
ing
with
the unwinding
point
and
synthesizing
the leading
strand
continuously.
The
other unit
is moving
"backward"
relative
to
the DNA,
along
the
exposed
single
strand.
Only
short
segments
of template
are
exposed
at
any one
time.
When
CHAPTER
18 DNA
Replication
Ettt
ase6rl
Aq pa)ull
elv sluauberl
rlezelg
ZI'8I
'raqtaSot
sluatu8ery
DIpze>lO uroI o]
ge;
.daql JSneJJq sluelnru
-Bt1
u1
tslsrad
sluaru8e:;
peDauuof,un
'satoLrelna
pue
salo,{relord
qroq
ur
luasa,rd
are
sase8tl
'ase8q
y11q
aruzkua aqt
qtlm
sJrl
>lJru sqr
8ut
-lpJS
roJ Llqrqrsuodsal JqJ
'1ueu8er;
snourard
aq1
Jo
pua
ateqdsoqd-,S aq] ol
luarelpe
sI
luelu
-3ery
auo
Jo
pue
HO-,€
aql
'raqtaSot
pJ>luII Jq
lsnu
slueru8er;
qezelg
tuarelpe
aq1
'pareldar
pue pJ^orueJ
uaJq seq
vNu
Jr{]
eJuo
'JruouJE
eql
ul suoll
-errldnp
ro suorlalap asne;
r(eru
leql
saJntJnJls
Surteraua8
ruorl
VN(
1o
sdeg
sluanard
tl lPql
sl
INgd;o
aruesodrut
praua8
aq1
'(97'9
arn
-8r4
aas) sleadar
Jo
rJqlunu
aql aSueqr
z(eru
seJntlnJls
asaql
'sutdrleq
IuJoJ
uer saruanbas
rnuorpurled
:ateldurat
eqt
qllM pau8tlesrur
pue pareldsrp aq ueJ
sleadar
1JaJICI
'saruanbas
paleadar;o suofar
ur saruanbasuor
luBlrodrur
aleq
ue)
.{ptder deg
e anourJJ
01 arnlled
'[PrJJlPru
1ua;e[pe
:nouar ol
,{t!
-ArDP
eSPaIJnUOXJ
S1I esn
ueql
puP
Jplloellnu
pareldsrp
e ot
txau
alpalJ.deru
tNgd
'suoIlJPJJ
rredar
vN61
uI
'z(tr.rrpe
esealJnuoxJ
,€-,5
e setq-
osp
1r 1nq'JSPJIJnuopue
ue
se suolpunJ
INad
'uorlJpJJ
srql uI
'INiI{
aru,{zua
eqt
,{q
pJ^eJIJ
sr de1; aql
Jo
espq
aql
reql
sMoqs
ta.g1
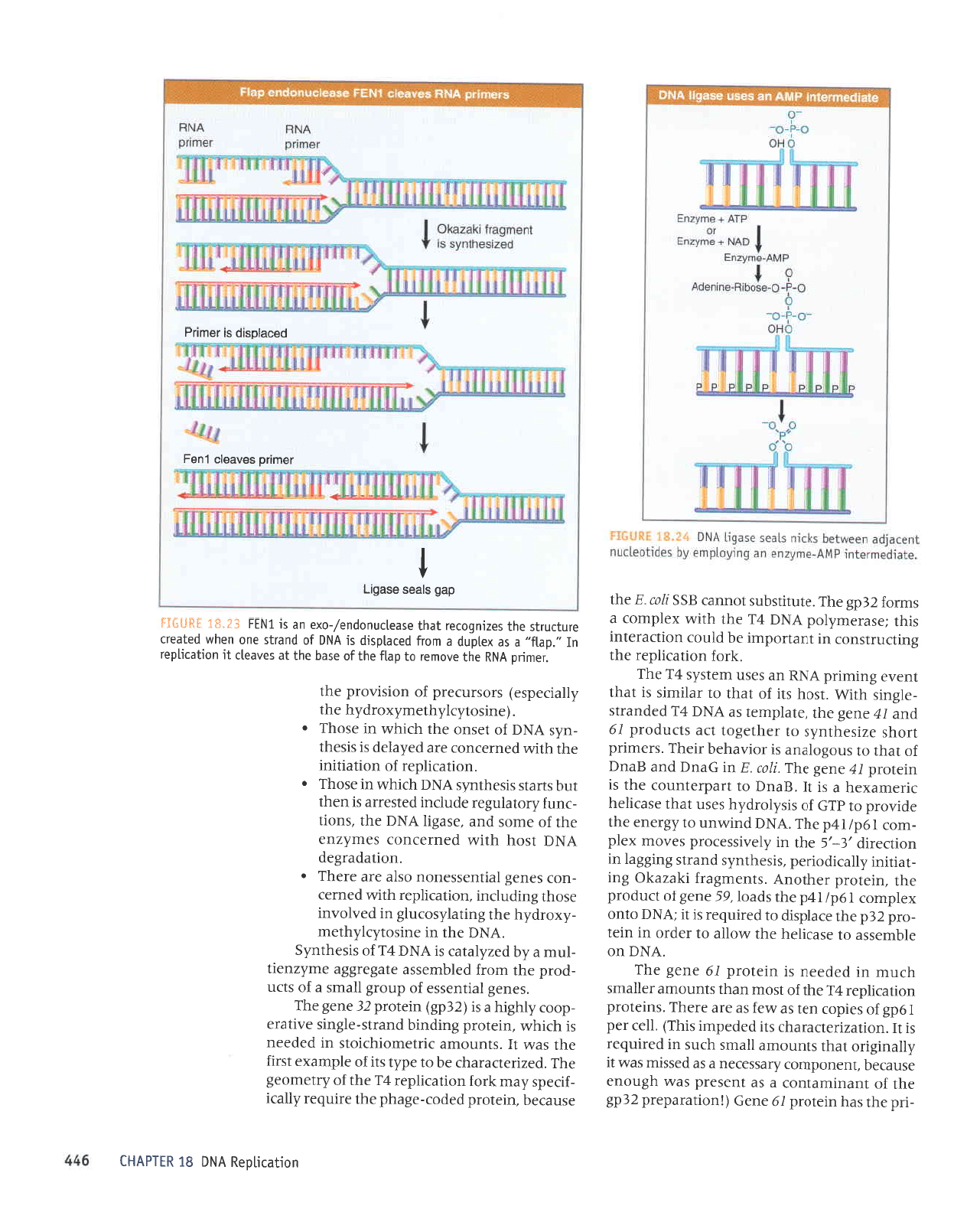
3Hn$Ii
,,'deg,,
e
Jo
ruJoJ
el{t ul
luaru8erl
Supal
-ard
aqt
yo
rarutrd
vNU
eql
sateldsrp
luaurEer;
r>leze>lo ue
Jo
slseqlu,{5
'ssarord
dals-oanl e
,(q
panauuot
are
sluaru8eJJ
DIPZP>IO
'(dlrrrrDe
JSpJlJnuoxJ
,€-,s
e JAeq
tou
saop
:seraurz(1od
VNC
eq1 araqa,r)
srualszi.s
uellelulueru
uI
'YNCI
Jo
luJrudes
P
ueql
rJqlPr
vNU
Jo
qJlJJls e sareldar
vN(
MJU
aql
leqr
ldalxa
'uoIlPISuPJl
>lJIu 01
lualeatnba
sr
srql
'luaru8er;
DIezDIO
txau
aqt
Jo
pue
HO-,€
Jql ruorl
pepuJlxe
aruanbas
YNq1
e
qlyvr
t1
3ut
-re1dar,{lsnoauellnuls
ellqM
rarutrd
YNU
aql
sa.l,oruar
LlrzlllJp
JspJlJnuoxe
,€-,s
aq;
.dlradord
sluaurEe.rl
rlpzeIO laqt
u1oIot
ye1
sruenas
ypd
11 ase:arudlod
VNq
rtq
pailU
sr de8
aql
'deE
e aq
IIrM
eJJql
'pelorueJ
sr
raturrd
aql uJqM'lueur
-3er;
Surparard
aqt;o
rauud
VNU
aql
Jo
uPls
aql
eJoJeq
tsnI
sateunurat
luaru8erJ
pl€ze>lo
ue
Jo
srsJrllu.,{.s
leqt
s1sa38ns
arn8r;
aq;
'sluaru8er;
DIPZP>IO
luare
[Pe
;o
Surlutt
luelelo)
JI{l
pue
'VNO
Jo
qJlJrls
e
.dq
luaruareldar
slt tarurrd
y51g
Jqt
Jo IeAoIueJ
'VNq
qll^
uolsuelxJ
slr
'rarutrd
VNU
Jo
sIS
-aqlur(s
sAIoAuI
lsnru
1I lnq'ulelrJJun
sI sluJ^e
Jo
rJpJo alaldruor
eqJ
'eg's{
36fi$IJ
uI
pJleJl
-snllr
sp
'sluaru8er;
DIezP>lO
Sututolut
paAIoAuI
suorlJe
Jql
Jo
,l,rJIA Jno
puedxa
Mou ueJ aM
'lueuDe.u
uau
aq1
1o
Euruur6aq
,g
aql
o1
lueuLbe4
DlPze)g auo
Jo
puo
,€
eql spauuol
leql
puoq eql salPul
eseott
y1'19
r
'uoqelsuPrl
Ilru
solquesel
leql
uoqle
ue ut
vNo
qlw
ll
serelder
pue
taurud
aql
senoual
1
aselauAlod
y16
.
'1uau6er;
lxau
aql ato;aq
sdols
pue
rauud
p
qlvv\
silels
luaLuEe4
tlezPl0
qlPl
.
oseErl
,tq
pa)utl erv
'1ede
anou saseleufilod
puells
6uL66e1
pue
6urpeal
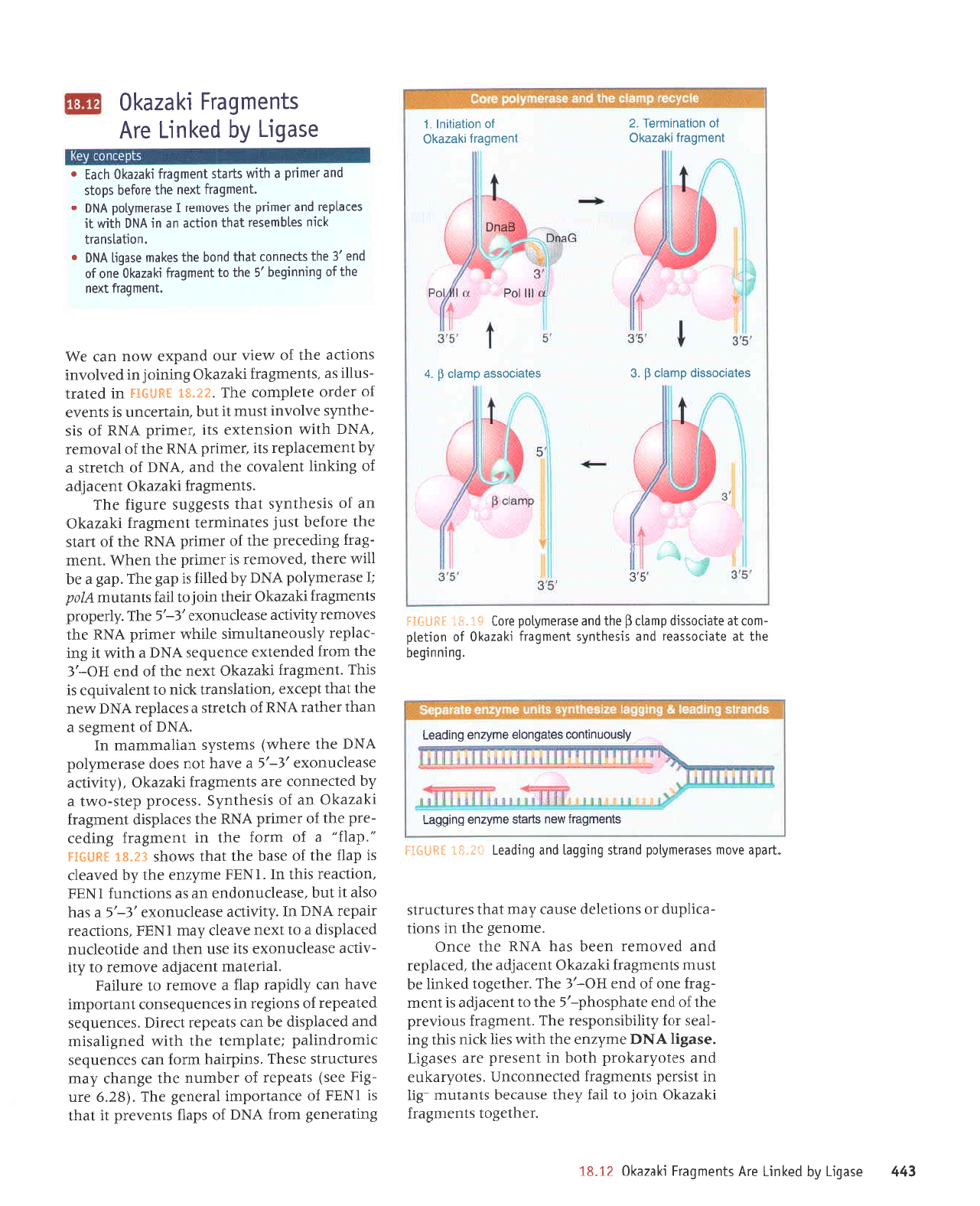
*t't! S$fi$:j
slueru6ell
areu spels eultzue
6ul66e-l
I;snonurluoc sele6uola
eu.dzue 6ulpee-l
'6u
Lu u rDaq
eql
lp
eleoossear
pup
stsaqlur\s
luaubery
l)pze)0 Jo
uotlald
-uorle
oleoossrp duLelr$
eql
pue
asereufi1od alo]
#l'S;.
jsdl*Ii
solercosse duep
$
'p
lueuOerl
ryezelO
lo
uortEuruuol'z
lueuber;
t)iezqo
Jo
uorlerlrul
I
9e
al
solercossrp durelc
d
'e
ilt tod
quauberl
Dlezel0
'VN)d
pue
f
-{U
'sulaloJd
JJqlo
o^{t I{]I^t
uouJe
-relq
stl uolJ
sllnsJJ
ltrnrssarord
sU
'puprts
8ul
-peel
Jql
sazrsaqtuz(s
[lsnonurtuor
1eq1
au,,{zua
anrssarord
[tq8tq
p
sr g eserJru.{1od
y1qq
'xaldruot
uorlerlrur
Jql
Jo
sluJuodruor
lera
-,Las
Suoue
suorlJpJJlur
senlolur
11
'qrtr,ns
1od
rqt
pJIIeJ
sr
IUJAJ
slql'g
aseraru,{1od
VNO
sr srql
'puerls
Surpeal
eql
uO
'ureqJ
rqt
puJtxr
IIrM
tpql
aru,,lzua
ue z(q
pareldJr
uaqt
sr
U
.(VN(r
prlle)
sarurlaruos)
VNC
Io
srseq
0€
ot
0Z
^q prMolloJ
VNU
Io
sJseq
0I-
Jo
SurtsrsuoJ pupJts
lJoqs
e sazrsaqlu,{.s pue
ur8rro
aql
le
xalduro)
uortpll
-rur
aql
01 spurq
aruLzua
aseurrdTn
1od
aq1
'aserurrdTn
1od
pa11er
sJrurlJluos
sr
lr
'surpqf,
puJlxe
pue
aurrd
ol
.{1r
-Jedpl
pnp
slr Surpauau
'.{lrrrrlre
aseruud
e apr.L
-ord
leqt
suratord
tJIIeus
oMl
pue
.ri.lqurasse
roy Lressarau
sreadde
teqt
trunqns
g
Jqt qllm
pJleDossp
sr
qJrqM
'llunqns
It,ileter
cI-0gI
e
Jo
Surlsrsuor
xalduor
p
se
slsrxe
auz(zua
aq1
'spuerls
3u63e1 pue
Surpeal
rqt qtoq
rlelllul
ol
pJsn
sr
tI'pupJts
Mau
p
Jlerttur
01 ^UIlqp
Jqt seq
lr
JsnpJJq
Jpnsnun
sr
D
JSeJJur,{1od
ygq
'sJIor
J3qlo
spq osle
tnq
'srseqtu,{s
puels
3ur33e1
ur
pJAIoAur
aq Leu
a
aserauLlod
VNC
.
'puerls
bur
-peal
eqt sale8uola
g aseraru.dlod
VNO
.
'spueJls
A^au
Jo
srs
-aqtu,{s
aql sJtertrur
n aseraru,{1od
VNq
.
:uort)unJ
luJJeJJrp
P seq
saserrldar
VN(
leJIJnu
JaJr{t
Jql
Jo
r{teg
'1"
aseraur,,{1od
VXO
,(q
ue>leuJpun
sr
'uorlerrldar
se
IIJM
se suortJpeJ
uorleurqruof,Jr pue
rredar
Surpnpur,srsaqtuLs
VN(
IelJpuoqlotpu
IIV
'sJleJ
Jorra
ralear8 qrnru
a^pq
sJJqlo
arlt
Jo IIp
:saseltdar
aql
Surqreordde
.{tlapg
e
seq
$
ase.raru.d1od
ygq,{1uo
treder
ur
pJAIoAur
sarudzua
aql
y6
'(sau^,{zua
rredar
raqlo
;o
xalduor
e
Jo
txJtuoJ
Jqt ur
uorlrun; .deu
l1
q8noqrle)
trunqns
)rJJruouoru
a13urs e;o
tsrs
-uoJ
uauo r{Jlq,lr'sJJnlJnJls
raldrurs qlnu
aleq
saseraru.dlod
rredar
aq1'aru,,lzua
IprrpuoqJotrru
xaldruor
ssal .{1tq8r1s
rqt
seop
se .{rqaply q8lq
qll1lt
VN(
ele)lldJr
11e
sauLzua
asaql
'3urpeaJ
-yoord
.ro ,{.trarssalord
'Surulrd
se
q)ns
,suorl
-run;
drelpue
qlrM
pJuJJJuo)
eJp
sJJqlo
eql
pue
'srs.{.p]el
roy dlqrqrsuodsar
aqt
spq strunqns
Jql
Jo
euo
'aseJ
qJeJ
u1
'sauLzua
f,rJJruerlJloJJ
-taq
a8rel
a-re sasetrldar
rpalf,nu
rqr
Jo IIe
teql
s1\Aor{s
::ir"i{t
iiii::ji:i
'lerrelelu
paSeuep
areldar
ot
vNO
Mau
Jo
sJqJtJtts
Surzrsaqtu.ds
qtru
pauJJJuoJ
are
saseraru.{.lod
VNq
JeJI)nu
JJqlo
Jql
IIV
'3
pue
'g
',n
seseraur,(lod
vN(
sarrnbar
uorlerrldar
vN(
JpJpnN
'y5q
paSeruep
rreda.r
ot
lprJJtpru
Suursaqlu^ds
ur
pJ^lolur
esoqt
pue
uor.lPlltoau
vNo
8I
utldvHl
uorlerrldar
elrlenlJSuolrues
JoJ
parmbar
asoql
olur
pJpr^rp
zllpeorq
eq uel .daq1
'saseraru,{1od
VN61
Io
JJqunu
a8rel
e eneq slleJ rrlodrelnE
'puetls
6ur66e1
eq1 sale6uola
a
aserau{1od
VN6
lo
g
aseraurf1od
VN6
puoras
p
pue
puerls
burpeal
aql salebuola g
eserauAlod
yp6
.
'spuetls
VN0
qloq
1o
srsaqlufis
aq1
salerlrur
xaldLuor
aseuud/n aseteLu[1od
VN0
agl
r
'z
to/pue g
aserauflod
y116
1o
saxelduol
oml
pue
eseuud/n
aseretur{1od
y1r;6
;o
xelduor
auo spq
uo1
uoqerLldat
y
o
uoqPbuoll pue
uoqeqrul
a)Puapun
sasPrau^lo6
vNo
lllofue1n3
alPrPdas
awy
Jqr
pue
aru.{zua aqt
Sursealar
'>lrru
Jr{l
Jo
snururel
Ho-,€
Jql r{lr^1.
paruJo}
sI
puoq
rrlsJlpoqdsoqd
e uaqt
pue
'>lJru
aql
Jo
aleqdsoqd-,S
Jqt ot
pJqJelle
saruof,Jq
xalduor
au,{zua
Jqt
Jo
dwv
JqJ
(g1y
sasn aru,{zua
tI
Jql
seJJaqM
'Jol)pJoJ
e se
[aprtoalJnurp
JuruJpp
eprueurloJru]
CVN
sasn aru^dzua
tTot
'A
eqJ
'sJolJeJoJ
tuJJaJJrp
:sn saru,{.zua
7J
pue
qot'g
aqTl'xalduor
4ryy-au.dzue
ue sJAIoAur
leql
uollJpJr
dals-orr,rl
p
e>lelJJpun
saru,{zua
QloB'rij'Si
ttf ii:i:'j
uI
pJleJlsnl[
se
'tutural
aleqd
-soqd-,S
pue
HO-,€
a^eq
teqt
s>IJru Suqeas;o
,{.uadord
rqt
erpqs sase8rl
V;.pue
qo
'A
JqJ
'luaLubetl
tlezpl0
lxau
aql
azrsaqlufs o1
y116
qlrM
saleoosse
lrun
eufzua Mau
e
pup
'1uaube4
rlpzqg
up selalduol
lr
uaqM saleoosstp
aserauAlod
puerls
6u16
-6e1
aq1
leql
sr
lepou
lamol aq1
'1uauberl
lxeu
oql azrsaql
-ufs
o1
uorltsod ,ueu
p
ol sa^ou
1r'1ueu6et1
rlpzplg
auo
salalduor
ltun
auAzua
ue uaqM
1eq1
sr asetaurtlod
puerls
6uL66e1
Jo
uorllp eql roJ
lapou
raddn
aq1
rn-*t
3i*{i*IS
vw
pasn
sr
ltunqns
crp{;e1ec
}uetaillp
V
posnoJ
sr
lrunqns
cr{1e1ec
otles oLlf
The roles
of RF-C and PCNA are analogous
to the E. coliy clamp loader
and
B
processivity
unit
(see
Section 18.10,
The Clamp Controls
Association
of Core
Enzyme
with DNA). RF-C
is a clamp
loader
that catalyzes the loading
of
PCNA on to DNA. It binds
ro the 3'end of the
iDNA and
uses
ATP-hydrolysis
to open the ring
of PCNA so that it can
encircle the
DNA. The
processivity
of DNA
polymerase
6
is
maintained
by
PCNA, which
tethers DNA
polymerase
6 to
the template.
(PCNA
is called
proliferating
cell
nuclear antigen for historical
reasons.) The crys-
tal structure of PCNA closely resembles
the
E. colip subunit: A
trimer forms a ring that sur-
rounds the DNA. The
sequence and subunit
organization are different from the
dimeric
p
clamp;
however,
the function is likely
to be
similar.
We
are
less
certain about events
on the
lag-
ging
strand. One
possibility
is that DNA
poly-
merase 6 also
elongates the lagging strand. It
has
the capability to dimerize, which
suggests
a model analogous to
the behavior of. E. coli
replicase
(see
Section 18.9, DNA Polymerase
Holoenzyme Has Three
Subcomplexes). There
are, however,
some indications that DNA
poly-
merase
s
may
elongate
the
lagging
strand,
although
it
also has been identified
with other
roles.
A
general
model
suggests that a replica-
tion
fork
contains one
complex of DNA
poly-
merase c,/primase
and two other DNA
poly-
merase complexes.
One is DNA
polymerase
6
and the other is
either a second DNA
polymerase
6 or
may
possibly
be a DNA
polymerase
e. The
two complexes of DNA
polymerase
6/e behave
in the same way as
the two complexes
of
DNA
polymerase
III in the E.
coli replisome: one
syn-
thesizes the
leading
strand, and the other
synthesizes Okazaki fragments
on the lagging
strand. The exonuclease
MFI removes
the
RNAprimers
of Okazaki fragments.
The enzyme
DNA ligase I is
specifically required
to seal
the nicks
between the
completed Okazaki
fragments.
Phage
T4 Provides
Its
Own
Replication Apparatus
r
Phage T4
provides
its
own reptication
apparatus,
which consists
of
DNA
poLymerase,
the
gene
j2
55B. a heUcase,
a
primase,
and accessory
proteins
that
increase
speed
and
processivity.
fg$#R[
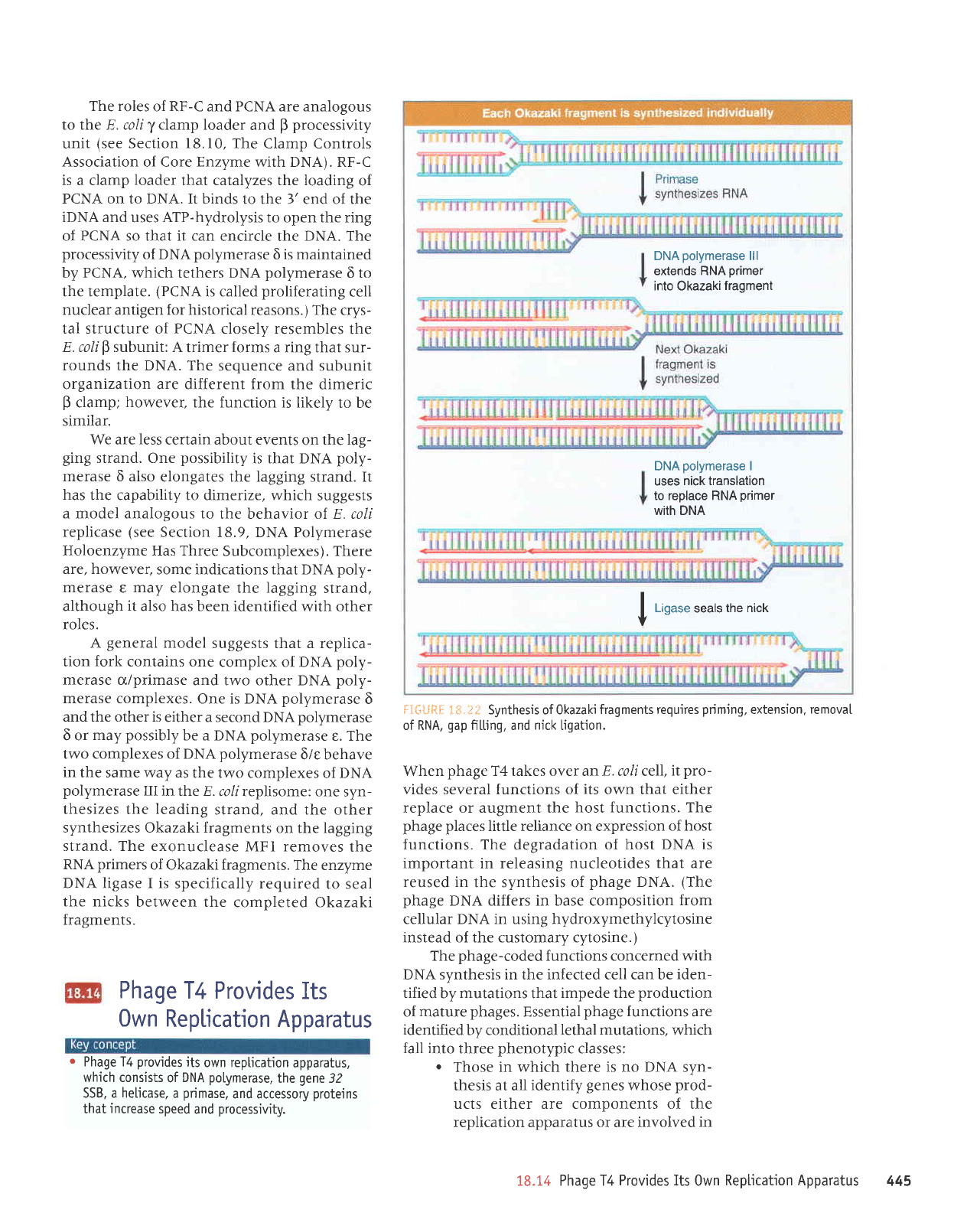
i*.tt Synthesis ofOkazakj
fragments requires
prim'ing.
extension,
removaI
of RNA,
gap
fitting, and
nick
ligation.
When
phage
T4 takes over an E. coli cell,
it
pro-
vides
several functions of
its
own that either
replace or augment the
host functions. The
phage places
little reliance on expression
of host
functions. The
degradation
of host DNA
is
important in releasing
nucleotides that are
reused in
the synthesis of
phage
DNA.
(The
phage
DNA differs in base composition
from
cellular DNA in using hydroxymethylcytosine
instead of the customary cytosine.)
The
phage-coded
functions concerned with
DNA synthesis in the infected cell can be
iden-
tified
by
mutations
that
impede the
production
of mature
phages.
Essential
phage
functions
are
identified
by conditional lethal
mutations,
which
fall into
three
phenotypic
classes:
.
Those in which there is no DNA syn-
thesis at all identify
genes
whose
prod-
ucts either are components of the
replication apparatus or are involved in
DNA
polymerase
lll
extends RNA
primer
into Okazaki
fragment
DNA
polymerase
I
uses
nick translation
to replace RNA
primer
with DNA
|
,,n"."
seals
the nick
18.14 Phage T4
Provides
lts Own
Reptication Apparatus
445
-rrd
er{l serl
urJlord
/9
Jua)
(
iuorle.redar
d
TgdB
Jql
Jo
lueuruetuoJ
p
se
tuJsJrd
seaa
q8noua
JSneJJq
'luauoduror
fuessarau
p
se
pJSSrru
se.!r
tt
,{lleur8rro
leqt
slunorue
IIetus
qJns
ur
parnbar
sl
lI'uollezlrrlrprpqr su
prprdul
srql)
.11ar
rad
19dB
1o
sardor
uJt
sp ,trJJ
se
Jre eJJqJ
.suratord
uorlerqdar
7I
aqt
Jo
tsotu
ueqt
stunorxe
Jellerus
qrnu
ur
papaJu
sr
uratord
19
aua8
aql
'vNo
uo
alqruJsse
ol Jse)llJq
eql
A{olle
o1 JJpJo
ur ural
-ord
7gd
aq1
areldsrp
ot
pallnbJr
sr
tr
:VNO
otuo
xaldruor
IgdlWd
eql
speol
,6E
aua8;o
nnpord
aql
'uralord
JJqlouV
'stuaru8erJ
rlezelg
Bur
-leltlul
[lerrporrad
'srsaqluds
puerls
BurBBel
ur
uoIlJJJrp
,€-,5
erql
ur z(lanrssaro.rd
sanoru
xald
-ruor
1 9d7
17d
aqt
'VNC
purmun
or .d8raua
aqt
apnord
01
dJg
;o
srsz(1orpz(q
sJSn
teql
JSeJrIrq
JrrJruexJq
e sr
lI
'geuc
ol
trpdJJlunof,
er{l
sr
uratord
1V
aua8
eql'qn'E
ur
)puq
pup
gpue
Jo
leqt
ot
snoSoleue
sr Jor^eqJq
rraql
.srarurrd
uoqs
Jzrseqlu,{s
o1
raqla8ol
pe
spnpord
19
pue
t,
auaS
aql
'a1e1drua1
se
vNq tJ
prpuerls
-a13urs
rlll1v\
.lsoq
slr
Jo
leql
o1 lelr{urs
sr
lpql
luarra
8u[urJd
VNU
ue
sesn
ruats.{s
tI
a:{J
'>lJoJ
uorteJlldal
aql
Surlrnrlsuor
ur
luelrodrur
Jq
plnoJ
uorlJeJalur
slqt lJsereru,{1od
y51q
tJ
rql qtun
xaldruor
e
suroy
TgdB
aql'atntusqns
touupJ
gSS
4or.E
rql
uor.lPrltoau
vNC
gI
ulldvHl
JSnEJJq
'ulatord
papor-a8eqd
aql armbar
dlelr
-;nads
^deru
1ro;
uorlerrldar
7J
Jqt
Io
d,rlaruoa8
Jr{J
'pJzrrJlJpreqJ
eq
or adz(t sll
Jo
elduexr
tsJIJ
Jql Se/!t
lJ
'Slunoup
JrrlJurorqJrols
uI
pepeJu
sr
qJrqm
'ulJlord
Surpurq puerls-a13urs
JArleJe
-door
Ilq8rq
e sr
(3gd8)
uralord
7g
aua8 aq1
'saua8
Iertuasse;o
dnorS
Ileus
p
Jo
sDn
-pord
aqr
uoJJ
palqruasse
ate8arSSe
aruz(zuar1
-lntu
p
Aq
paz,{1e1et
sl
yN(
7;;o
srsaqludg
'VNO
Jql
ur aursor{r1,{qlau
-dxorp,{q
aqt
SurtBlz(sorn13
ur
panlolur
asoqt
Surpnpur
'uorlerrldar
qtru pauJJ)
-uor
saua8
lerluassJuou
osle
eJp JrJef
o
'uorteperBap
VNq1
lsoq
qtlM pJuretuol
sarulzua
Jql
Jo
Jruos
pue
'ase8q
yN(
eql
'suorl
-run;.dro1e1n8ar
apnpur pelsaJJe
sr uJr{l
lnq
suels
srsaqluz(s
vN(I
qJIqM
ur ssoQf
r
'
uorletrld:.r
Jo
uotletlru I
Jql qlr,tr pJurJJuoJ
are
p:,{e1ap
sr
stsJql
-u,{s
y111q
Jo
lJsuo
Jqt
r{Jrr{M
ur esor{J
r
'
(
aursolzbl,{q1aru,(xorpdq
aqr
z(11eoadsa)
srosrntard
;o
uorsnord
aql
'lauluo
VNU
eql
a^oulel
ol dq1
aqlJo
aseq aql
lp
sa^eall
1r
uorlerqdar
ul,'de;,,
e
se
xaldnp
e uo4
pareldslp
sl
VN6
Jo
puplls
auo
uaqM
polpel)
arnllnrls
aq1
sazru6orat
lpql
ospaltnuopua/-oxa
ue
sl
lNll
{t"gi
EHnS:j
de6 s;ees
eseOr-1
I
poceloslp
st lou.lud
JOIIjUO
SgAeOlC
LUOJ
High fidelity replicases
Nuclear replication
350
kD tetramer
"
250 kD tetramer
"
350
kD
tetramer
Mitochondrial
200 kD dimer
replication
High
fidelity repair
Base
excision
repair
39 kD monomer
Low fidelity repair
Thymine dimer bypass heteromer
Base
damage
repair
monomer
Required in meiosis
monomer
Deletion
and
base substitution
monomer
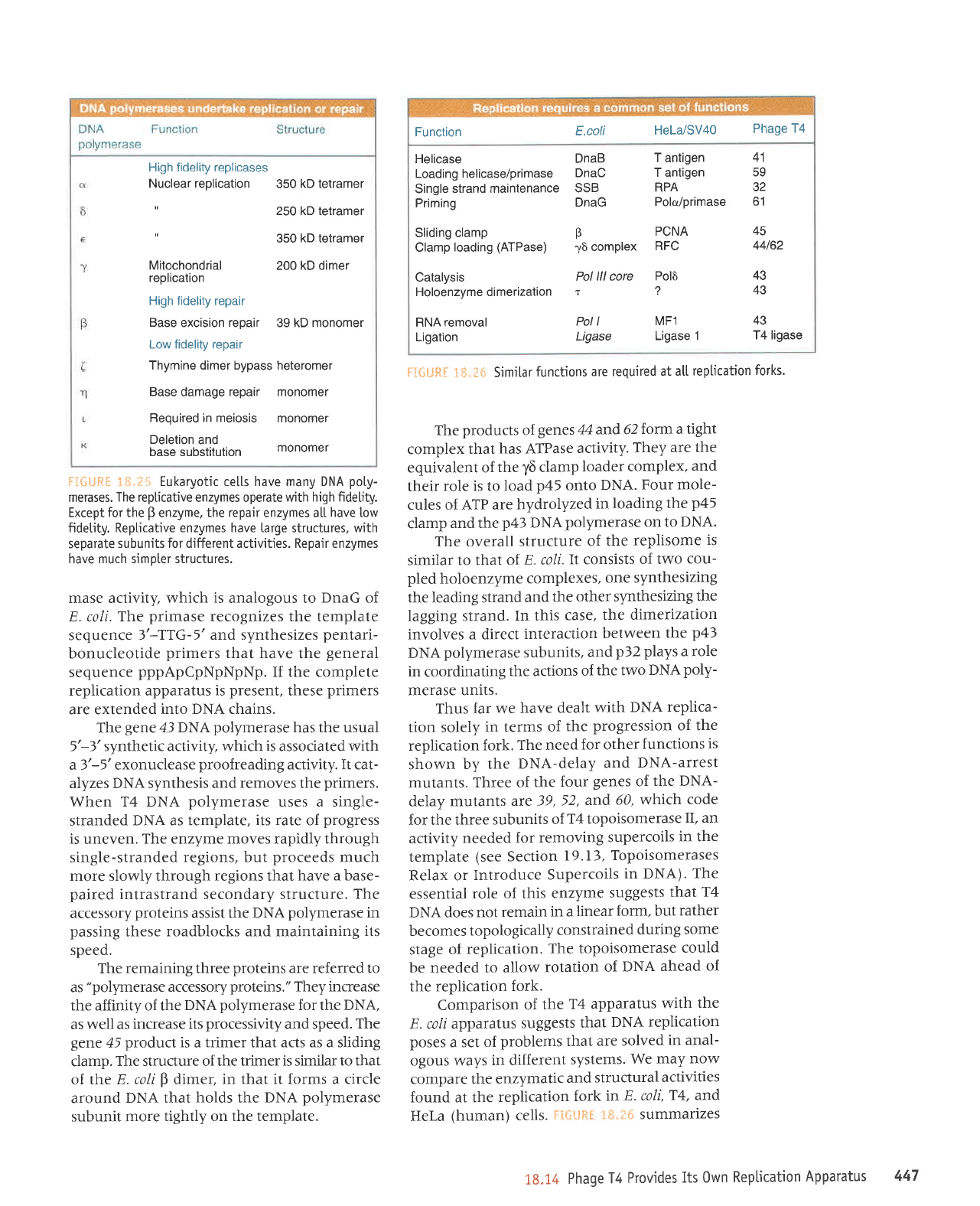
Function
E.coti
HeLa/SV40
Phage
T4
Helicase
DnaB
T antigen
41
Loading helicase/primase
DnaC
T antigen
59
Single
strand maintenance
SSB
RPA
32
Priming
DnaG
Polo/Primase
61
Sliding clamp
P
PCNA
45
Clamp
loading
(ATPase)
16
complex
RFC
44162
Catalysis
Pol lll core
Pol6
43
Holoenzymedimerization
r
?
43
RNA
removal
Pol I
MF1
43
Ligation
Ligase
Ligase
1
T4 ligase
f;IilLlRg
1$"fi* SjmiLar
functions
are required
at a[[
replication
forks'
FtSil*e 1**i5 Eukaryotic ce[[s have many DNA
poty-
merases.
The repticative
enzymes operate with
high fidetity.
Except
for
the
p
enzyme, the
repair
enzymes a[[
have
low
fidel.ity. Replicative enzymes have [arge
structures,
with
separate
subunits for different activities. Repair enzymes
have much simoter structures.
mase activity, which
is
analogous to
DnaG of
E. coli. The
primase
recognizes the template
sequence
3'-TTG-5' and synthesizes
pentari-
bonucleotide
primers
that have
the
general
sequence
pppApCpNpNpNp.
If the complete
replication apparatus
is
present,
these
primers
are extended
into DNA chains.
The
gene
43 DNA
polymerase
has the usual
5'-3'synthetic activity, which
is
associated with
a 3'-5' exonuclease
proofreading
activity. It cat-
alyzes DNA synthesis and
removes
the
primers.
When
T4 DNA
polymerase
uses a single-
stranded
DNA as template,
its rate
of
progress
is uneven.
The enzyme moves rapidly through
single-stranded
regions, but
proceeds
much
more slowly through regions that
have a base-
paired
intrastrand secondary structure.
The
accessory
proteins
assist the DNA
polymerase
in
passing
these
roadblocks and maintaining
its
speed.
The remaining three
proteins
are referred to
as
"polymerase
accessory
proteins."
They
increase
the affinity of the
DNA
polymerase
for the DNA,
as well as
increase its
processivity
and speed.
The
gene
45
product
is
a
trimer
that
acts as a sliding
clamp.
The structure of the trimer
is
similar
to that
of
the E. coli
$
dimer,
in that it forms a circle
around
DNA that
holds
the
DNA
polymerase
subunit more
tightly on the template.
The
products
ol
genes 44 and 62
form a tight
complex
that
has ATPase
activity.
They
are the
equivalent of
the
y6
clamp
loader complex,
and
their
role is to load
p45
onto
DNA.
Four mole-
cules of ATP are
hydrolyzed
in loading
the
p45
clamp
and the
p43
DNA
polymerase
on to DNA.
The overall structure
of
the replisome
is
similar to that
of.
E. coli.It
consists
of two
cou-
pled
holoenzyme
complexes,
one synthesizing
the leading strand
and
the
other slmthesizing
the
lagging strand.
In this
case,
the dimerization
involves a direct
interaction
between
the
p43
DNA
polymerase
subunits,
and
p32 plays
a role
in
coordinating
the actions
of
the two
DNA
poly-
merase units.
Thus far we
have
dealt
with
DNA
replica-
tion solely
in terms
of
the
progression of the
replication
fork.
The need
for other
functions
is
shown
by the
DNA-delay
and
DNA-arrest
mutants.
Three of
the
four
genes
of
the DNA-
delay
mutants
are 39,
52,
and
60, which
code
for the three
subunits
of
T4 topoisomerase
II, an
activity
needed for
removing
supercoils
in the
template
(see
Section
19.l),
Topoisomerases
Relax or
Introduce
Supercoils
in DNA).
The
essential
role of this
enzyme
suggests
that
T4
DNA does
not
remain
in a
linear
form, but
rather
becomes
topologically
constrained
during
some
stage
of replication.
The
topoisomerase
could
be needed to
allow
rotation
of
DNA
ahead
of
the
replication
fork.
Comparison
of
the
T4 apparatus
with
the
E. coli apparatus
suggests
that
DNA replication
poses
a set
of
problems that
are solved
in anal-
ogous ways
in different
systems.
We
may
now
compare the
enzymatic
and
structural
activities
found at the
replication
fork
in E.
coli,
T4, and
HeLa
(human)
cells.
Fe&Lift[
"I*.td
summarizes
18.14
Phage
T4
Provides
Its Own
Replication
Apparatus
447
the functions
and assigns
them to individual
proteins.
We
can interpret
the known
proper-
ties of replication
complex
proteins
in
terms of
similar functions
that involve
the unwinding,
priming.
catalytic, and
sealing reactions.
The
components
of each
system interact
in restricted
ways,
as shown
by the fact
that
phage
T4 requires
its
own helicase, primase,
clamp,
and so on, and
by the fact
that bacterial
proteins
cannot substi-
tute for
their
phage
counterparts.
@
Creating
the Reptication
Forks
at an Origin
r
Initiatjon
at onC requires
the sequentiaI
assembly
of a large
protein
comptex.
o
DnaA
binds to short repeated
sequences and forms
an otigomeric
comptex
that mel.ts DNA.
o
Six DnaC
monomers
bind each hexamer
of
DnaB.
and
this complex
binds to the
origin.
.
A hexamer
of
DnaB
forms
the reptication
fork.
Gyrase and
SSB are also required.
Starting
a cycle
of
replication
of duplex DNA
requires
several
successive
activities:
.
The
two strands
of DNA
must suffer
their
initial
separation.
This is, in
effect,
a
melting
reaction
over a
short region.
.
An
unwinding point
begins
to move
along
the DNA;
this marks
the
genera-
tion of the replication
fork,
which con-
tinues
to move
during
elongation.
.
The
first nucleotides
of the new
chain
must
be synthesized
into
the
primer.
This action
is required
once
for the lead-
ing
strand,
but is repeated
at the start
of
each
Okazaki fragment
on the lagging
strand.
Some events
that
are required
for initiation
therefore
occur
uniquely
at the
origin; others
recur
with the initiation
of each
Okazaki frae-
ment
during
the
elongation phase.
Plasmids
carrying
the E
coli oriC
sequence
have
been
used
to develop
a cell-free
system
for
replication.
Initiation
of replication
at oriC in
vitro
slarts
with
formation
of a complex
that
requires
six
proteins:
DnaA,
DnaB,
DnaC,
HU,
Gyrase,
and
SSB. Of the
six
proteins
involved
in prepriming,
DnaA
draws
our attention
as the
only one
uniquely
involved
in
initiation
vis-)-
vis
elongation.
DnaB/DnaC
provides
the
"engine"
of initiation
at
the origin.
The
first
stage in
complex
formation
is
bind-
ing
to
oriC
by DnaA
protein.
The reaction
CHAPTER
18
DNA
Reptication
involves
action at two types
of sequences:
9 bp
and l3
bp
repeats.
Together
the 9 bp
and l3 bp
repeats
define the limits
of the 245
bp minimal
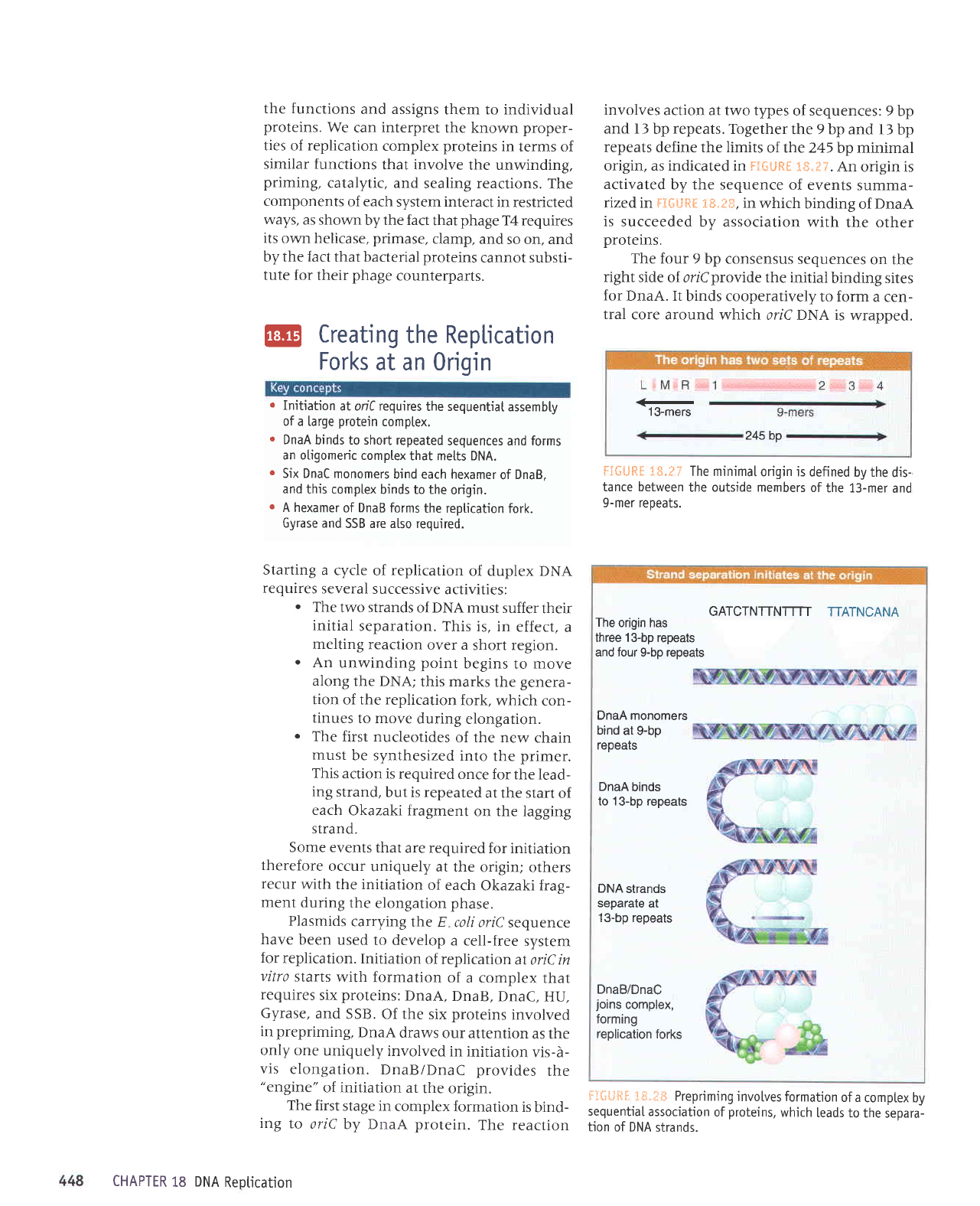
origin, as indicated in F?GtJftf,
3S.:T.
An
origin is
activated
by the sequence
of events
summa-
rized in fgGilft* 1S.iltr,
in which
binding
of
DnaA
is
succeeded by association
with
the other
proteins.
The four 9
bp consensus
sequences
on the
right
side of oriCprovide the
initial
binding sites
for
DnaA. It
binds cooperatively
to form
a cen-
tral
core around which
oriC DNA is
wrapped.
13-mers
245 bp
F€##ftf, i*.P?
The minimaI
origin is defined
by the
dis-
tance between
the outside members
of
the 13-mer
and
9-mer repeats.
f lfi{if,{il
3ii"ii* Prepriming
involves formation
of a
comptex by
sequentiaI
association
of
proteins,
which
leads
to the separa-
tion
of DNA
strands.
GATCTNTTNTTTT
TTATNCANA
The
origin has
three 13-bp repeats
and four 9-bp repeats
DnaA monomers
bind
at
g-bp
repears
DnaA
binds
to 13-bp repeats
DNA
strands
separate
at
13-bp repeats
DnaB/DnaC
joins
complex,
forming
replication
forks
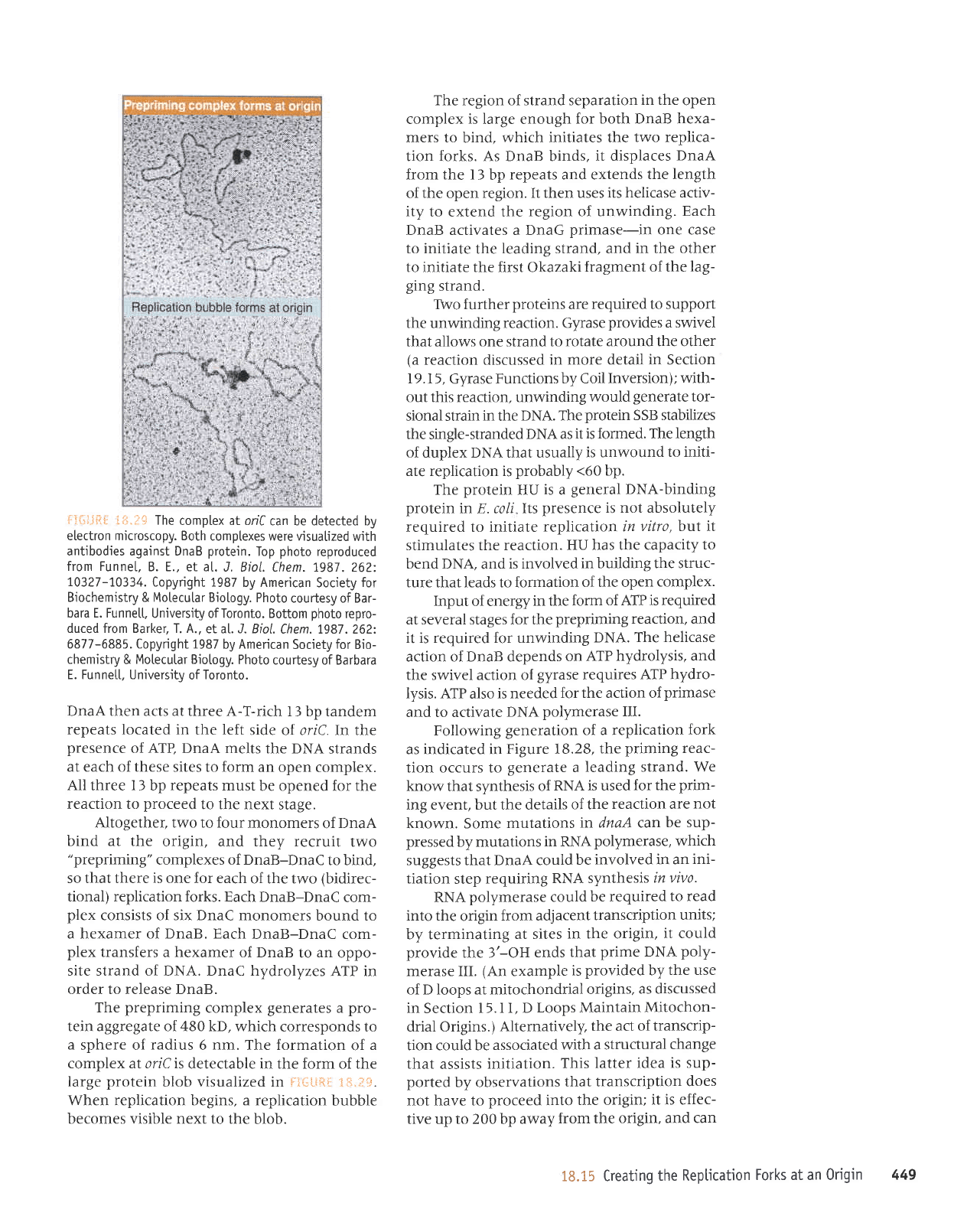
:::.{!LJfi{
ii:1.1+
The
complex at onC
can be detected by
etectron microscopy. Both
comptexes were visuatized
with
antibodies against DnaB
protein.
Top
photo
reproduced
from Funnet,
B. E., et al.
J. Biol. Chem.1987.262:
1.0327-70334.
Copyright 1987
by American
Society
for
Biochemistry & Motecu[ar
Biotogy. Photo
coudesy of Bar-
bara E. Funnett,
University ofToronto. Bottom
photo
repro-
duced from Barker, T. A.,
et at. J. Biol.
Chem. 7987.262:
6877-6885. Copyright 1987
by American
Society
for
Bio-
chemistry
&
Motecutar Biotogy.
Photo
courtesy of Barbara
E. Funne[t, University
of
Toronto.
DnaA then acts at three A-T-rich
l3 bp tandem
repeats located in
the left
side oI oriC.In the
presence
of ATP, DnaA melts
the DNA
strands
at each of these
sites to form an
open complex.
All
three
l3
bp
repeats
must
be opened for the
reaction to
proceed
to the next
stage.
Altogether, two to four
monomers
of
DnaA
bind at the
origin, and they recruit
two
"prepriming"
complexes of DnaB-DnaC
to bind,
so that there
is
one for each
of the two
(bidirec-
tional) replication forks.
Each DnaB-DnaC
com-
plex
consists of six DnaC monomers
bound to
a hexamer of DnaB. Each
DnaB-DnaC com-
plex
transfers
a
hexamer
of DnaB
to
an
oppo-
site strand of DNA. DnaC hydrolyzes
ATP in
order to release DnaB.
The
prepriming
complex
generates
a
pro-
tein aggregate of 480 kD,
which corresponds to
a
sphere of
radius
6
nm.
The formation
of
a
complex ar lric is
detectable in the form of the
large
protein
blob visualized in
F:*Liq{
?*.,?+.
When replication begins, a replication
bubble
becomes visible next to
the blob.
The region
of
strand separation
in the
open
complex is large enough
for both DnaB hexa-
mers to
bind,
which
initiates the
two replica-
tion forks. As DnaB binds,
it displaces
DnaA
from the l3
bp
repeats and extends
the length
of
the
open region.
It then uses
its helicase activ-
ity
to extend the
region of unwinding.
Each
DnaB activates a DnaG
primase-in
one case
to initiate
the
leading strand, and
in the other
to initiate the first Okazaki
fragment of the
lag-
ging
strand.
TWo further
proteins
are
required to support
the unwinding reaction.
Gyrase
provides
a swivel
that allows one strand to
rotate around the other
(a
reaction discussed
in more detail in Section
19.15,
Gyrase Functions
by Coil
Inversion);
with-
out this reaction, unwinding
would
generate
tor-
sional strain
in
the
DNA. The
protein
SSB stabilizes
the single-stranded
DNA as
it is formed. The length
of duplex DNA that usually
is unwound
to initi-
ate replication is
probably <60 bp.
The
protein
HU
is
a
general DNA-binding
protein
in E. coli
Its
presence is not absolutely
required to initiate
replication in
vitro, but ir
stimulates the reaction.
HU has the capacity
to
bend DNA,
and
is involved
in building the struc-
ture that leads to formation
of the open
complex.
Input
of energy
in the form
of ATP is required
at several stages for the
prepriming reaction, and
it is required for unwinding
DNA.
The helicase
action of DnaB depends
on
ATP hydrolysis, and
the swivel action of
gyrase
requires
AIP hydro-
lysis. ATP
also
is needed
for the action of
primase
and to activate
DNA
polymerase III.
Following
generation of a replication
fork
as indicated in
Figure 18.28,
the
priming reac-
tion occurs to
generate
a
leading strand. We
know
that synthesis
of
RNA is used
for the
prim-
ing event, but the details
of the
reaction are
not
known. Some
mutations
in dnaA can
be sup-
pressed
by mutations
in RNA
polymerase,
which
suggests that DnaA could
be
involved in an
ini-
tiation
step
requiring
RNA synthesis
invivo.
RNA
polymerase
could
be required
to read
into the
origin
from adjacent
transcription
units;
by terminating
at sites
in the origin,
it could
provide
the 3'-OH
ends
that
prime
DNA
poly-
merase III.
(An
example
is
provided
by
the use
of D loops at mitochondrial
origins,
as discussed
in
Section
l5.l l, D
Loops Maintain
Mitochon-
drial Origins.)
Alternatively,
the act of
transcrip-
tion could be
associated
with a structural
change
that
assists
initiation.
This
latter idea
is sup-
ported
by observations
that transcription
does
not have to
proceed
into
the origin;
it is effec-
tive up to 200 bp away
from
the origin,
and can
18.15 Creating
the Reptication
Forks at an 0rigin
449
use
either strand of DNA as template in
vitro.
The
transcriptional event is inversely related to
the requirement for supercoiling invitro, which
suggests that it
acts
by
changing the
local DNA
structure so as to aid
meltins
of DNA.
the events at lric, this does
not
necessarily imply
that the RNA
provides
a
primer
for the leading
strand.
Analogies between the systems
suggest
that
RNA
synthesis could be
involved
in
pro-
moting some structural change in the region.
Initiation requires the
products
of
phage
genes
O and.B as well
as
several
host
functions.
The
phage
O
protein
binds to the lambda ori-
gin;
the
phage
P
protein
interacts
with
the
O
protein
and with the bacterial
proteins.
The
ori-
gin
lies within
gene
O, so the
protein
acts close
to
its
site of synthesis.
Variants of the
phage
called Xdv consist
of
shorter
genomes
that carry all
the
information
needed
to
replicate,
but
lack infective
functions.
l,dv
DNA survives in the bacterium as
a
plas-
mid, and can be replicated in vitro
by a system
consisting of the
phage-coded proteins
O and
P
together with bacterial
replication
functions.
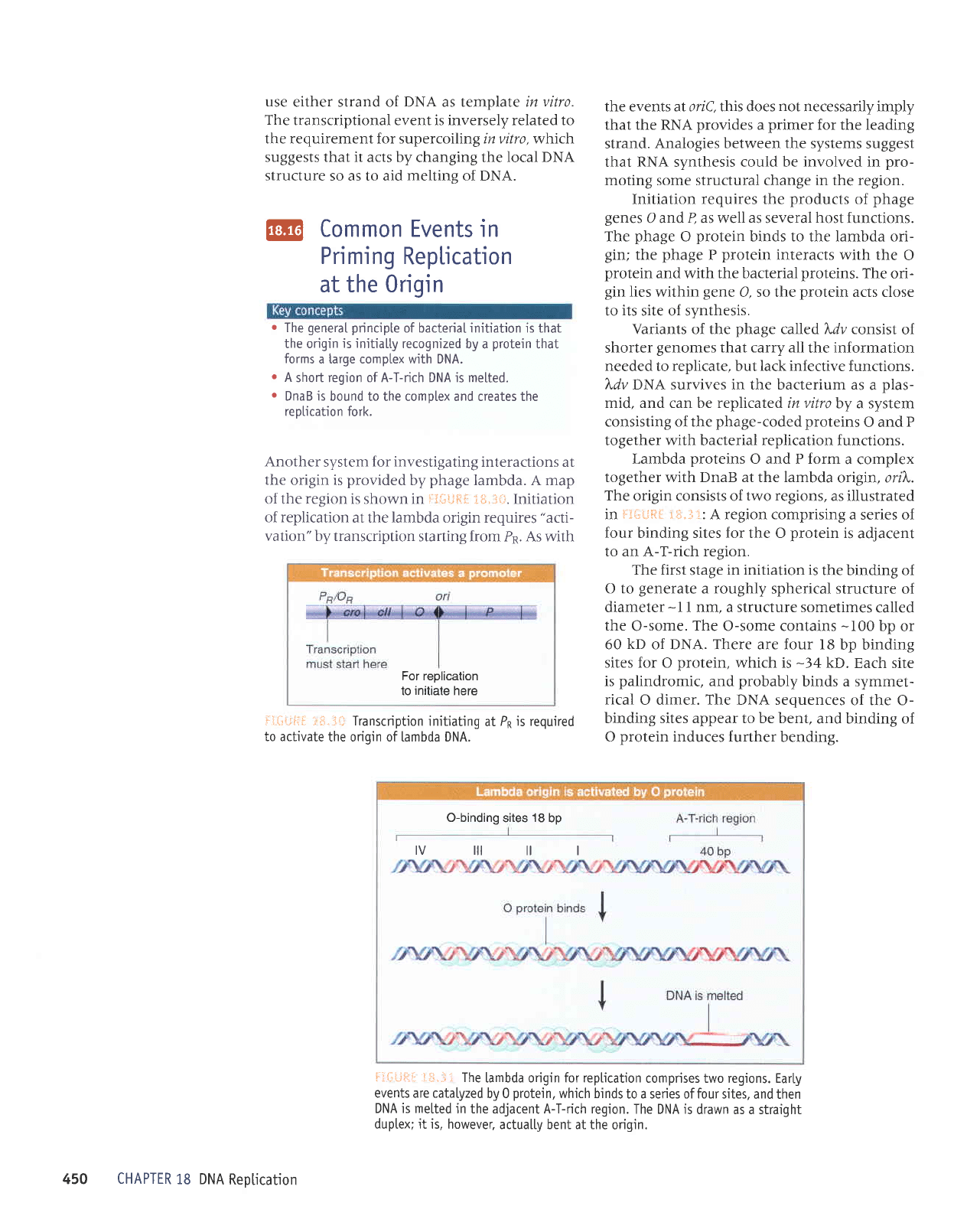
Lambda
proteins
O and P form
a complex
together with DnaB at the lambda
origin, ori)".
The
origin consists of
two regions,
as illustrated
in
F3*iiill
i*.-i1:
A region comprising
a series of
four
binding sites
for
the O
protein
is
adjacent
to an A-T-rich region.
The first
stage
in initiation is
the binding of
O to
generate
a
roughly
spherical structure
of
diameter
-l
I nm, a structure sometimes
called
the
O-some.
The
O-some contains
-100
bp
or
60
kD
of DNA. There are four I8
bp binding
sites for O
protein,
which
is
-34
kD. Each site
is
palindromic,
and
probably
binds a symmet-
rical
O dimer. The DNA sequences
of the O-
binding sites appear to be bent.
and binding of
O
protein
induces further
bending.
liit-;i;1;i J.*,-ii
The
[ambda origin for reptication
comprises two regions. Early
events are catalyzed
by 0
protein,
which binds
to a series of four sites,
and then
DNA is metted
in the adjacent A-T-rich region.
The DNA is
drawn as a straight
duptex; it is, however,
actua[[y bent at the
origin.
@
Common Events in
Priming
Reptication
at the 0rigin
.
The
generaI principte
of bacteriaI initiation is that
the origin
is initiaLty
recognized
by a
protein
that
forms
a large comptex with DNA.
o
A
short region of A-T-rich DNA is melted.
.
DnaB is
bound to the complex and creates the
reotication fork.
Another
system
for investigating
interactions at
the
origin is
provided
by
phage
lambda. A map
of the region is shown in
:rT*t-i*i:
:9. ii:.
Initiation
of replication
at the lambda origin requires
"acti-
vation" by transcription
starting from Pp. As with
For replication
to
initiate
here
i:i:r-ii:i
:iS.-ii"r Transcription
initiating
at
Pq is required
to activate
the origin of Lambda DNA.
O-binding
sites
18
bp
rvilt
[|
450 CHAPTER
18 DNA
Reolication
