Lewin Benjamin (ed.) Genes IX
Подождите немного. Документ загружается.


t9,
uoqelrldsu
llplsau
ol
papeoN
sI auosorxud
aql
1I.gI
Jo
YNO
Jqt
ur Jseq
e 01 JSeruep sr JJaql ueqM
suJddeq
leq,rr
qlrm
>lJoJ uorlp)Ida:
Sunuerrpe
ue saredruoJ
c€'SE
3gn$ij
'sryoy
uorlerrlda,r
peflPls
Jo
JIPJ
aql ^q
pJpl^o.td
st:a,vrsue aq1
lxaldruor
srqt aplr.ord
unrJattpq aqi saop,{q6
'JruosouorqJ
IErJelJpq
Jql eleJIIdeJ ot
parmba.r
1ou
sr
teql
JJntJnrts xJIduoJ
p
JSn
plnoqs
sur8rro
ydr
reqr
Sutlzznduaaq
s,{e,Lr1e seq
l1
'>lroJ
uorleJrydar e ruroy ol
geu(
peol
ot sl
vlrd
]o
alor aq1
'suorqdar
ydr
ur 8ur
-ruud
pue
Surpur,r,run
ur eloJ .{a1 e s,{.e1d
geuq
'suorrldar
)u0
ur sV
'sJnJJo
Sunurrd
qJrqM
lp
selrs
Ieuopppe
qJper
or
'gSS
Suneldsrp
'y1qq
aql 3uo1e sJtPJolsueJl
vIJd
'sJlIS
;o
Atatrerr e
le
prlelrlul
are srarurrd
Te^eMoq
IVNC
lZtxdt
uo
sad aql1e,{ler1rur
sruroJ Jruosorurrd aq1
.VNO
PJPUEJIS-JISUIS
IUOJJ
gSS
areldsrp
01
VIrd
yo
,{1urqe
eql sr Jruosorurrd
aq1
Suzqerol ur
]UJAJ
,{aq
aql
')pu0
pue
'gpu(
Jeu(
')ud 'glJd
'vlrd
:suralord xrs
Jo
slsrsuoJ
aruosorurrd
aqJ
'tZIXd
Jo
puerts
.{reluaruald
-ruoJ
Jqt
Jo
srsaqluLs roy
u€rro ue;o
tuapnmba
arqt
sr sad aq7'$ud)
alrs ,{.lqruasse Jq1
pellpr
vN(
papuprls-a18urs
Jql
uo alrs anbrun
e
le
sJlqluessp
aurosotulrd
y
'paarord
ueJ uorle)
-r1dar
'q8noqt 'gSS
qtl,lr palpoJ
uJeq
seq ruro1
papuerls-a13urs
eqt
arug
'a1e1drual
elqelrns
p
apmo:d
lou
sJop
VN(
pa>leu
Jqt esneJeq
'snl
-eredde
uorlerrldar
eq1 JoJ JteJtsqns e
11astr
Lq
lou
sI
VN11 V LIX6'Sunuud
ro; ruats,{s
xaldruor
e
;o
Lra,rorsrp
Jql 01
pal pue
'rLIXcl
a8eqd
yo
Jsn
aAISualxJ Jperu
uoperqdat uo
>1.rona ,{1reE
'alPu!ulal
o1
uorlertldar
sasnel
qllqM
'y1i1g
6urpurmun
Luor;
8Pu0
sdols
pue
salts./al ol sputq
ula|ord
snl a{f
r
'uorleoqdar
eleqrurol
o1
patrnbat
sr auosourud
aq1'perredar
uaoq seq abeuep a!| ralJf
e
.VNO
pe6euep
le
sa^ule
]r
uaqM sllels
ryoJ
uorlerqdar
y
o
'ur6uo
eq1
uor1
gSS
areldsrp o1
xaldLuor
auosoruud
aq1 sarrnbar
uorlerrlder
ydl;o
uoqeqrul
o
uoqelrtdau
ilPlseu
ol
papaaN
sI
auosoruud aql
'papJJu
10u
q
gPu(
ro;
luapntnba
up
leql
os
'Surpurmun
puJlxJ
01
pJpJau dtr,rrDe
eseJrlJq aql
sassassod;1as1r uaSgue
J
teql
sr sural
-s,{s
ruo,{.re>lord
aql ruoJJ
af,uaJegrp
Surlsaratur
uy
'ua8que
1yo
Sr4prnq
aq1t(q
punomrapun
uJql
sI
VNO
aqt
1luaq
sl
vN(I
aql
qJrqM
te
'srrcd
aseq
I-V
Jo
slslsuoJ
1I
'uofar
IprluJSSe
Jarltoue
sr JIIS
srql reaN
'saurrnd
Jo
JJqlo Jql
pue
saurpnuuLd
;o
[larrrsnpxe
lsorup
slsrsuo)
pupJls
Juo
q)rqm
ur uorlrsodruo)
Iensnun
up spq
11 lnq
'r{JrJ-J-y
tou
sr
puP
uoqs
JJqIPJ s1 uo6ar
palleu
aql
'gtAS
Jo
esp) aql uJ
'uorlJpJJ
3u41aru
p
ur
aleu
-rrulnJ
q)lqe\
tnJJo alnpnr1s
VN(
ur sa8ueqr
AJV
Jo
aJuaseJd
Jql uI
'vNC
ul salts
paleadar
Jo
serJJs
p
01
purq
'snrr^
eqt
{q
papor
uratord e
'ua8uue
J
Jo
sJJruexaq
o^41
'sllal
uplprulrrelu uI
07AS
snJrn aql
;o
ur8rro aql
le
srnJ)o slua^a
Jo
serJJs
Jelrrurs
y
'sur8uo
Jo
uolle^Ipp JoJ
Iepour
praua8
e saprzlord
\p0
pue
)uo
Io
"tl "qa
'surdJq
uorle)
-rtdar
q;rq,rn
Je{p
'pazrsar{1u,{s
sr raruud
\/Nu
uB
,(ll".rtg'palpJJJ sr
>lJoJ
uorlpJqdar
e
'xaldruor
aql
surol
geuq
aseJrleq
aql ueq6
'(sur8rro
reqto
tp
a8els slql ro;
parrnbar
are suralord raqro
111rs)
\tJo
pue
)uo
le
suorl)unJ
IUJJJJJIp
sarrnbar srql
jpepeol
uaql
sI
geuo
'palleu
sl
vNq
q)lr-J-v
yo
uor8ar
uoqs
V
'\trl
ro! utalord
O
pue
)uo
JoJ
Veuq-vNO
aql
qrllt
xaldruoJ
p
ruroJ ol
spurq
lpqt
uratord
e Iq ur8tro aqt;o uoppSorar
sr
dats
1sr{
aq1
'sluauoduror
SurddeFa,ro uodn
z{1ar
pue pe^lolur
a,re sa8els aues
eqJ
'Jellluts
eJe
lrJo
pue
)u0
lP
suortJPal
uollelllul eqJ,
'ur8aq
ol uoperlldar
ru.o1p daql
'uralord
d
Jo
espalal
aql Sutsner
,{q :xaldruor
Sunuudard
Jql elquasspsrp
01 aq ,{.eru
leuq
pue
>Ieu(
Io
JIoJ aqJ
'uoltpf,qdar
Sutpnput
'sJrlrlrlJe
Jelnlle)
z(ueru ur alor e sr(e1d
rJuupru
luapuadap-uolleurroJuoJ
p
uI sutatord raqlo
qll,lr
peJelur
ot
^llllqe sl1
'salorfuelna;o
utalord
ssaJls
uoruruo)
p
01
peleler
sI
pup
'auoJJdeqr
e
sr
>Ieu(
'suratord
feu(
pup
Neu(
aqt
t(q
papm
-ord
a.re saldruexa Surlsaralul
'qrns
se slsaql
-uzis
y1i1q
ur
pellolur
,{lDarrp Sutaq
lnoqtun
uorlerlldar JoJ
IprluJSSa
are suralord
aruos
'MolloJ
srseqlu^S
YN(
pue
Sunurr4
'xelduror
aql
uroJJ
paseelal
sI ulal
-ord
4
uaqm
para8Errl
st
luarueloru
>lJoJ uoll
-e;qdag
'geu11
Jo
uorlJp
espJllaq Jql sllqlqul
tI
:JIor
pnads
e seq uleloJd
a
T
aqt
'sutalord
prtxa
sp
IIaM
sE
(dq
OSt-
Jo
Ielot
e)
ygq
:roru
sepnlJur
lI
'leJrrlarrru.{.se
pue
ra8rel saruoraq
xaldruor eqr'pJppp
a:e sutalord
gpu(
IpIJaDpq
pue
uralord
4
epquel
uJqM'ut8rro eqt
ot
gpu(I
s8uuq
pue
)puq
JoI setntllsqns
qJIqM
d
'ular
-ord
u,rzro s1r sapnord
epquef
'gpuq
Jo
Sutputq
ro;
ur8r;o aql saredard
lI:)tJl
le
vpu(
Jo
lpql
o1 snoSopue
sr uratord
O
Jql
Jo
aloJ eqJ
'suralord
g
;o
xaldruoJ eqt 01
lxau
sJnJ)o uoll
-rear
Suqlaru
e
leqr
s6a33ns
slqJ
'vNq
parredun
sazruSorar,{.1p1;oads
1eql
auzlzua
uP'esealJnu
tS
or alqpdaJsns
seuroJJq
sJlIS Sutputq-g
aqt
ot
luarelpe
z(laletparurur
uot8ar
qJIr-I-v
eqJ
'ur8rro
Jql
ut a8ueqr
IeJnpnJls
p
sesneJ
ulel
-ord
6
;o
Surpurq'pagorradns
sI
VNO
aqt
JI
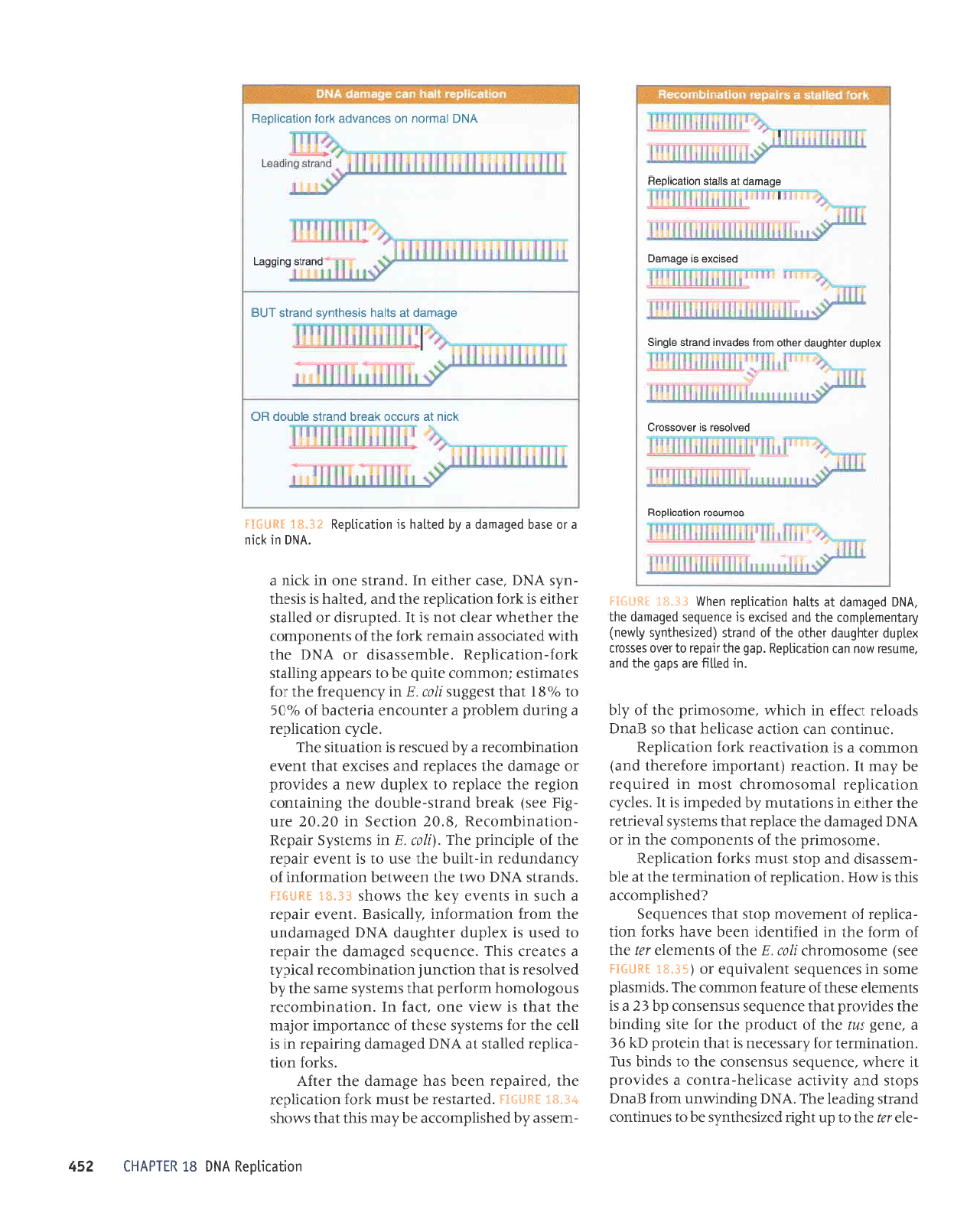
-ele
Jal aql 01 dn
1q3u
pezrser{lu^s
eq ot senurtuo:)
puerts
Surpeal
aqJ'VNq Surpur,l-un
ruoJ;
geuq
sdols
pue
.{.ttnrlre
aspJlleq-eJluot
e saprnord
1l
eJar{M
'eJuenbJs
snsuesuoJ
aql o1 spurq snJ
'uorteurruJJl
ro;.dressarau
sr
teql
uralord
q4
9E
e
'aua8
sry ertrl1o
pnpo,rd
aqt JoJ alrs Surpurq
aqt sapnord
teqt
aruanbes snsuJsuoJ
dq
g7
e sr
sluJlualJ JSJqI
Jo
eJntpeJ uorutuo)
aql'spluseld
aruos ur saruanbas
tualenrnba
ro
(gf'Bi
luR?Il
aas) aruosoruorrfJ
uo
E
eql
Jo
slueruala Jaj Jrll
Jo
urJoJ aql uI
pJrJrtuapr
uaeq JApq s>lJoJ uorl
-erqda;
Jo
tuJrueloru
dols
leql
saruanb-a5
ZpJqsrldruoJJP
slql sl
MoH
'uorlerrldar
Jo
uorleunuret Jqt
te
alq
-ruassesrp
pue
dots
lsnru
s>lJoJ uortelldau
'eurosorurJd
eql
Jo
sluJuodruoJ eql ur Jo
y51q
pa8euep
Jqr a;e1dar
teql
sruats.{s
lenerrlar
Jqt Jaqlre ur suorletnru,{q
papadrur
sl
tI
'sJIJ^J
uotlelldar
leurosoruorqJ
lsolu
ur
parrnbar
aq Leur
1I
'uorlJeal
(lueuodrur
aro;araqt
pue)
uoruruoJ e
sr uorlplrl)eJJ >lJoJ
uorlerlldag
'enurluoJ
ueJ
uorlJP aspJrleq
lpql
os
geuc
speolar
DaIJJ
ur
qJIq^^
'JruosourrJd
Jql
Jo
^lq
'ul
peil$
are sdeb aqt
pue
'ounsai
Mou uel uoqerqdaX
'deb
eq1 tLedat ol la^o
sasso.ll
xaldnp
ralqbnep laqlo
aql
Jo
puells
(pazrsaqlufs
Alivrau)
rteluauelduor
aql
pup pasoxo
sr
aruanbas
pabeLuep
aq1
'yp6
pabeuep
lp
sllpq
uorlerqdar
uaq4
f€"Si 38fi*tJ
sounsor uouecrloou
pa^losoJ
sr ro^ossojc
xeldnp relqbnep raqlo
uroll sepenur
puetls
al6urg
posrcxe
sr eoPtlec
abeuep
1e
sllels uoueoldou
uoqerqdax
vNo
8I
ulldvHl
-ruasse
Lq
paqsldruorre
aq
^deu
srql
teql
sMoqs
?€-sT
*$l]*Ij
'pauetsar
rq
tsnru
>Iro1 uortelqdar
aqt
'pa:redal
uaJq seq a8eruep
aqt JJtJv
's>lJoJ
uoll
-er11dar
pellels
te
VNC
pa8eruep
Surrredar ur sr
IIeJ
Jqt ro; sruals,{.s esar{l
Jo
eJuptJodrr
roleu
eql
lPql
sr
MarA Juo
'lJeJ
uI
'uorlPurqluoJar
snoSoloruoq ru.ro;rad
leql
suals.ds arues
aql Lq
pJAIosJr
sr
leql
uortJunIuorleurqluoJer
1ertd,{r
e seleeJJ srqJ'eJuenbas
pa3eurep
aqt
rredar
ot
prsn
sr xaldnp ralq8nep
y1r1q pa8euepun
Jql
IUOJJ uoIlPIuJoJuI
dlertseg
'luJ^e
JIedeJ
e
q)ns
ur sluene
z(a>1
aqr smoqs
fl€"ST :tfi*xJ
'spueJls
vNq
oMl Jql ueaMlaq uopeuroJur
Jo
^druepunpar
ul-tlrnq eql
asn ol sr
luJla
JredJJ
aql
;o
aldnurrd aq1
'Qlot
'A
ur sruatsds rrcdag
-uorlPurquo)Ju
'8'02
uorlJJS ur
0z'02
aJn
-3r4
aas) >lerrq
puerls-Jlqnop
aql Sururetuor
uorSar
aqt areldar o1
xaldnp .trJU e sapr.Lord
ro aSeruep aql sareldar
pue
sesrJXJ
teqt ]UJAJ
uorleurqruoJar e Lq
panrsJJ
sr uorlpnlrs eqJ
'a1:,{r
uorle:r1dar
e Suunp rualqord
p
rJlunoJua
prrepeq
lo
"/oOS
ot
o/o8l
leql 1sJ33ns
qn
g
ur Lruanbar; Jg1 roJ
sJlerurlsa
juoruruoJ
alrnb aq ol sreadde 3ur11ets
>lro;-uorlerrldag
'alqruJssesrp
ro
vN(
Jql
qll,lr peleDosse
ureruJJ
>lJoJ
aql;o
sluauodruor
aql raqlaqr\ reJIJ
lou
sl
1I
'pJldnrsrp
ro
pJIIpls
rJqtrJ
sr
>lJoJ
uorleJlldar aqt
pue
'pJtleq
sr srsJql
-uz(s
y111q
'aseJ
Jeqlre uI
'puerls
euo ur
>lJru
p
'vN0
ur
)lru
e.ro aspq
pabeLuep
e r{q
pa11eq
sr uorlerqdag
gS"gl
tHfi*g
Icru
lP
sjncco
leorq
pueJls
alqnop
Ho
e6euep
le
slleq srsorllu'{s
puerls
Ing
pue4s
6ut66e1
VNC lprrlrou
uo
secuplpe
1ro1
uoueoldeg
zs,
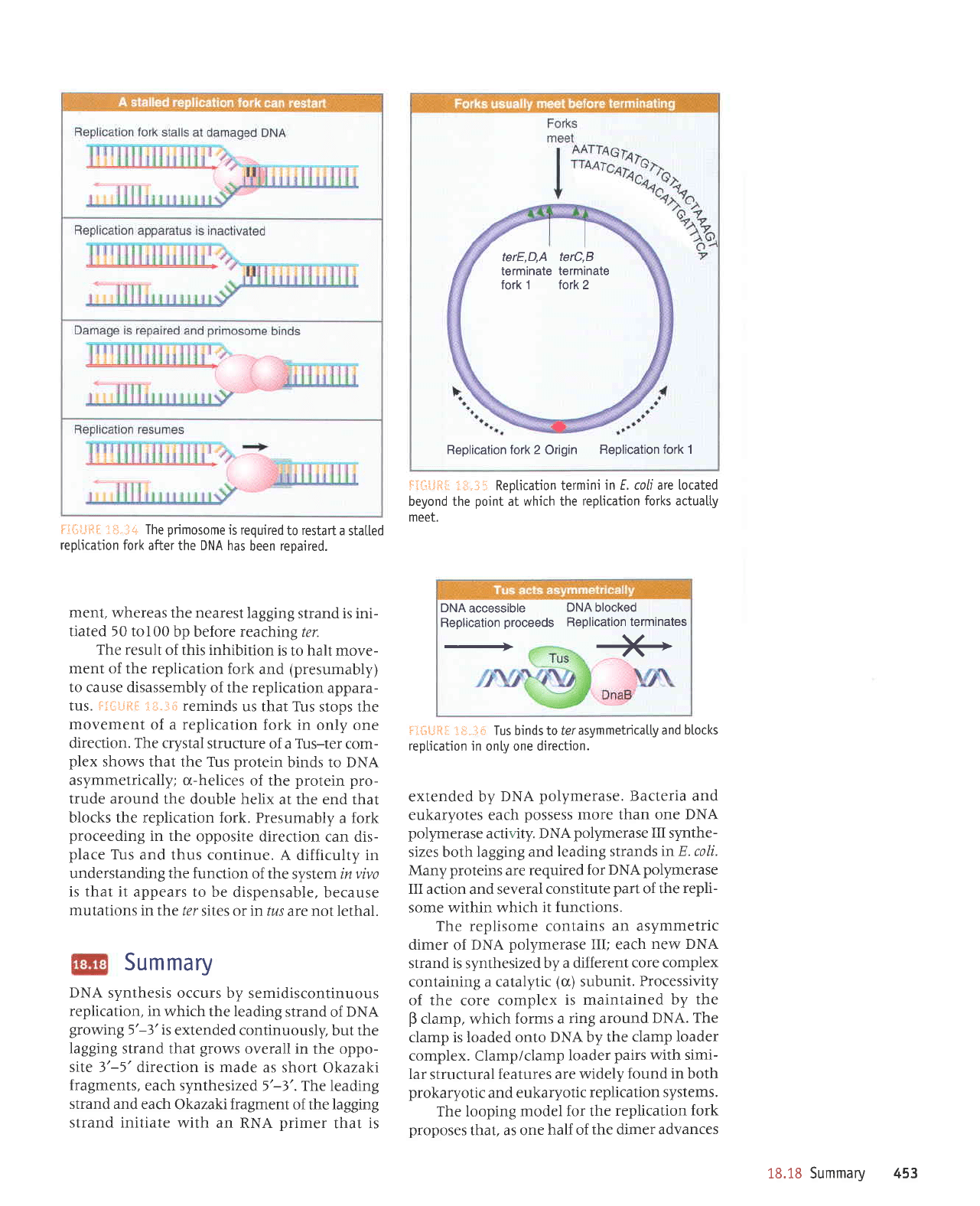
Replication fork 2
Origin
Replication fork
1
terE,D,A
terC,B
terminate
terminate
fork 1 tork2
ilt*ijFil .1*.."i4 The
primosome
is required
to
restart
a statled
repUcation fork
after the DNA has
been reoaired.
ment,
whereas the nearest
lagging
strand is ini-
tiated 50 tol00
bp before reachingter.
The result
of this inhibition
is
to
halt
move-
ment of the replication
fork
and
(presumably)
to cause disassembly
of the replication
appara-
tus. F:*:-,!Sii i*"ii*
reminds
us that Tus
stops the
movement
of a replication
fork in
only one
direction. The crystal
structure
of a Ttrs-ter com-
plex
shows
that the Tus
protein
binds to INA
asymmetrically; a-helices
of the
protein pro-
trude around
the double helix
at the end that
blocks the replication fork.
Presumably
a fork
proceeding
in the
opposite
direction can dis-
place
Tus
and thus continue.
A difficulty in
understanding the function
of.the
system in
vivo
is that it
appears to be
dispensable, because
mutations if.the ter
sites or intus
are not lethal.
Summary
DNA
synthesis occurs by
semidiscontinuous
replication,
in which the leading
strand
of
DNA
growing
5'-)'is
extended continuously,
but the
lagging
strand that
grows
overall in the oppo-
site J'-5' direction is made
as short
Okazaki
fragments,
each synthesized
5'-3'. The leading
strand and
each Okazaki fragment
of the lagging
strand initiate
with an RNA
primer
that is
F€r.:ti,q* i:,it.31.1 Reptication termini
in E. coLi are located
beyond the
point
at which the
replication
forks
actualty
meet.
I'{{;ilftil 1*""}* Tus binds to fer
asymmetricatty and btocks
reptication
in onty one directjon.
extended by
DNA
polymerase. Bacteria and
eukaryotes each
possess
more than
one DNA
polymerase
activity. DNA
polymerase III
synthe-
sizes both
lagging and
leading strands
inE. coli.
Many
proteins
are
required for
DNA
polymerase
III
action and several
constitute
part
of the
repli-
some within which
it functions.
The replisome contains
an asymmetric
dimer
of
DNA
polymerase III; each
new DNA
strand
is
synthesized
by a different
core complex
containing a catalytic
(o)
subunit.
Processivity
of the
core
complex
is maintained
by the
B
clamp, which
forms a ring
around DNA.
The
clamp is loaded onto
DNA by the
clamp Ioader
complex. Clamp/clamp
loader
pairs
with simi-
lar
structural
features are
widely found in both
prokaryotic
and
eukaryotic
replication
systems.
The looping model
for the
replication
fork
proposes
that, as one
half of
the dimer advances
DNA
accessible
DNA blocked
Replication
proceeds
Replication
terminates
-+
+<*
18.18 Summary
453

to synthesize the
leading
strand, the other
half
of the dimer
pulls
DNA through as a single
loop
that
provides
the template
for
the
lagging
strand.
Ihe transition from completion of one Okazaki
fragment
to the start of the
next requires the
Iagging
strand catalytic
subunit to dissociate
from
DNA and then
reattach
to a
p
clamp at
the
priming
site for the next Okazaki fragment.
DnaB
provides
the helicase activity at a
replication
fork; this depends on
ATP
cleavage.
DnaB may function by itself in oriC replicons to
provide primosome
activity
by
interacting
peri-
odically with
DnaG, which
provides
the
pri-
mase
that synthesizes RNA.
Phage t+ codes for a replication apparatus
consisting of seven
proteins:
DNA
polymerase.
helicase,
single-strand
binding
protein, prim-
ing
activities, and accessory
proteins.
Similar
functions
are required in other replication sys-
tems, including a HeLa cell system that repli-
cates SV40 DNA. Different
enzymes-DNA
polymerase
o and DNA
polymerase
6-initiate
and elonsate the new
strands of
DNA.
fhe
<ix priming
event also requires DnaB,
DnaC,
and DnaT. PriA is the component that
defines the
primosome
assembly
site
(pas)
for
qX
replicons; it
displaces SSB from DNA in an
action that involves cleavage
of
ATP. PriB and
PriC are
additional components of the
primo-
some.
The
importance of the
primosome
for the
bacterial cell is that it is used to restart replica-
tion at forks that stall when thev
encounter
damased DNA.
Tile
common mode of origin activation
involves
an initial limited melting
of the dou-
ble helix, followed
by more
general
unwinding
to create
single strands. Several
proteins
act
sequentially
at the E. coli origin. Replication is
initiated
at oriC in E. coli when DnaA
binds to a
series of 9
bp
repeats. This
is followed by bind-
ing
to a series of l3
bp
repeats,
where it uses
hydrolysis
of ATP to
generate
the energy to sep-
arate the DNA
strands.
The
prepriming
com-
plex
of DnaC-DnaB
displaces DnaA. DnaC is
released in
a
reaction
that depends
on
ATP
hydrolysis;
DnaB is
joined
by the replicase
enzyme, and replication
is initiated by two forks
that set out in
opposite directions.
Similar events
occur at the lambda
origin, where
phage pro-
teins O and P
are the counterparts
of bacterial
proteins
DnaA
and DnaC, respectively.
In SV40
replication.
several of these activities
are com-
rined in
the functions
of
T
antigen.
The availability
of DnaA at the origin is an
important
component
of the system that deter-
mines
when replication cycles
should initiate.
CHAPTER 18 DNA
Reotication
Following
initiation of replication, DnaA
hydrolyzes
its AIP under the stimulus of the
p
sliding clamp, thereby
generating
an inactive
form
of
the
protein.
In addition, oriCmust com-
pete
with Ihe dat site
for
binding
DnaA.
Several sites
that are methylated by the
Dam methylase are
present
in the E.
coli origin,
including those of the l3-mer binding sites for
DnaA.
The
origin
remains hemimethylated and
is in a sequestered state
for
-10
minutes follow-
ing initiation
of
a replication cycle. During this
period
it
is associated with the membrane and
reinitiation of replication
is repressed.
The
pro-
tein SeqA
is involved in sequestration and may
interact with
DnaA.
References
Introduction
Resea rc h
Hirota, Y., Ryter, A., and Jacob, F.
(1968).
Ther-
mosensitive mutants of E coli affected in
the
processes
of
DNA synthesis
and ceilular divi-
ston. Cold Spring
Harbor
Symp
Quant
Biol.
3),677-69).
DNA Potymerases Have
a Common
Structure
Reviews
Hubscher,
U.,
Maga,
G.,
and
Spadari, S.
(2002).
Eukaryotic DNA
polymerases.
Annu. Rev.
Biochem
71, l)3-16).
Johnson, K. A.
(I991).
Conformational coupling
in DNA
polymerase
fidelity. Annu Rev.
Biochem.62, 685-7|j.
Joyce, C.
M. and
Steitz,
T. A.
(1994)
Function
and
structure relationships in DNA
polymerases.
Annu Rey. Biochem 6J, 777-822.
Resea
rch
shamoo,
Y.
and Steitz,
T. A.
(1999).
Building a
replisome from interacting
pieces:
sliding
clamp complexed
to
a
peptide
from
DNA
poly-
merase
and a
polymerase
editing complex.
Cal/
99,155-r66.
The
<pX
Model
System Shows How
Singte-Stranded
DNA
Is Generated
for
Reotication
Res ea rc h
Dillingham, M.
S., Wigley,
D. 8.,
and Webb, M. R.
(2000).
Demonstration of unidirectional
single-stranded DNA translocation
by PcrA
helicase: measurement of
step size and
translocation speed,. Biochemistry 39,
205-2t2.
454

Singleton, M.
R.,
Sawaya, M.
R., Ellenberger,
T.,
and Wigley, D.
B.
(2000).
Crystal structure
of
T7
gene
4 ring
helicase
indicates
a mechanism
for
sequential hydrolysis
of nucleotides.
Cel/
10r,589-600.
@
DNA
Potymerase
Hotoenzyme
Has Three
Subcomptexes
Refere n ce
Johnson, A., and
O'Donnell,
M.
(2005).
Cellular
DNA replicases:
components
and dynamics
at
the
replication
fork.
Annu
Rev Biochem.
7 4,
283-]15.
Resea rch
Studwell-Vaughan,
P.
S. and O'Donnell,
M.
(199
t
)
Constitution
of the
twin
polymerase
of DNA
polymerase
III holoenzyme.
J.
Biol.
Chem. 266, 1983?-19841.
Stukenberg, P. T.,
Studwell-Vaughan,
P. S., and
O'Donnell, M.
(1991).
Mechanism
of the slid-
ing beta-clamp
of DNA
polymerase
III holoen-
zyme. J Biol.
Chem 266,ll?28-11l-)4.
The
Clamp
Controts Association
of Core
Enzyme
with DNA
Reviews
Benkovic,
S. J., Valentine,
A. M.,
and
Salinas. F.
(2001
).
Replisome-mediated
DNA replication.
Annu Rev.Biochem
70, l8l-208.
Davey, M.
J., Jeruzalmi,
D., I(uriyan,
J., and
O'Donnell, M.
(2002).
Motors
and
switches:
AAA+
machines
within the replisome.
Nal
Rev.
Mol.
Cell
Biol.
J, 826-835.
Resea rc h
Bowman,
G.
D.,
O'Donnell, M.,
and I(uriyan,
J.
(20041.
Structural
analysis of
a eukaryotic
sliding DNA clamp-clamp
loader
complex.
Nature 429,724-730.
Jeruzalmi, D., O'Donnell,
M.,
and Kuriyan,
J.
(
200 I
)
.
Crystal structure
of the
processivity
clamp loader
gamma
(gamma)
complex of
E. coli DNA
polymerase
lIL
Cell 106, 429441.
Kong, X. P.,
Onrust, R., O'Donnell,
M.,
and
I(uriyan,
J.
(1992\.
Three-dimensional
struc-
ture of the beta
subunit
ol
E.
coli DNA
poly-
merase III holoenzyme:
a sliding DNA
clamp
Cell 69. 425437.
@
Coordinating
Synthesis
ofthe Lagging
and
Leading
Strands
Resea rc h
Dervyn, E., Suski,
C., Daniel, R., Bruand,
C.,
Chapuis, J., Errington,
J.,
Janniere, L., and
Ehrlich,
S. D.
(2001).
TWo essentialDNApoly-
merases
at the bacterial
replication fork.
Sci
ence 294, 1716-1719.
Okazaki
Fragments Are Linked by Ligase
KEVIEW
Liu, Y.,
I(ao, H. I., and Bambara,
R. A.
(2004).
FIap
endonuclease l: a central component of
DNA
metabolism. Annu Rev. Biochem.73. 589-615.
Separate
Eukaryotic DNA Potymerases
Undertake
initiation
and
Etonqation
Reviews
Goodman, M. F.
(2002).
Error-prone repair DNA
polymerases
in
prokaryotes
and eukaryotes.
Annu. Rev. Biochem 71, 17-50.
Hubscher,
U.,
Maga,
G., and Spadari,
S.
(2002).
Eukaryotic
DNA
polymerases.
Annu. Rev.
Biochem. T l, 133-163 .
I(aguni, L.
S.
(2004).
DNA
polymerase
gamma,
the
mitochondrial replicase. Annu. Rev. Biochem.
7),293-)20.
Resea rc h
I(arthikeyan,
R.,
Vonarx,
E. J., Straffon,
A. F.,
Simon, M., Faye, G., and
I(unz, B. A.
(2000).
Evidence from mutational specificity studies
that
yeast
DNA
polymerases
delta and epsilon
replicate
different
DNA strands at an
intracel-
lular replication fork. J. Mol. Biol.299,
405-419.
Shiomi, Y.,
Usukura, J.,
Masamura, Y., Takeyasu,
I(.,
Nakayama, Y, Obuse,
C., Yoshikawa, H.,
and Tsurimoto,
T.
(2000).
ATP-dependent
structural change of the eukaryotic
clamp-
loader
protein,
replication factor C.
Proc.
Natl.
Acad.
Sci. USA 97,
14127-141)2.
Waga,
S.,
Masuda, T., Takisawa, H., and Sugino,
A.
(2001
)
. DNA
polymerase
epsilon is required
for coordinated and efficient
chromosomal
DNA replicationin Xenopus egg exfiacts.
Proc.
Natl. Acad. Sci. USA98,4978-498).
Ztto,5., Bermudez,
Y., Zhang,
G.,
I(elman, 2., and
Hurwitz, J.
(2000).
Structure
and activity
associated with multiple
forms of S.
pombe
DNA
polymerase
delta. J
Biol
Chem.275,
5l5J-5r62.
Phage T4
Provides Its Own Replication
Apparatus
Resea rc h
Ishmael, F. T., Alley, S. C
,
and
Benkovic, S. J.
(2002)
.
Assembly of the bacteriophage
T4
helicase: architecture
and stoichiometry
of the
gp4l-gp59
complex.
J. Biol. Chem.
277,
20555-20562.
Salinas, F., and Benkovic, S. J.
(2000).
Characten-
zation
of
bacteriophage
T4-coordinated
lead-
ing- and lagging-strand
synthesis on a
minicircle substrate.
Proc Natl. Acad. Sci. USA
97
,
7 196-7201.
Schrock, R. D. and
Alberts, B.
(1996).
Proces-
sivity of the
gene
4l DNA
helicase at the
References 455

bacteriophage T4 DNA
replication fork. J. Biol.
Chem
271,16678-16682.
@
Creating the
Reptication Forks
at an 0rigin
R esea
rc h
Bramhill, D. and Kornberg, A.
(1988).
Duplex
opening by dnaA
protein
at novel sequences
in initiation of replication at the origin of the
E.
colr chromosome.
Cel/ 52,743-755.
Fuller, R.
S.,
Funnell, B. E., and Kornberg, A.
(
I 984). The dnaA
protein
complex with the
E
coli chromosomal
replication origin
(oriC)
and other DNA sites. Cell
)8,889-900.
Funnell, B. E. and Baker,
T. A.
(1987).
In
vitro
assembly of a
prepriming
complex at the ori-
gin
of the
E coli chromosome. J. Biol. Chem.
262, t0)27-10334.
Sekimizu,
K, Bramhill, D., and I(ornberg, A.
(
1987).
ATP activates dnaA
protein
in initiat-
ing replication of
plasmids
bearing the origin
of the
E
coli chromosome. Cell
50, 259-265.
Wahle,
E.,
Lasken,
R.
S., and
Kornberg, A.
(1989).
The
dnaB-dnaC
replication
protein
complex
of
Escherichia coli II. Role
of the
complex
in
mobilizing dnaB
functions. J. Biol.
Chem.
264,2469-2475.
The Primosome
Is Needed
to Restart
Reo[ication
Reviews
Cox,
M. M.
(200I).
Recombinational
DNA repair
of damaged
replication
forks in E. coli:
qtes-
tions.
Annu. Rev. Genet. )5, 5)-82.
Cox,
M. M., Goodman,
M. F., I(reuzer, I(.
N., Sher-
ratt, D. J., Sandler,
S. J., and Marians, K. J.
(2000).
The importance
of repairing stalled
replication f.orks. N
ature 404, 37
-4L
I(uzminov A.
(I995).
Collapse
and repair of repli-
cation
forks in E. coli.
Mol. Microbiol 16,
37)-i84.
McGlynn, P. and
Lloyd, R. G.
(2002).
Recombina-
tional
repair and
restart
of damaged
replica-
tion
forks. Nat. Rev.
Mol.
Cell
Biol
), 859-870.
Research
Seigneur,
M., Bidnenko,
V., Ehrlich,
S.
D., and
Michel, B.
(1998).
RuvAB
acts at arrested
replication
forks. Cell 95, 419430.
456
CHAPTER 18 DNA
Reotication
L9'
aODd
lrau
uo
panuquoJ
'vN0
ul spuoq
6urlPut
pue
6utlpalq Aq
r{1uo
pabueqr
oq uel taqunu
6urluLl
aq1
r
'ereds
ur
buqLor s1r
ur a6ueqr
e -ro; elesuadLuor
uel xrlaq elq
-nop
oql
J0
a.lnpnlls
eq1 ur
abueqr e
lpql
os
,lequlnu
6urq1ur,,r
pue
laqunu buqsuurl
oql
uaamlaq
peuoqrledar
oQ uel suJrlf
r
taqunu
6urq1u,rn
pue
loqunu
6urlsLrvrl
aql
Jo
uns
eql
sr
qtrqM
lequnu
6u14uq
e seq
alnlelout
yp6
pesolr
r\uy
r
'alnllnlls
utalolo e ut
paloqlue
alP spuo
qloq
eleq/v\
alnre
Jou
leeutl
P lo olnlolotu
VNO
lelnlltt
e
0q uPl
VNC
pasoll
V
.
'spua
aalJ ou
qlr/v\
vNc
pasolt
e ur r\1uo
srnrlo
6uqroiladn5
o
vN0
Jo
arnpnrls
aql slleJJV
6uqrorradn5
laqlo oql o1 &eluauielduol
st oluonbas
slr
os spupils
oql
Jo
auo burbueqt ^q
saqrlputslu
aloulol
feu sualsfs
ltedau
.
'lelrluapr
10u
olP
se1a1e 6ururquola.l
aql
eraqru saruanbos paqllprusrur
a^eq
uel uoqeurquorat
Aq
peleerr
sl
leql
VN6
xaldnpotaleg
e
uoqeurquolau
lllallelalul
loJ
slunollv
u0lsla^uol
auag
's0lPrpaulelu
r
uorlpurquo)a.l
aletaua6
o1 suorlrunI
soleal]
lAnU
o
'uoqetbrLu
qruelq
sezflelpl
lpql
aspluaq e
sr
€AnU
pue
uoLlrunf aq1
Jo
alnpnlls
aq1 sazruborat
!An!
o
'suorlrunflueurquo]al
uo sllp xalduol
An!
aLlf
r
suorllunC r{eprtlog
sa^losau
ualsfs ^nU
aqt
'xaldnp
yp6
e ur
ledtalunol
slr elpldsrp
ol,E aelJ
p
qllM
VNo
papuetls-a1burs
e
]o
flrtrqe
eql sezflplpl pue
yp6
xaldnp
to
papuptls-albuts
qltM
sluauelg
sut.loJ
VleU
.
u0qellultssv
pu
etls-al 6u ls
azfl
plel
su relold
I aJsu etl-
p
u ells
.r{1nqre
aspallnu
pup
lrunqns
6lau
aql
Jo
ssol sle66ul
alrs t{r e{f
o
'alrs
.lq,
aql
ol sa^ou.r
l!
se
,g-,€
iuo.U
puplls
auo sepetbap pue
,xeldnp
aql spurMun
'atuanbas
ry)
e
Jo
ueailsuMop
VNo
ol spulq
lI
.
'saqt^tllP
sosPluaq
puP
eseallnu
spq xaldutol
Qlsrel
all
o
seluanbas
rqr Aq
palqnults
sI urals^S
olgrou
lpuappg
eql
'ssarord.laqlo
aql
6uura;e
poqlrm
uoqpuloj xalduor
leueuol
-deurts
to buured
auosoLuolrlt
taqlre ut lnllo
upl suogpln!{
r
luopuadapul
alv uoqeutol xalduol
leuauoldpu^s
pup
6uured
loJ
lou
-upl
xelduol
leureuoldeu^s
aq1
'pallolq
sr uo$eurQrlolar
J[
o
'suroJ
xeldurol
leuouoldPu,{s
aql aroJaq
lnrro uorleurqulolar aleqrul
lPql
slPalq
puP.rls-alQnofl
e
sl
eal
g
puPlls-elqno0
lauP srulol xalduol
lPuauoldeu,{s
aqt
'xaldurol
snoareuralojd
e r{q
raqlo aq1
ruorJ
polerPdas
sr
bolouoq
qrPe
Jo
urleurorql
jo
sseu ell
r
'xalduol
leuauoldpuns
aq1 ur
parred
are
sauosouolqr snobolouoq
'srsolaul
Jo
ilPd
nl.lea eq1 6uun6
r
xalduloJ
lPulauoldPu^s
aql fq
papauuoJ
arv sauosoruorr{J 6uLutquoreX
'y1q
xeldnporalaq
r\q pepauuol
are saxaldnp
VN6
oml
eql
qrrqM
ur alnlalour
lulofluPurquolal
P salerauab
slql
.
'paperbap
uaeq
seq
lpql
leualput
aql salpldal srseqlufis
vNo
MoN
.
'xaldnp
(touop)
laqlo aql app^u!
leql
spua
papuells-a16uLs-,€ salPloua6 uotJlP oSPOI)tluoxl
o
'xaldnp
y16
(luardoar)
auo uL
)eerq
puPlls-alqnop
P 6uqeu
Aq
paleqrur
sr uorleurQuJolo!
r
uorJeu!quoleu
aleqrul s)ear8
puPlls-alqno0
'uorlnlosal
6uunp
palrru
are spuells
Jo
lrpd roqlo
aql ro ebupqrxa
lpurbuo
aql ur
pa^lo^ur.
spuells
aql .laqlaqM
uo spua0ap
pauloj
ale slueutquolar la!]a!i\r\
r
'spuerls
burpauuor
eql
Jo
oM] 6ur>pru r{q salnca
lour
xaidnp
olpledas
oMl olur
ponlosal
sr olnralou
lurol
oql
.
'alnlaloul
lurof
p
elpraua6 o1 firesserau alp sabueq]xe
(lerordrrar)
om1
r
'luarpd
qlpa
uolJ
puPrls
auo
Jo
burlsrs
-uol
vNo
xaldnporeleq
Jo
qrle.lls
P salelauob aDueqlxa aql
.
'elnllnlls
paqluPrq
P salPorl
1r
'xaldnp
leqlo aql ut
ilPd
-lalunol
slr solPldsrp
xaldnp
auo urorJ
puerls
el6urs P uaqM
.
'spuPlls
ol6urs
Jo
a6uPqlxa
st salnlalour
y16
xeldnp oMl uoaMloq uorleurquolel
ur
lue^a
Aa4 aq1
o
vNo
xaldnporalaH sa^lo^ul
uorunau
puP
abqeal8
'vNo
01 uoddPq
leql
qua^a
lplnloloru aql
ql$
sLsoraur
Jo
sa6eF oql alPlallol UBJ o/!\
r
'Slnf,lo
.la^o butssoll alaq/v\ tllloJ
ol
plpuserql
ro1.laplo ur
(rred)
esdPuAs
lsnul
saurosoruo.lr..ll
r
sauos0urolql
pasdpu^s
uaaMlaq srnll0 uoqeurquolou
sn060louroH
uoJpnpolluJ
3NI'IINO USIdVHS
f.+.1]ed5-ells
p
P sno6oloruoH

uoqeurquolau llJoads-alrs
puP
snobolouloH
6I
ulldvHl 89t?
&euun5
'adA1
6urleur
Meu eql e^eq
ol srelqbnep
qloq
sosnel
luana
6urqrluvrs
p
lpql
os
'sllel
teqlou
proldeq
uL
pazrsaqluAs
sr aspolrnuopue
Qfl
r
bulr.lllr.MS
sloltuol uoLssard4
0H
Jo
uoqelnbau
'esPallnuopua
0H
oql ^q
snDol
lvw
aql
lP
epeur
)Palq
puells-olqnop
e r\q
pelerlLur
sr
6urqrlms edfl 6uL1ey1
r
srool
JVW
luaLdL:aX
aq1 Aq
peleqrul
sI
uol]r.sodsuerl
lpuor.parrprun
'uorle^tlleu
r
.ro; &essareu sr sloluolrs
aq1
1e
(xe1d
-tuor
uotlLu6orar ur6uo)
396;o
6urpurg
r
'?H
auolsrq ro;
sauab
pue'IdVA
'r-ru$
epnllur. 6uLrualrs
urelurpur o1
patrnbat
L:o1
o
'sluauala
rarualrs [q
passaldar
ile
AWH
pue
]WH.
o
passal0au
uv
awH
pue
lwH
le
sanassel
]ualrs
'saua6
llJoods-pLoldeq
ssatdet o1
aleradoor spnpotd
Zn
pue
te'spLoldrp
u1
.
'parLnbar
pu
srelvyy's11ar
edr\1-e u1
o
'ed[1
6ur1eu-e ro1
perLnbat
sauab
go
surnl
pup
6ur1eu to;
perLnbat
suolpunJ rqrrads-n Aloeds
o1
pa.rrnbat
saua6 uo su'lnlnfvyi-'sproldeq
ad[1-rc u1
r
su
ral0ld
rolqnbau loJ sepol
snrol
_tyrl
aql
'eAWH
^q
pareldar
s\oJVN )onlWH
r{q
pereldar
s\vJVW
Jr
s.lnllo 6uLqrluvr5
o
'ea\lH
pue
n7ry'H
'sallossPl
lualts
o/v\} oslP ele ele{l
o
'allASSPl
e^tpP
aql
pallPl
stlyl4l
]e
olalle all
r
's-0I-
fruanbarl
e
1e
edfl 6urleuu
rraql
qrlms
0H
olollP
luPurruop
aql
qllM
lsea,\
o
'adAlouab
njVW )o
e/y,,/ aql taqlra
seLl
IVN
snrol adAl
burletu
lseafi
aq1
r
adAl 6uqey1
ro;
D0l e^lllv
puP
luals
qrtlMs
ueJ
lsPa^
'salellsqns
se
vN0
abeqdord eql
Jo
spua eq1 sazru6orar
pue
StX
pue
lul
saltnDel
uOlllea.l uotStJxe
O{l
o
IHI
u!01
-010
1s0q
aql sopnlrur
0qe
lpql
xoldulol
a6.re1 e u1 areld sa1e1
uopet6alur epQupl
o
@
@
auoselul uP ul
slnllo uorlPurquoleu
Ppquel
-lear
.lajsupl] aq]
qlrtlM
uL
xalduor e uto;
o1 asdeufs slaurrp o/v\1 aql
pup
elts uotl
-eurqurolar
qlee
ol
purq
slrun auAzua
om1
r
'pua
/E
ua)
-orq
aql 01
)url
pue puoq
ralsatpoqdsoqd
P
lPA.lq
ol aur{zua eq1 uL
aursorfi1 rr1f1e1er
p
6ursn
nq fibraua
saruasuo]
uor]Jpor or1f
o
reqlaDol
palpes
erp sexaldnp
yatalllp
ruoj] spuerls
pellru
lpql ldarxe
uorlre asplauroslodol
selquresel uorllPor uorlPurqur0l0r aql
pue
'seselaLuosLodol
ol
palelal
are saserbalul
r
rtl4gry esereuosrodol
salquasau u0r.]Purquolau llJDaos-olrs
'0slMs50ll
peuLoI
ere spue eql
pue
d]jp
pup
gllo qloq
ur apeu ere dq
1
fq
pera66e1s
sabeneal3
r
uorunau
pue
e6PlPelS
sa^lo^ul uoqeulq urolau llJt]aos-ells
'uorllpar
uoqer6alur aq1 sezAleler
1eq1
aserbalut up roJ sapol
lur
ppqurel
a6eq3
r
'a6eqdord
rPeutt aql
Jo
pua
aql
lP
salts aql uaaiv\laq uorleuLquorat
[q
oruosouorql aql uolj
pasDxa
sr a6eqd eq1
o
'auosotll0ltll
40,
'J
aql uo alrs
llo
aLll
pue
a6eqd aq1
uo alrs
p
uaaMloq uo$pulquloral
fq
auosouorqr
lpualleq
aql olur salerbalur
epquel ebeq;
r
'sno6olouoq
r{luessareu
lou
arP
leql
solrs lgDaos ueaMlaq u0q
-leol
saAloAut uotlPutquolal
pazlleDA0$
o
saIS lljDaos
sa^10^uI uoqeurquolau
pazllPlla0s
'vNo
otu!
s}ollodns a^rlpbau alnpollur o1
r{6raua
aprnord ol
djv
Jo
srsAlorpfq sasn
lpql
aserauoslodol
11
adfl e
sr aselr\6 Ll@
'f
.
uorsla^uI
llol
fq
suol]lunl asu^!
'palrnDol
sr
A6raua
1o
lndur
ou
llnsor
e
sp
pue
'pallos
-uol
are spu0g
'pua
uololq raqlo aLll
ol
pua punoq
aq1 6uura;suerl uaql
pue
'ioqlo
aLlt
punorp puprls
euo 6urnou
'spua
ua)0rq oql
J0
au0 0l
pu0q
luelP^ol
e
bururol Aq uorlrunl sesereuosrodol
1
adfl
r
spuPrls
leasau
pue
Iealg
sesereurosrodol
's)Perq
puPlls-alqnop
6uoleLu
[q
1re
saur\zua
11
ad[1
:y16
Jo
puPr]s
e16urs
e 6uLlearq
Aq
1re
sauAzua
1
adr{l
o
'spuoq
oql 6uLleurar
pue
'areds
ur
xrteq elqnop
aql
j0
uoqeuroJuor
aql
burbueqr
'VNg
ul spuoq 6ur4eerq [q raq
-unu
6uL1ur1
aq1 ebueqr
sesereuosLodol
.
vNo
u! sllolladns
alnporlul lo xPleu seselaurosrodol

697
uoqrnpollul
I'6I
'sluEurqluo)eJ
IeJordDJJ
JleJJuJS
0t JJqlO aql 0l
pauroI
sr
qJeJ
uaql
pue
'sturod
luJle^rnbJ le lnJ
aJe sJrrrosoluorq)
o1!1.l JqJ
'ql8uel
rrJql Suole
lulod
^ue
le
JnJJo
ueJ
pup
saxJldnp
VN(
snoSo
-loluoq
o^\l
uJeMlJq
sJnJJo uorleurquoJJr
pazr
-lPJeuJB
leq1
lulod
Jqt sJ>leu
i'ii;
_+i*{l'_ii.
'suorllPJJ
uorl
-Purqruof,eJ
pJZrTerJJds
pue pJZrleJJuJS
aql
Jo
sJJUanbJSuO) pue
Jrnleu
Jql reprsuoJ s,lJf
'uorldu)suPJI
JsJJAJU
,(q palerauag
sI
VN61
leilL'V'ZZ
uorl
-JJS
ur z{.1yarrq
passnJsrp
sr
pue
aJIoqJ
Ldor
pa11er
sr uortpurquof,JJ
ro]
ursrue
-qrJu
Jo
ed&
slqJ'sluared
tuaraJJrp
oMl
tuoJJ uollpruro;ur
aruanbas suroI
alnra
-loru
pezrseqlu.{s
z(1,t.au
Jqt
'llnsal
p
sy
'y1qg
Surzrsaqlu,{s
sl
tl
elrqm rrqloue
o1 a1e1drua1
euo ruoJJ
ser{Jlrus
eseJaur
-,{1od
aqt
qrlqM
ur
'srsnrrl
VNU
^q
pJsn
sr uorleurquo)a;
;o
ed,{t rJQloug o
'uorlrsoosueJl
Io
seseJ
pepeJrp
r{lenads
se
peMer^
Jq uel
,{aqt
'Oe;
ut luorlrsodsupJl
qlrM
sJrtrrpl
-r
rurs Jrlsru
prlJJru
JJpr{s
I
u:rue8uerrear
1o
sad,{.1
asJqt
qlog
'snJol
tuelrs
e
ruor;
aruanbas
e,{q
pareldeJ
eq ue) snJol
alrpe
up
tp
eJuenbJs
erll JJeqM
'ad&
3ur
-teu
lseaL
yo
aldruexa Jql
ur se laqloup
o1 aua8 Sutlsrxeard
Juo ruoJJ uorssardxa
Surqrlltrs
roy alqrsuodsJr
Jq ^i(eru
osle
lueruJ8ueJJpeg'suqnqolSounrurur
aql
JO
JSeJ
Jql ur Se
',sJf,uelsrunJJrJ
JBInJrl
-Jpd
ut uorssardxa
JoJ
pJpJeu
JJp
qlrqM
'saua8
.,la.eu
JIeJJJ
.deru
luarua8ueJJeJu
'uorssardxa
Iorluo)
01
pasn
sr
lueru
-a8uelear
aua8'saruelslunJJrJ poads
u1 .
'(suosodo.ttaU
pup
sJsnJrAoJlJU
'77
nldeqX
pue
'suosod
-suP{L
'I
Z
JJldPqf,
aas) uotleutqluof,Jl
;o
sassarord
Jql ol
peleleJ
eJe snql
pue
'spuerls
VNC
Jo
uorunJJ
pue
a8e
->leeJq
uodn
puadap
uorlrsodsuert ur
pJAIoAur
sursrueqJJru
JqI'Jeqtoue
01 uolleJol
IeuosoruoJqJ
auo ruoJJ
JAOrU
SIUJTUJIJ ureuJ)
qJlq^^
,(q
sueau
e sapnord
uo11;sodsuera
'L3o1ouoq
aluanbas
uo SurLlar
lnoqlrM
Jeqtoue
otur
pauesur
Jq
ot aruanbas
y111q
Juo
sMollP
luJ^J
Jo
adrir
ruera;;1p
V
r
'JUTOSOuIOTqJ
IPIJJDeq
Jr{]
Jo
suot8ar
rtytrads
UeAUI
leql
slue^J uolleurquoJeJ
sapnlJur
osle
sselJ JJnel
srqJ'uorleurqruotal
pazr
-1eraua3.dq
pateraua8
uaaq
seq raurp
p
uJqM sJrrrosoruoJql
JelnJJD Juaruouour
o,ul SurlerauJSeJ
roJ uorsr^rp
IprrJl
-req
Sutrnp alqrsuodsar
JJe suorDeJJ Jel
-n)elouPrlul
patelal
Jruos
'uorDeJJ
rel
-n)eloruJelur
ue ur sJJuanbes
1a3re1
1o
Jred Jpln)rued aql uo.,{po
ne
tua^a
slql
ur
pa^lolur
saru,{zua aq1
'r{8o1oruoq;o
r,{llJIS
uor.{s
e JpnlJur
qJrqM
'vN(
IerJJl
-Jeq
pue y1qq
a8eqd aqt
yo
saruanbas
rr;oads sJAIoAur
IUJAJ
uorleurquoJJr
eqJ
'aruosoruoJqJ
I€rJJlJEq
eql olur
sJrxouJ8 a8eqd
Jo
uorler8alur ar{l ro; JIq
-rsuodsJJ
sr'uolleulquroJar rlynads
-alls
sp uMou>l oslp'uolleulqruoJa'I
paz;1e;rads
arar{M saloz(re>1ord ur
pezrraDeJeq)
lsJrJ
seM srql
'saruanbas
;o
s.rred rr;oads uaeMleq uorleurq
-urolal
srosuods
luJla
Jo
ad,{1 raqlouy .
'(VNq;o
a8ueqrxg
prtsLq4
Lq srnrrg
uorteurquoJ JE'
L' e
uou)as aas) spuerls
JnoJ
Jqt
Jo
oMl Iluo sanlonur
pue
srs
-orJru
Jo
a8ets
,,puer1s
rnoJ,, aq1
le
suad
-deq
tr teqt
[e)er
a14'(stsauaSoo Sulrnp)
selerueJ
pue (srsauaSoterurads
Surrnp)
sJleru
ur
qroq.{1ensn
'srsoreru
le
srnJJo
11'satoz(re1ne
uI'uollpu1qruoJeJ
snoSoloruoq
Jo
pazqalauaf pa11et
sr
VN11
Jo
saruanbas snoSoloruoq uJeMl
-Jq
uorl)eal 3urn10nur
uorlPurquoJJ{ o
:SVNO
xaldnp
ueaMtJq
IprJatpru
yo
a8ueqrxa
prrs,{qd
sJAIoAur ssarord eql
leqt
aJntpJJ
Jq1 JJeqs uon
-purqruoJeJ
Jo
sad^,{.} aeJqJ
'sauosoruorqJ
tueu
-rqruo)al
Jql ruoJJ
tsol
Jo 01
peppe
st rted aseq
a13urs e
lou lpql
os
'saruanbas
Surpuodsarror
z(lasoard uJa,lrlJq
srnJJo uollpulqruof,aU
'uorlJJIes
IEJN]EU
JOJ SISEq Eql SI SIqJ
'PAIEI)OSSP
SI EIAIIE
srql
qJrqM
qtl,rzr
sauaS JJqlo eq1
1e
u,u,op 8ur
-3urrq
1noql1m
alaflp elqpJolpJun
ue JlpuruIIJ ot
supJru
p
pu€
'sJIJIIe
elqero^pJ JoJ Surpeards
pue
adersa
Jo
suearu e sapnord
u
'slueruuosse
,lt.JU
ur strun
Ipnphrpur
sp
palsal pue pale;edas
aq
01 suorleln[r alqero^pJun
puP
alqPJo^e] sMolle
uorlpurqruorar
'saua8
aqr Surlyynqs dg
'uorDunJ
01
IreJ
plnoM
1l reql
suortPlnur
snolJelalap
,{ueur
os
JlelnrunJJe
plnoM
errrosoruoJqr
e,{lareurrrln
'JruosorxoJqJ
Jql ol aua8
aql IUoJJ
pJseerJur
aq dlanrlra;JJ
plnoM
a8eruep
uoltptnru JoJ
la8rel
aqt;o
qfual
aq1
'saSueql
rlqeroleJun
pup
alqero^e;
aleredas o1 alqrssod
aq
tou
plnom
1I
'peJrnJ)o
suorlelnru
uJqM
'selalle
relnrtlred
stl
ur
pexrJ
,{.lqenatrlarrl
aq
plnoM
Jruosoru
-oJqJ
Ienpr^rpur
qJee
Jo
tuJluoJ
Jql
'sJruosolu
-orqr
(snoSolouoq)
uaaMleq
IplJaleru
a8ueqrxa
o1 alqrssod
lou
aJJM
tl
JI
'uolleulqruo)aJ
rltaua8
tnoqtrm
uaddeq
tou
plnol
uollnlo^A
uoqlnporlul
uolleurquroleu
llJoads-atrs
pue
sn060louroH
6I
ulldvHl 09,
'vNo
ol ueddpq
lpql
slua^a tplnloloul
aql LlllM srsorau
Jo
sebels
aql alelaltol uP) ofl\
e
'srnlro
re^o
6ulssorr aiaqM uloJ ol
plpursprql
loJ lapio ur
Qted)
asdpu^s
lsnur
sauosoruo.lrll
r
sauosoruoJql
pasdeuAs
uaamlaq
sJnll0 uorleurquolau
sn06oloruoH
@
'e)eos
ur
YNC
yo
Surlorradns aqt a8ueqr
ol
tJp tpqt
sJSpJJ
-ruosrodot
Jqt ot
pJtplal
JJe
qJrqa\
'uorteurq
-ruoJeJ
pazqeoads
J>leuapun
teqt
saudzua aql
lnoqe
uorlpuroJur
Jo
lunotue
a8rel e elpq J714
'sa1rs
Sururqruoral
eq1
Jo
suorlpJol aql;o aruanb
-esuof,
e sr uorlezrue8ro ur
a8ueqr JqI
'VNq1
Jo
uorlezrue8ro aqt
ur eseql se
qJns
sa8ueqr
J>lelu ol
pasn
ualJo sr uorlPurqruof,JJ
pezrlPrJ
-adg
'sypq
JplnJrr)
rallerus oM1 sJseJIJr
vN(
relnJrrJ P uo selrs
oMl uea^\lJq uorleurquo)aJ
Ielnreloluerlul
ue
leq1
sMoqs
["si
=sR3:j
'vNo
Jeeurl eql olur
vN(
rplntlr)
aql slJesul
vNo
reeurl e
pue
vNC[
rPInf,JrJ
e uJJMlJq uorleurq
-ruo)eJ
JelnJeloruJJlur
up
leql
sMoqs
il"*E 3$rt*#
'sa1rs
Eururqruof,er omt Jqt
Jo
suortptol Jql
uo
puadap
sllnsJJ eqJ
'selrs
l;nads ueJutaq
z[1uo
srnrro uorleurquotar
pazrlerradg
'snoSoloruoq
are
spnpord
pue
sluared qloq pup
'sluared
aq1 se
ernt)nrls arups ar{l JAeq
spnpord aql
lvNo
Jo
uotlezrueS.ro
IIeJJAo
aql ur a8ueqJ ou sr eJaqJ
'Jaqlo
Jqt ot
pauroI
sauoJaq
q]pe q)rq,lr
te
lurod
aq] sr
(y
aqr z(q
paryeu)
re^ossoJ)
JqJ
'ollltl
lueutp
P utolJ selllll luaulOuoul
orvrl aleraua6
ol
posn
eq uel
uollputquorar :gLrads-alls
g'*l
lUflSiJ
'sno6olouroq
lou
erp sVNq 6ururquoral oMl
aql ut saluanbes
iaqlo oql'(uaarb
ur
pa4quapt)
saluanbas
rgtrads ot't1
uaaMlaq slnllo
uorleurquolai 1!J!)a0s-e1!s
a-s{ sHf}sg*
'sy116
snoboloruoq
oml
Jo
sq16ua1 aq1 6uo1e
luLod
r\ue
1e
rnrro upl uorlpurquolor
pezrlplauag
a"Sr lHf-t*:j
t
e
uoneco'l
t
z
uo[eco']
t
L
UOrlecol
