King M.R., Mody N.A. Numerical and Statistical Methods for Bioengineering: Applications in MATLAB
Подождите немного. Документ загружается.

a few diff erent values of α. Choose α
max
such that zero and α
max
bracket the
minimum along the direction of the search. Perform this exercise using the
starting point (8, 2) and use your chosen value of α
max
for all iterations.
(b) Write a function m-file that performs the multidimensional minimization using
steepest ascent. You can use the steepestparabolicinterpolation func-
tion listed in Program 8.10 to perform the one-dimensional minimization.
(c) Perform three iterations of steepest ascent (maxloops = 3). Use the following
tolerances: tolerance for α ¼ 0:0001; tolerance for krf k¼ 0:01.
8.5. Friction coefficient of a n ellipsoid moving through a fluid Many microorganisms,
such as bacteria, have an elongated, ellipsoidal shape. The friction coefficient ξ
relates the force on a particle and its velocity when moving through a viscous
fluid – thus, it is an important parameter in centrifugation assays. A lower friction
coefficient indicates that a particle moves “easier” through a fluid. The friction
coefficient for an ellipsoid moving in the direction of its long axis, as shown in
Figure P8.2, is given by
ξ ¼
4πμa
lnð2a=bÞ1=2
;
where μ is the fluid viscosity (for water, this is equal to 1 μg/(s μm)) (Saltzman, 2001).
For a short axis of b =1μm, find the value of a that corresponds to a minimum in the
friction coefficient in water. Start with an initial guess of a = 1.5 μm, and show the
first three iterations.
(a) Use the golden section search method to search for the optimum.
(b) Check your answer by using Newton’s method to find the optimum.
8.6. Nonlinear regression of chemical reaction data An experimenter examines the
reaction kinetics of an NaOH:phenolphthale in reaction. The extent of reaction is
measured using a colorimeter, where the absorbance is proportional to the concen-
tration of the phenolphthalein. The purpose is to measure the reaction rate constant
k at different temperatures and NaOH concentrations to learn about the reaction
mechanism. In the experiment, the absorbances A
i
are measured at times t
i
. The
absorbance should fit an exponential rate law:
A ¼ C
1
þ C
2
e
kt
;
where C
1
, C
2
, and k are unknow n constants.
(a) Write a MATLAB function that can be fed into the built-in function
fminsearch to perform a nonlinear regression for the unknown con stants.
(b) Now suppose that you obtain a solution, but you suspect that the fminsearch
routine may be getting stuck in a local minimum that is not the most accurate
solution. How would you test this?
8.7. Nonlinear regression of microfluidic velocity data In the entrance region of a micro-
fluidic channel, some distance is required for the flow to adjust from upstream
conditions to the fully developed flow pattern. This distance depends on the flow
conditions upstream and on the Reynolds number (Re ¼ Dvρ=μ), which is a
Figure P8.2
b
a
537
8.7 Problems

dimensionless group that characterizes the relative importance of inertial fluid forces
to viscous fluid forces. For a uniform velocity profile at the channel entrance, the
computed length in laminar flow (entrance length relative to hydraulic diameter =
L
ent
/D
h
) required for the centerline velocity to reach 99% of its fully developed value
is given by
L
ent
D
h
¼ a exp b ReðÞþc Re þ d:
Suppose that you have used micro-particle image velocimetry (μ-PIV) to measure
this entrance length as a function of Reynolds numb er in your microfluidic system,
and would like to perform a nonlinear regres sion on the data to determine the four
model parameters a, b, c,andd. You woul d like to perform nonlinear regression
using the method of steepest descent.
(a) What is the objective function to be minimized?
(b) Find the gradient vector.
(c) Describe the mini mization procedure you will use to find the optimal values of a,
b, c,andd.
8.8. Using hemoglobin as a blood substitute: hemoglobin–oxygen binding Find the
best-fit values for the Hill parameters P
50
and n in Equation 2.27.
(a) Use the fminsearch function to find optimal values that correspond to the
least-squared error.
(b) Compare your optimal values with those obtained us ing linear regression of the
transformed equation.
(c) Use the bootstrap method to calculate the covariance matrix of the model
parameters. Create ten data sets from the original data set. Calculate the stand-
ard deviation for each parameter.
References
Boal, J. H., Plessinger, M. A., van den Reydt, C., and Miller, R. K. (1997) Pharmacokinetic
and Toxicity Studies of AZT (Zidovudine) Following Perfusion of Human Term
Placenta for 14 Hours. Toxicol. Appl. Pharmacol., 143, 13–21.
Edgar, T. F., and Himmelblau, D. M. (1988) Optimization of Chemical Processes (New York:
McGraw-Hill).
Fournier, R. L. (2007) Basic Transport Phenomena in Biomedical Engineering (New York:
Taylor & Francis).
Nauman, B. E. (2002) Chemical Reactor Design, Optimization, and Scaleup (New York:
McGraw-Hill).
Rao, S. S. (2002) Applied Numerical Methods for Engineers and Scientists (Upper Saddle
River, NJ: Prentice Hall).
Saltzman, W. M. (2001) Drug Delivery: Engineering Principles for Drug Therapy (New York:
Oxford University Press).
538
Nonlinear model regression and optimization

9 Basic algorithms of bioinformatics
9.1 Introduction
The primary goal of the field of bioinformatics is to examine the biologically impor-
tant information that is stored, used, and transferred by living things, and how this
information acts to control the chemical environment within living organisms. This
work has led to technological successes such as the rapid development of potent
HIV-1 proteinase inhibitors, the development of new hybrid seeds or genetic varia-
tions for impr oved agriculture, and even to new understanding of pre-historical
patterns of human migration. Before discussing useful algorithms for bioinformatic
analysis, we must first cover some preliminary concepts that will allow us to speak a
common language.
Deoxyribonucleic acid (DNA) is the genetic material t hat is passed down from
parent to offspring. DNA is stored in the nuclei of cells and forms a complete set of
instructions for the growth, development, and functioning of a living organism. As
a macromolecule, DNA can contain a vast amount of information through a
specific sequence of bases: guanine (G), adenine (A), thymine (T), and cytosine
(C). Each base is attached to a phosphate group and a deoxyribose sugar to form a
nucleotide unit. The four different nucleotides are then strung into long polynu-
cleotide chains, which comprise genes that are thousands of bases long, and
ultimately into chromosomes. In humans, the molecule of DNA in a single chro-
mosome ranges from 50 × 10
6
nucleotide pairs in the smallest chromosome, up to
250 × 10
6
in the largest. If stretched end-to-end, these DNA chains would extend
1.7 cm to 8.5 cm, respectively! The directional orientation of the DNA sequence
has great biological significance i n the duplication and translation processes. DNA
sequences proceed from the 5
0
carbon phosphate group end t o the 3
0
carbon of the
deoxyribose sugar end.
Aspresentedinalandmark1953paper,WatsonandCrickdiscovered,from
X-ray crystallographic data combined with theoretical model building, that the
four bases of DNA undergo complementary hydrogen binding with t he appro-
priate base on a neighboring strand, which causes complementary sequences of
DNA to assemble into a double helical structure. T he four bases, i.e. the basic
building blocks of DNA, are classified as either purines (A and G) or pyrimidines
(T and C), which comprise either a double or single ring structure (respectively) of
carbon, nitrogen, hydrogen, and oxygen. A purine can form hydrogen bonds only
with its complementary pyrimidine base pair. Specifically, when in the proper
orientation and separated by a distance of 11 A
˚
, guanine (G) will share three
hydrogen bonds with cytosine (C), whereas adenine (A) will share t wo hydrogen
bonds with thymine (T). This phenomenon is called complementary base
pairing. The two paired strands that form the D NA double helix run anti-parallel
to each other, i.e. the 5
0
end of one strand faces the 3
0
end of the complementary

strand. One DNA chain can serve as a template for the assembly of a second,
complementary DNA strand, which is precisely how the genome is replicated
prior to cell division. Complementary DNA will spontaneously pair up (or
“hybridize”) at room or body temperature, and can be caused to separate or
“melt” at elevated temperature, which was later exploited in the widely used
polymerase chain reaction (PCR) m ethod of DNA amplification in the
laboratory.
So how does the genome, encoded in the sequen ce of DNA, become the thousands
of proteins within the cell that interact and participate in chemical reactions? It has
been determined that each gene corresponds in general to one protein. The process
by which the genes encoded in DNA are made into the myriad of proteins found in
the cell is referred to as the “central dogma” of molecular biology. Information
contained in DNA is “transcribed” into the single-stranded polynucleotide ribonu-
cleic acid (RNA) by RNA polymerases. The nucleotide sequences in DNA are
copied directly into RNA, except for thymine (T), which is substituted with uracil
(U, a pyrimidine) in the new RNA molec ule. The RNA gene sequences are then
“translated” into the amino acid sequences of different proteins by protein/RNA
complexes known as ribosomes. Groups of three nucleotides, each called a “codon”
(N =4
3
= 64 combinations), encode the 20 different amino acids that are the
building blocks of proteins. Table 9.1 shows the 64 codons and the corresponding
amino acid or start/stop signal.
The three stop codons, UAA, UAG, and UGA, which instruct ribosomes to
terminate the translation, have been given the names “ochre,” “amber,” and
“opal,” respectively. The AUG codon, encoding for methionine, represents the
translation start signal. Not all of the nucleotides in DNA encode for proteins,
however. Large stretches of the genome, called introns, are spliced out of the
sequence by enzyme complexes which recognize the proper splicing signals, and
the remaining exons are joined together to form the protein-encoding portions.
Major sequence repositories are curated by the National Center for Biotechnology
Information (NCBI) in the United States, the European Molecular Biology
Laboratory (EMBL), and the DNA Data Bank of Japan.
9.2 Sequence alignment and database searches
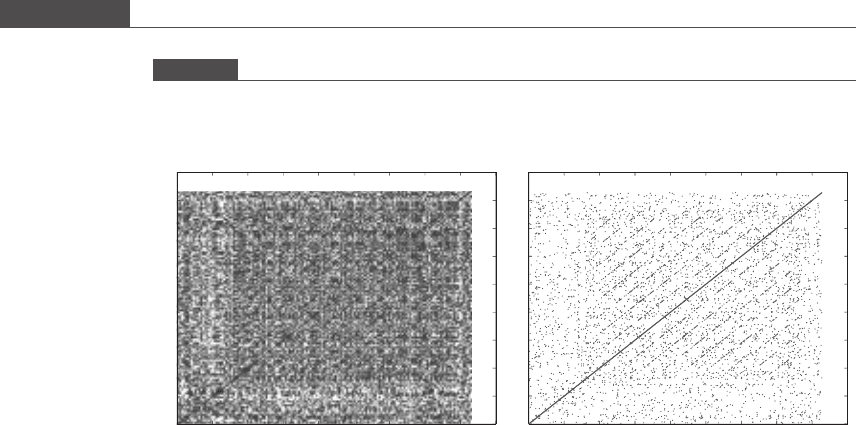
The simplest method for identifying regions of similarity between two sequences
is to produce a graphical dot plot. A two-dimensional graph is generated, and a
dot is placed at each position where the two compared sequences are identical.
The word size can be specified, as in MATL AB program 9.1, to reduce the
noisiness produced by many very short (length = 1–2) regions of similarity.
Identity runs along the main diagonal, and common subsequences including
possible translocations are seen as shorter off-diagonal lines. Inversions,wherea
region of the gene runs in the opposite direction, appear as lines perpendicular to
the main diagonal, while deletions appear as interruptions in the lines. Figure 9.1
shows the sequence of human P-selectin, an adhesion protein important in
inflammation, compared against itself. In the P-selectin secondary structure,
it is known that nine consensus repeat domains
1
exist, and these can be seen as
1
A consensus repeat domain is a sequence of amino acids that occurs with high frequency in a
polypeptide.
540
Basic algorithms of bioinformatics
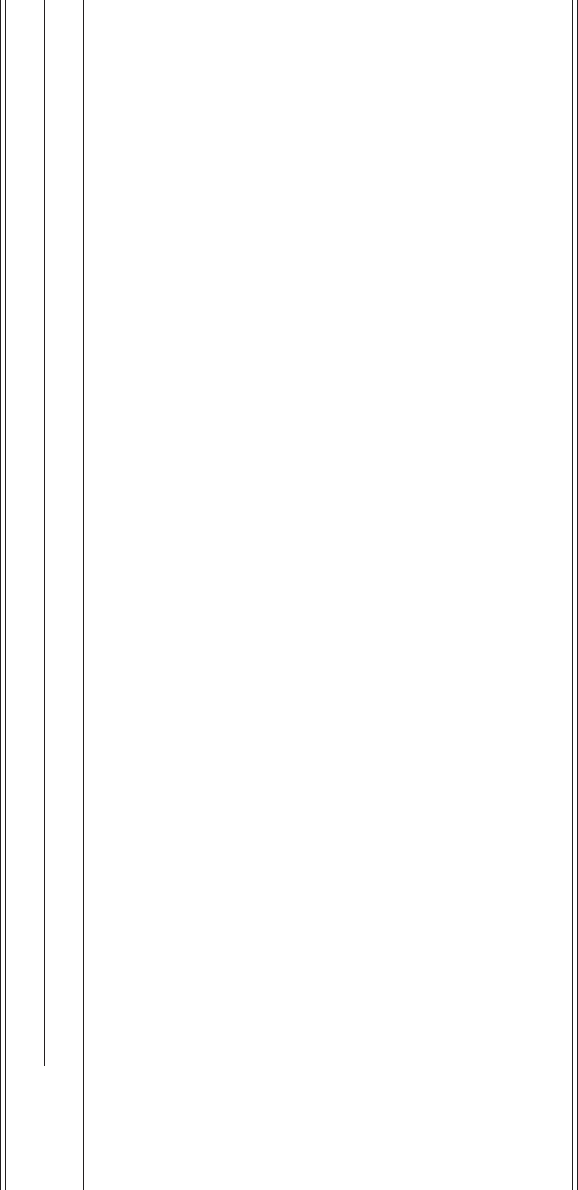
Table 9.1. Genetic code for translation from mRNA to amino acids
2nd base
UCAG
1st base
U UUU: phenylalanine (Phe/F)
UUC: phenylalanine (Phe/F)
UUA: leucine (Leu/L)
UUG: leucine (Leu/L)
UCU: serine (Ser/S)
UCC: serine (Ser/S)
UCA: serine (Ser/S)
UCG: serine (Ser/S)
UAU: tyrosine (Tyr/Y)
UAC: tyrosine (Tyr/Y)
UAA: stop (“ochre”)
UAG: stop (“amber”)
UGU: cysteine (Cys/C)
UGC: cysteine (Cys/C)
UGA: stop (“opal”)
UGG: tryptophan (Trp/W)
C CUU: leucine (Leu/L)
CUC: leucine (Leu/L)
CUA: leucine (Leu/L)
CUG: leucine (Leu/L)
CCU: proline (Pro/P)
CCC: proline (Pro/P)
CCA: proline (Pro/P)
CCG: proline (Pro/P)
CAU: histidine (His/H)
CAC: histidine (His/H)
CAA: glutamine (Gln/Q)
CAG: glutamine (Gln/Q)
CGU: arginine (Arg/R)
CGC: arginine (Arg/R)
CGA: arginine (Arg/R)
CGG: arginine (Arg/R)
A AUU: isoleucine (Ile/I)
AUC: isoleucine (Ile/I)
AUA: isoleucine (Ile/I)
AUG: start, methionine (Met/M)
ACU: threonine (Thr/T)
ACC: threonine (Thr/T)
ACA: threonine (Thr/T)
ACG: threonine (Thr/T)
AAU: asparagine (Asn/N)
AAC: asparagine (Asn/N)
AAA: lysine (Lys/K)
AAG: lysine (Lys/K)
AGU: serine (Ser/S)
AGC: serine (Ser/S)
AGA: arginine (Arg/R)
AGG: arginine (Arg/R)
G GUU: valine (Val/V)
GUC: valine (Val/V)
GUA: valine (Val/V)
GUG: valine (Val/V)
GCU: alanine (Ala/A)
GCC: alanine (Ala/A)
GCA: alanine (Ala/A)
GCG: alanine (Ala/A)
GAU: aspartic acid (Asp/D)
GAC: aspartic acid (Asp/D)
GAA: glutamic acid (Glu/E)
GAG: glutamic acid (Glu/E)
GGU: glycine (Gly/G)
GGC: glycine (Gly/G)
GGA: glycine (Gly/G)
GGG: glycine (Gly/G)

off-diagonal parallel lines in Figure 9.1(b) once the spurious dots are filtered out
by specifying a word size of 3. Website tools are availabl e (European Molecular
Biology Open Software Suite, EMBOSS) to make more sophisticated dot plot
comparisons.
Box 9.1 Sequence formats
When retrieving genetic sequences from online databases for analysis using commonly available web
tools, it is important to work in specific sequence formats. One of the most common genetic sequence
formats is called Fasta. Let’s find the amino acid sequence for ADAM metallopeptidase domain 17
(ADAM17), a transmembrane protein responsible for the rapid cleavage of L-selectin from the leukocyte
surface (see Problem 1.14 for a brief introduction to selectin-mediated binding of flowing cells to the
vascular endothelium), in the Fasta format. First we go to the Entrez cross-database search page at the
NCBI website (www.ncbi.nlm.nih.gov/sites/gquery), a powerful starting point for accessing the wealth
of genomic data available online. By clicking on the “Protein” link, we will restrict our search to the
protein sequence database. Entering “human ADAM17” in the search window produces 71 hits, with the
first being accession number AAI46659 for ADAM17 protein [Homo sapiens]. Clicking on the AAI46659
link takes us to a page containing information on this entry, including the journal citation in which the
sequence was first published. Going to the “Display” pulldown menu near the top of the page, we select
the “FASTA” option and the following text is displayed on the screen:
>gi|148922164|gb|AAI46659.1| ADAM17 protein [Homo sapiens]
MRQSLLFLTSVVPFVLAPRPPDDPGFGPHQRLEKLDSLLSDYDILSLSNI
QQHSVRKRDLQTSTHVETLLTFSALKRHFKLYLTSSTERFSQNF
KVVVVDGKNESEYTVKWQDFFTGHVVGEPDSRVLAHIRDDDVIIRI
NTDGAEYNIEPLWRFVNDTKDKRMLVYKSEDIKNVSRLQSPKVCGYLKVDN
EELLPKGLVDREPPEELVHRVKRRADPDPMKNTCKLLVVADHRF
YRYMGRGEESTTTNYLIHTDRAN
This is the amino acid sequence of human ADAM17, displayed in Fasta format. It is composed of a
greater than symbol, no space, then a description line, followed by a carriage return and the sequence in
single-letter code. The sequence can be cut and pasted from the browser window, or output to a text file.
We may also choose to output a specified range of amino acids rather than the entire sequence. If we
instead select the “GenPept” option from the Display pulldown menu (called “GenBank” format for DNA
sequences) then we see the original default listing for this accession number. At the bottom of the
screen, after the citation information, is the same ADAM17 sequence in the GenPept/GenBank format:
ORIGIN
1 mrqsllflts vvpfvlaprp pddpgfgphq rlekldslls dydilslsni
qqhsvrkrdl
61 qtsthvetll tfsalkrhfk lyltssterf sqnfkvvvvd gkneseytvk
wqdfftghvv
121 gepdsrvlah irdddviiri ntdgaeynie plwrfvndtk dkrmlvykse
diknvsrlqs
181 pkvcgylkvd neellpkglv dreppeelvh rvkrradpdp mkntckllvv
adhrfyrymg
241 rgeestttny lihtdran
//
Note the formatting differences between the GenBank and Fasta sequence formats. Other sequence
formats used for various applications include Raw, DDBJ, Ensembl, ASN.1, Graphics, and XML. Some
of these are output options on the NCBI website, and programs exist (READSEQ; SEQIO) to convert from
one sequence format to another.
542
Basic algorithms of bioinformatics
MATLAB program 9.1
% Pseldotplot.m
% This m-file compares a protein or DNA sequence with itself, and produces a
% dot plot to display sequence similarities.
% P-selectin protein amino acid sequence pasted directly into m-file
S=‘MANCQIAILYQRFQRVVFGISQLLCFSALISELTNQKEVAAWTYHYSTKAYSWNISRKYCQN
RYTDLVAIQNKNEIDYLNKVLPYYSSYYWIGIRKNNKTWTWVGTKKALTNEAENWADNEPNNK
RNNEDCVEIYIKSPSAPGKWNDEHCLKKKHALCYTASCQDMSCSKQGECLETIGNYTCSCYPG
FYGPECEYVRECGELELPQHVLMNCSHPLGNFSFNSQCSFHCTDGYQVNGPSKLECLASGIWT
NKPPQCLAAQCPPLKIPERGNMTCLHSAKAFQHQSSCSFSCEEGFALVGPEVVQCTASGVWTA
PAPVCKAVQCQHLEAPSEGTMDCVHPLTAFAYGSSCKFECQPGYRVRGLDMLRCIDSGHWSAP
LPTCEAISCEPLESPVHGSMDCSPSLRAFQYDTNCSFRCAEGFMLRGADIVRCDNLGQWTAPA
PVCQALQCQDLPVPNEARVNCSHPFGAFRYQSVCSFTCNEGLLLVGASVLQCLATGNWNSVPP
ECQAIPCTPLLSPQNGTMTCVQPLGSSSYKSTCQFICDEGYSLSGPERLDCTRSGRWTDSPPM
CEAIKCPELFAPEQGSLDCSDTRGEFNVGSTCHFSCDNGFKLEGPNNVECTTSGRWSATPPTC
KGIASLPTPGVQCPALTTPGQGTMYCRHHPGTFGFNTTCYFGCNAGFTLIGDSTLSCRPSGQW
TAVTPACRAVKCSELHVNKPIAMNCSNLWGNFSYGSICSFHCLEGQLLNGSAQTACQENGHWS
TTVPTCQAGPLTIQEALTYFGGAVASTIGLIMGGTLLALLRKRFRQKDDGKCPLNPHSHLGTY
GVFTNAAFDPSP’;
n=length(S);
figure(1)
% For first figure, individual residues in the sequence are compared
% against the same sequence. If the residue is a “match”, then a dot is
% plotted into that i, j position in figure 1.
Figure 9.1
Dot plot output from MATLAB program 9.1. The amino acid sequence of human adhesion protein P-selectin was
obtained from the NCBI Entrez Protein database (www.ncbi.nlm.nih.gov/sites/entrez?db=protein) and compared
against itself while recording single residue matches (a) or two out of three matches (b).
0
(a)
100 200 300 400 500 600 700 800 900
0
100
200
300
400
500
600
700
800
900
0
(b)
100 200 300 400 500 600 700 800 900
0
100
200
300
400
500
600
700
800
900
543
9.2 Sequence alignment and database searches

for i=1:n
for j=1:n
if S(i)==S(j)
plot(i,j,’.’,‘MarkerSize’,4)
hold on
end
end
end
figure(2)
% For the second figure, a more stringent matching condition is imposed. To
% produce a dot at position (i, j), an amino acid must match AND one of the
% next two residues must match as well.
for i=1:n-3
for j=1:n-3
if S(i)==S(j) & (S(i+1)==S(j+1) | S(i+2)==S(j+2))
plot(i,j,’.’,‘MarkerSize’,4)
hold on
end
end
end
The alignment of two or more sequences can reveal evolutionary relationships and
suggest functions for undescribed genes. For sequences that share a common ancestor,
there are three mechanisms that can account for a character difference at any position:
(1) a mutation replaces one letter for another; (2) an insertion adds one or more letters to
one of the two sequences; or (3) a deletion removes one or more letters from one of the
sequences. In sequence alignment algorithms, insertions and deletions are dealt with by
introducing a gap in the shorter sequence. To determine a numerically “optimal”
alignment between two sequences, one must first define a match score to reward an
aligned pair of identical residues, a mismatch score to penalize an aligned pair of non-
identical residues, and a gap penalty to weigh the placement of gaps at potential
insertion/deletion sites. Box 9.2 shows a simple example of the bookkeeping involved
in scoring proposed sequence alignments. The scoring of gaps can be further refined by
the introduction of origination penalties that penalize the initiation of a gap, and a
length penalty which grows as the length of a gap increases. The proper weighting of
these two scores is used to favor a single multi-residue insertion/deletion event over
multiple single-residue gaps as more likely from an evolutionary point of view.
Alignment algorithms need not weigh each potential residue mismatch as equally
likely. Scoring matrices can be used to incorporate the relative likelihood of different
base or amino acid substitutions based on mutation data. For instance, the tran-
sition transversion matrix (Table 9.2) penalizes an A↔GorC↔T substitution less
severely because transitions of one purine to another purine or one pyrimadine to
another pyrimadine (respectively) is deemed more likely than transversions in which
the basic chemical group is altered.
While common sequence alignment algorithms (e.g. BLAST) for nucleotides utilize
simple scoring matrices such as that in Table 9.2, amino acid sequence alignments utilize
more elaborate scoring matrices based on the relative frequency of substitution rates
observed experimentally. For instance, in the PAM (Point/percent Accepted Mutation)
matrix, one PAM unit represents an average of 1% change in all amino acid positions.
544
Basic algorithms of bioinformatics
ThePAM1matrixwasgeneratedfrom71protein sequence groups with at least 85%
identity (similarity) among the sequences. Mutation probabilities are converted to a log
odds scoring matrix (logarithm of the ratio of the likelihoods), and rescaled into integer
values. Similarly, the PAM250 matrix represents 250 substitutions in 100 amino acids.
Another commonly used scoring matrix is BLOSUM, or “BLOCK Substitution
Matrix.” BLOCKS is a database of about 3000 “blocks” or short, continuous multiple
alignments. The BLOSUM matrix is generated by again calculating the substitution
frequency (relative frequency of a mismatch) and converting this to a log odds score.
The BLOSUM62 matrix (Table 9.3) has 62% similarity, meaning that 62% of residues
Box 9.2 Short sequence alignment
The Kozak consensus sequence is a sequence which occurs on eukaryotic mRNA, consisting of the
consensus GCC, followed by a purine (adenine or guanine), three bases upstream of the start codon
AUG, and then followed by another G. This sequence plays a major role in the initiation of the translation
process. Let’s now consider potential alignments of the Kozak-like sequences in fluit fly, CAAAATG, and
the corresponding sequence in terrestrial plants, AACAATGGC.
2
Clearly, the best possible alignment
with no insertion/deletion gaps is:
CAAAATG
AACAATGGC (alignment 1)
which shows five nucleotide matches, one C–A mismatch and one A–C mismatch, for a total of two
mismatches. If we place one single-nucleotide gap in the fluit fly sequence to represent a “C” insertion
in the plant sequence, we get
CAA–AATG
AACAATGGC (alignment 2)
which shows six matches, no mismatches, and one gap of length 1. Alternatively, if we introduce a gap
of length 2 in the second sequence, we obtain the following alignment:
CAAAATG
AACA––ATGGC (alignment 3)
which results in five matches, no mismatches, and one gap of length 2. Note that we could have placed
this gap one position to the left or right and we would have obtained the same score. For a match score
of +1, a mismatch score of –1, and a gap penalty of −1, then the three proposed alignments produce
total scores of 3, 5, and 3, respectively. Thus, for this scoring system we conclude that alignment 2
represents the optimal alignment of these two sequences.
2
Note that we are considering the DNA equivalent of the mRNA sequence, thus the unmethylated
version T (thymine) appears instead of U (uracil).
Table 9.2. Transitio n transversion matrix
ACG T
A 1 −5 −1 −5
C −51−5 −1
G −1 −51−5
T −5 −1 −51
545
9.2 Sequence alignment and database searches
Table 9.3. BLOSUM62 scoring matrix
CSTPAGNDEQHRKMI LVFYW
C9−1 −1 −30−3 −3 −3 −4 −3 −3 −3 −3 −1 −1 −1 −1 −2 −2 −2
S −1 4 1 −1101000−1 −10−1 −2 −2 −2 −2 −2 −3
T −114 1 −1101000−10−1 −2 −2 −2 −2 −2 −3
P −3 −117 −1 −2 −1 −1 −1 −1 −2 −2 −1 −2 −3 −3 −2 −4 −3 −4
A 01−1 −1 4 0 −1 −2 −1 −1 −2 −1 −1 −1 −1 −1 −2 −2 −2 −3
G −301−206 −2 −1 −2 −2 −2 −2 −2 −3 −4 −40−3 −3 −2
N −310−2 −206 100−100−2 −3 −3 −3 −3 −2 −4
D −301−1 −2 −116 20−1 −2 −1 −3 −3 −4 −3 −3 −3 −4
E −400−1 −1 −2025 2001−2 −3 −3 −3 −3 −2 −3
Q −300−1 −1 −20025 0110−3 −2 −2 −3 −1 −2
H −3 −10−2 −2 −211008 0 −1 −2 −3 −3 −2 −12−2
R −3 −1 −1 −2 −1 −20−20105 2 −1 −3 −2 −3 −3 −2 −3
K −300−1 −1 −20−111−125 −1 −3 −2 −3 −3 −2 −3
M −1 −1 −1 −2 −1 −3 −2 −3 −20−2 −1 −1 5 12−20−1 −1
I −1 −2 −2 −3 −1 −4 −3 −3 −3 −3 −3 −3 −314 210−1 −3
L −1 −2 −2 −3 −1 −4 −3 −4 −3 −2 −3 −2 −2224 30−1 −2
V −1 −2 −2 −20−3 −3 −3 −2 −2 −3 −3 −21314 −1 −1 −3
F −2 −2 −2 −4 −2 −3 −3 −3 −3 −3 −1 −3 −3000−1 6 31
Y −2 −2 −2 −3 −2 −3 −2 −3 −2 −12−2 −2 −1 −1 −1 −137 2
W −2 −3 −3 −4 −3 −2 −4 −4 −3 −2 −2 −3 −3 −1 −3 −2 −31211
