Iozzo Renato V. Proteoglycan Protocols
Подождите немного. Документ загружается.

418 Forsten and Nugent
3. Method
3.1. Experimental Protocol
1. Label and arrange 0.5-mL microcentrifuge tubes—1 per sample, 3 per condition.
2. Prepare incubation buffer—Buffer may be made in advance and filter-sterilized for later
use. No degassing is needed.
3. Add the needed incubation buffer to each vial—the volume should be 0.2 mL minus the
volume of
125
I-bFGF, inhibitor, and E-HS needed. Controls should be conducted with
vials containing only
125
I-bFGF, and
125
I-bFGF and the inhibitors, to determine the
non-proteoglycan–mediated adsorption of
125
I-bFGF to the filter. These values should be
substrated from the experimental points with E-HS to determine the level of
125
I-bFGF
bound to E-HS.
4. Add the E-HS, inhibitor, and
125
I-bFGF in that order. As a standard assay we have used
40 ng of E-HS and 0.2 ng of
125
I-bFGF in a final volume of 0.2 mL. Under these condi-
tions greater than 80% of the E-HS is retained with and without bFGF addition, and only
~5% of the bFGF will adsorb to the filter nonspecifically. Initial experiments should be
conducted with a range of
125
I-bFGF concentrations to determine the binding affinity and
capacity of each E-HS preparation before inhibitor competition data can be analyzed.
After each addition of
125
I-bFGF, vortex gently to mix and place in an incubation rack. If
the assay is done at a temperature other than room temperature (25°C), be sure that the
buffers are preequilibrated at the desired temperature and that vials are placed immedi-
ately in a temperature equilibrated rack.
5. Incubate for desired time (generally 60 min to reach a steady state in bound versus
unbound bFGF).
6. If incubation is shorter then 30 min, the cationic membrane should be prepared prior to
incubation (steps 3–5). The membrane is cut to fit the apparatus (cover all holes) and then
incubated on a rocking platform for at least 20 min in incubation buffer. Longer incuba-
tions (up to 1 h) do not adversely affect the assay.
7. The membrane is placed on the dot-blot apparatus and the top plate is tightened using a
criss-cross pattern. The apparatus is attached to a vacuum pump (GAST, model ROA-
P131-AA, Manufacturing Corp., Benton Harbor, MI) and the apparatus tightened again
under vacuum. Experiments should be done under a 25 mmHg vacuum.
8. Close off the apparatus to the vacuum and, using a multipipeter, add 0.2 mL of incubation
buffer to each well.
9. When the incubation is within 30 s of completion, turn on the vacuum and pull the wash
buffer through the membrane. Add samples (0.2 mL/well) in sets of three. Wash each
well of the triplicate set once with 0.2 mL of incubation buffer before adding the next set
of samples to the membrane. Allow all sample fluid to filter before adding the incubation
buffer. Repeat for each set of triplicate samples. Air bubbles may arise and interfere with
the filtration. Gently pipet the liquid near the surface of the membrane to mix, being
careful not to puncture the membrane.
10. After all samples have been added, do two additional washes with 0.2 mL of incubation
buffer per well.
11. Loosen the screws holding the unit in place and remove the top plate. Turn off the vacuum
and carefully remove the membrane.
12. Allow the membrane to dry sample side up on a paper towel and then cut out the individual
membrane dots corresponding to each individual well. Place each sample dot in a vial and
count the radioactive bFGF in a gamma counter. Sample data are shown in Fig. 2.
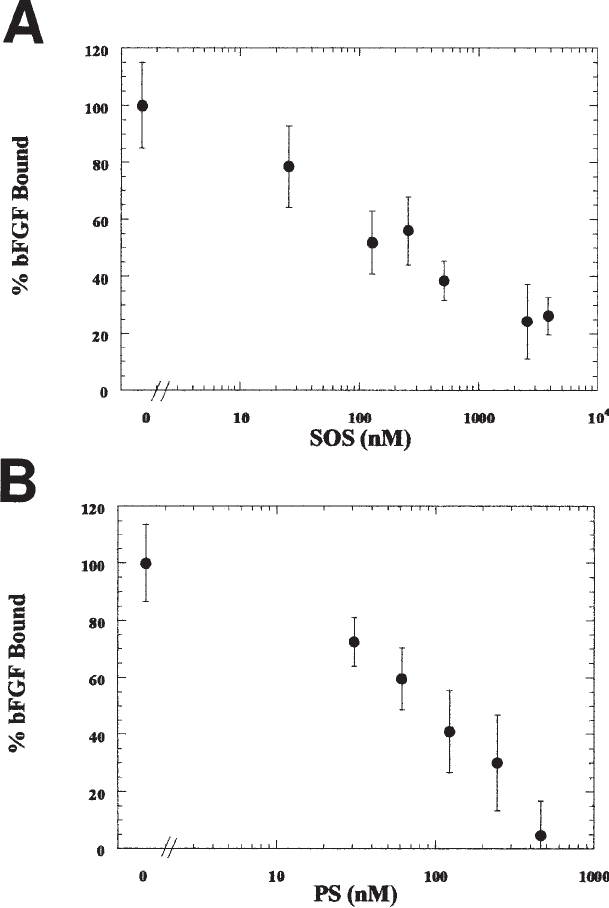
Growth Factor Binding 419
Fig. 2. Inhibitors of bFGF-E-HS Binding. E-HS (200 ng/mL), bFGF (0.056 nM), and inhibi-
tor ([A] sucrose octasulfate or SOS [B] protamine sulfate or PS) were incubated for 1 h and
then filtered across the cationic membrane. bFGF retention following incubation with SOS or
PS in the absence of E-HS was subtracted from total binding as nonspecific binding. 100%
bFGF bound corresponds to specific bFGF retention following incubation with E-HS in the
absence of inhibitor. The average ± standard error of at least triplicate samples is shown.

420 Forsten and Nugent
3.2 Analysis
3.2.1. Ligand–Proteoglycan Binding
1. The output data will represent the amount of bFGF retained on the filter for a given added
concentration. The amount retained in the absence of E-HS should be subtracted from
that with E-HS to generate data that represent specific E–HS bound bFGF.
2. Varying the concentrations of bFGF while holding constant the concentration of E-HS
(with no inhibitor), this data set should be analyzed to determine the binding capacity
(amount of bFGF bound per unit of E-HS) and the binding affinity (K
D1
), assuming a
simple monovalent binding reaction,
which, at steady state assuming negligible ligand depletion, is described by
where R
T
is the binding capacity, and L
0
is the concentration of FGF added. The
programming to determine K
D1
and R
T
is outlined below.
3. Mathematica software (Version 3.0, Wolfram Research) is launched, and a new notebook
page is initialized. Commands as outlined below are entered into the notebook page, and
the “shift” and “enter” keys are pressed simultaneously at the end of each instructional set
(symbolized in the procedure by “ ♦ ”)
4. The statistical package must be loaded by typing (see Note 2):
<<Statistics`NonlinearFit` ♦
5. Enter the volume of test solution (in liters) and the molecular weight of the growth factor
(bFGF) being investigated:
volume = 0.0002; ♦
mwF = 18000; ♦
6. Enter the amount of bFGF added per test well (L
0
) 〈ng〉
Fadd = {0.05, 0.05, 0.05, 0.05, 0.05, 0.05, 0.10, 0.10, 0.10, 0.10, 0.10, 0.10, 0.20, 0.20, 0.20,
0.20, 0.20, 0.20, 0.20, 0.20, 0.20, 0.20, 0.20, 0.20, 0.50, 0.50, 0.50, 0.50, 0.50, 0.50, 0.50, 0.50,
0.50, 0.75, 0.75, 0.75, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0, 2.0, 2.0, 2.0, 2.0, 2.0, 2.0, 2.0,
2.0, 2.0, 2.5, 2.5, 2.5, 4.0, 4.0, 4.0, 5.0, 5.0, 5.0, 5.0, 5.0, 5.0, 6.0, 6.0, 6.0, 10.0, 10.0}; ♦
7. Enter the measured values of bFGF retained (C
1
) 〈ng〉
Fbound = {.0025, .0035, .0034, .0013, .0055, .0033, .0067, .0069, .0076, .0028, .0033, .0035,
.017, .020, .018, .0063, .0076, .0063, .017, .016, .018, .016, .017, .017, .079, .034, .043, .029,
.034, .032, .021, .022, .020, .047, .056, .057, .080, .13, .099, .13, .071, .078, .037, .039, .086,
.088, .066, .070, .21, .24, .19, .17, .16, .16, .25, .20, .22, .32, .32, .30, .17, .37, .27, .22, .20, .17,
.51, .48, .45, .44, .39, .42}; ♦
8. Convert these values (L
o
and C
1
) from ng to nM and store as data sets 1 and 2:
dat1 = Fadd/(volume*mwF) ; ♦
dat2 = Fbound/(volume*mwF); ♦
C
1
R
T
L
0
K
D1
+ L
0
P + L
K
D1
C
1

Growth Factor Binding 421
9. Group the data (L
0
, C
1
):
data = {dat1, dat2}; ♦
binding = Transpose[data]; ♦
10. Perform the calculation for the nonlinear regression—ParameterCITable will output the
K
D1
and R
T
values at the end with the standard error and the confidence interval. The units
for each will be in nM:
ParameterCITable /. NonlinearRegress[binding, ligand*Rt/(Kd1+ligand),
{ligand}, {Rt,Kd1}] ♦
11. K
D1
is 2.3 ± 0.6 nM. R
T
is 0.23 ± 0.04 nM. The complete Mathematica program is shown
in Fig. 3. To convert R
T
to sites/ng E-HS (see Note 3):
Fig. 3. Mathematica program to solve for ligand–proteoglycan affinity and binding sites.
The code and data are input as shown. The ; following each line suppresses output and may be
eliminated if the user prefers. The output is displayed at the bottom and includes the asymptotic
standard error and the asymptotic confidence intervals.
sites = R
T
volume
PG
·
N
AV
422 Forsten and Nugent
where PG is the amount of E-HS added 〈ng〉, N
AV
is Avogadro’s number <6.02 * 10
23
sites mol>. R
T
for this system is 7.0 ± 1.2 × 10
8
sites/ng.
Although similar, the specifics for the ligand-binding inhibitor and the proteoglycan bind-
ing inhibitor do differ slightly and will each be outlined below. As example inhibitors, we
include data for sucrose octasulfate (ligand-binding inhibitor) and protamine sulfate
(proteoglycan-binding inhibitor)
3.2.2. Ligand-Binding Inhibitors (Sodium Sucrose Octasulfate)
An analysis of the inhibitor effectiveness is conducted based on steady-state data
acquisition with two independent first-order reactions occurring in solution.
(1)
(2)
where P is E-HS, L is the growth factor (bFGF), C
1
is the binding complex of E-HS
and bFGF, S is a growth factor binding inhibitor (SOS), and C
2
is the binding complex
of SOS and bFGF.
Assuming conservation of mass, the following equation at the steady state can be
solved for the K
D
value (K
D2
):
(P
0
– C
1
)(L
0
– C
1
)– (P
0
– C
1
)C
2
= K
D1
C
1
(4)
where
Knowing P
0
and K
D1
from steady-state analysis in the absence of inhibitor (see
Subheading 3.2.), experiments are run at a constant level of E-HS and bFGF (L
0
) and
various levels of inhibitor (S
0
). Retention is a measurement of C
1
. The programming
to determine K
D2
is outlined below.
1. Mathematica software (Version 3.0, Wolfram Research) is launched, and a new notebook
page is initialized. Commands as outlined below are entered into the notebook page, and
the “shift” and “enter” key are pressed simultaneously at the end of each instructional set
(symbolized in the procedure by “ ♦ ”)
2. The statistical package must be loaded by typing (see Note 2)
<<Statistics`NonlinearFit` ♦
3. The known parameters—P
0
and K
D1
—with both in units of nM:
P
0
= 0.23; ♦
K
D1
= 2.3; ♦
4. Enter the ligand concentration, L
0
〈nM〉, the reaction volume, volume 〈L〉, the molecular
weight of the inhibitor, mwI 〈g/mol〉, and the molecular weight of the growth factor,
mwF 〈g/mol〉:
L
0
= 0.056; ♦
volume = 0.0002; ♦
mwI = 1159; ♦
mwF = 18000; ♦
C
2
=
1
2
L
0
– C
1
+ S
0
+ K
D2
– L
0
– C
1
+ S
0
+ K
D2
2
–4S
0
L
0
– C
1
P + L
K
D1
C
1
S + L
K
D2
C
2
Growth Factor Binding 423
5. Enter the quantity of inhibitor (S
0
) added 〈ng〉:
s = {0, 0, 0, 0, 5, 5, 5, 25, 25, 25, 50, 50, 50, 100, 100, 100, 500, 500, 500, 750, 750, 750}; ♦
6. Enter the measured values of bFGF retained (C
1
) 〈ng〉:
c = {.018, .015, .022, .021, .02, .016, .013, .011, .0061, .011, .0082, .0061,
.013, .0094, .010, .0076, .0072, .0015, .0052, .0073, .0040, .0039}; ♦
7. Convert these values (S
0
and C
1
) to nM from ng and store as data sets A and B:
dat1 = s/(volume*mwI) ; ♦
dat2 = c/(volume*mwF); ♦
8. Enter K
D1
C
1
as a set of data:
dat3 = Kd1*dat2; ♦
9. Group the data (S
0
, C
1
, K
D1
C
1
):
data = {dat1, dat2, dat3}; ♦
SOS = Transpose[data]; ♦
10. Perform the calculation for the nonlinear regression—ParameterCITable will output the
K
D2
value at the end, with the standard error and the confidence interval. Entering an
estimate or initial guess for K
D2
will aid in the fitting process (the initial guess in our
example was 300; see Note 4). It should be noted that the C
2
value corresponds to one of
the roots of the quadratic equation (note the minus sign shown bold below)—the alternate
root yielded a negative value for K
D2
for this case.
ParameterCITable /. NonlinearRegress[SOS, (Po-C1)*(Lo-C1)-(Po-C1)*((Lo-C1+So+Kd2) –
((Lo-C1+So+Kd2)^2-4*So*(Lo-C1))^0.5)*0.5, {So,C1}, {Kd2,300}] ♦
11. K
D2
for SOS was 0.25 ± 0.08 µM.
3.2.3. Proteoglycan Binding Inhibitors (Protamine Sulfate)
Inhibitor effectiveness analysis is based on steady-state data acquisition with two
independent first-order reactions occurring in solution.
(1)
(2)
where P is E-HS, L is the growth factor (bFGF), C
1
is the binding complex of E-HS
and bFGF, PS is a proteoglycan binding inhibitor (protamine sulfate), and C
3
is the
binding complex of PS and bFGF.
Assuming conservation of mass, the following equation at the steady state can be
solved for the K
D
value (K
D3
):
(L
0
– C
1
)(P
0
– C
1
)– (L
0
–C
1
) C
3
= K
D1
C
1
(3)
where
C
3
=
1
2
P
0
– C
1
+ PS
0
+ K
D3
– P
0
– C
1
+ PS
0
+ K
D3
2
– 4PS
0
P
0
– C
1
PS + L
K
D3
C
3
P + L
K
D1
C
1
424 Forsten and Nugent
Knowing P
0
and K
D1
from steady-state analysis in the absence of inhibitor (see
Subheading 3.2.1.), experiments are run at a constant level of E-HS and bFGF (L
0
)
and various levels of inhibitor (PS
0
). Retention is a measurement of C
1
.
1. Mathematica software (Version 3.0, Wolfram Research) is launched, and a new notebook
page is initialized. Commands as outlined below are entered into the notebook page, and
the “shift” and “enter” keys are pressed simultaneously at the end of each instructional set
(symbolized in the procedure by “ ♦ ”)
2. The statistical package must be loaded by typing (see Note 2):
<<Statistics`NonlinearFit` ♦
3. The known parameters are then initialized—P
o
and K
D1
—with both in units of nM:
P
0
= 0.23; ♦
K
D1
= 2.3; ♦
4. Enter the ligand concentration - L
0
〈nM〉, the reaction volume, volume 〈L〉, the molecular
weight of the inhibitor - mwI 〈g/mol〉, and the molecular weight of the growth factor - mwF
〈g/mol〉:
L
0
= 0.056; ♦
volume = 0.0002; ♦
mwI = 6500; ♦
mwF = 18000; ♦
5. Enter the quantity of inhibitor (S
0
) added 〈ng〉:
s = {0, 0, 0, 40, 40, 40, 80, 80, 80, 160, 160, 160, 320, 320, 320, 600, 600, 600}; ♦
6. Enter the measured values of bFGF retained (C
1
) 〈ng〉:
c = {.018, .022, .019, .014, .017, .014, .009, .013, .014, .01, .012, .003, .001, .006, .011,
.003, .002, -.003}; ♦
7. Convert these values (PS
0
and C
1
) from ng to nM and store as data sets A and B:
dat1 = s/(volume*mwI) ; ♦
dat2 = c/(volume*mwF); ♦
8. Enter K
D1
C
1
as a set of data:
dat3 = Kd1*dat2; ♦
9. Group the data (PS
0
, C
1
, K
D1
C
1
):
data = {dat1, dat2, dat3}; ♦
Protamine = Transpose[data]; ♦
10. Perform the calculation for the nonlinear regression—ParameterCITable will output the
K
D2
value at the end or with the standard error and the confidence interval. Entering an
estimate or initial guess for K
D3
will aid in the fitting process (the initial guess in our
example was 75) (see Note 4). It should be noted that the C
3
value corresponds to one of
Growth Factor Binding 425
the roots of the quadratic equation (note the minus sign shown bold below)—the alternate
root yielded a negative value for K
D3
for this case.
ParameterCITable /. NonlinearRegress[Protamine, (L
0
– C
1
)*(P
0
– C
1
)– (L
0
- C
1
)
*
((P
0
–
C
1
+ PS + K
D3
) – ((P
0
– C
1
+ PS + K
D3
) ^ 2 – 4
*
PS
*
(P
0
- C
1
)) ^ 0.5)
*
0.5, {PS,C
1
},
{K
D3
,75}] ♦
10. K
D3
for protamine sulfate was 0.1 ± 0.02 µM.
3.3. Summary
Proteoglycan binding is a physiologically important means of regulating growth
factor activity and availability. This chapter details a fast and simple assay for
quantifying inhibitors of growth factor–proteoglycan binding. Although data and
the corresponding analysis focused on inhibitors of bFGF interactions, the method
should be easily transferable to other proteoglycan-binding growth factors and
molecules.
4. Notes
1. Many protocols for isolation of proteoglycans involve the use of denaturing agents such
as urea. These agents can interfere with protein–proteoglycan interactions and should be
removed from proteoglycan preparation prior to binding experiments.
2. When loading the Statistics packages, it is important to use ` (grave accent) as opposed to
‘ (open quote) in the statement: <<Statistics`NonlinearFit`.
3. In calculating the growth-factor binding sites per nanogram of proteoglycan, it is impor-
tant to enter the R
T
value in moles per liter, volume in liters, and proteoglycan in nano-
gram to generate binding sites per nanogram (see Subheading 3.2.1, step 11).
4. When fitting parameters using NonlinearRegress, a warning may be issued:
NonlinearFit::lmpnocon : Warning: The sum of squares has achieved a
minimum, but at least one parameter estimate fails to satisfy either an accuracy
goal of 1 digit (S) or a precision goal of 1 digit (s). These goals are less
strict than those for the sum of squares, specified by AccuracyGoal ->6 and
PrecisionGoal ->
If this occurs, it is best to rerun your regression fit with the output K
D
used as your new
initial guess. This may be easily done by highlighting the right sidebar and pressing ♦.
Repeat this procedure until the warning is not issued.
References
1. Conrad, E. (1998) Heparin Binding Proteins, Academic Press, San Diego, CA.
2. Rapraeger, A. C., Krufka, A., and Olwin, B. B. (1991) Requirement of heparan sulfate for
bFGF-mediated fibroblast growth and myoblast differentiation. Science 252, 1705–1708.
3. Fannon, M. and Nugent, M. A. (1996) Basic fibroblast growth factor binds its receptors, is
internalized, and stimulates DNA sythesis in Balb/c3T3 cells in the absence of heparan
sulfate. J. Biol. Chem. 271, 17949–17956.
4. Forsten, K. E., Courant, N. A., and Nugent, M. A. (1997) Endothelial proteoglycans inhibit
bFGF binding and mitogenesis. J. Cell. Physiol. 172, 209–220.
5. Dowd, C. J., Cooney, C. L., and Nugent, M. A. (1999) Heparan sulfate mediates bFGF
transport through basement membrane by diffusion with rapid reversible binding. J. Biol.
Chem. 274, 5236–5244.
426 Forsten and Nugent
6. Isner, J. M. (1996) The role of angiogenic cytokines in cardiovascular disease. Clin.
Immunol. Immunopathol. 80, S82–S91.
7. Rapraeger, A. and Yeaman, C. (1989) A quantitative solid-phase assay for identifying
radiolabeled glycosaminoglycans in crude cell extracts. Anal. Biochem. 179, 361–365.
8. Nugent, M. A. and Edelman, E. R. (1992) Kinetics of basic fibroblast growth factor bind-
ing to its receptor and heparan sulfate proteoglycan: a mechanism for cooperativity. Bio-
chemistry 31, 8876–8883.
9. Farndale, R. W., Buttle, D. J., and Barrett, A. J. (1986) Improved quantitation and dis-
crimination of sulphated glycosaminoglycans by use of dimethylmethylene blue. Biochim.
Biophys. Acta 883, 173–177.

Interaction with RTKs 427
427
From:
Methods in Molecular Biology, Vol. 171: Proteoglycan Protocols
Edited by: R. V. Iozzo © Humana Press Inc., Totowa, NJ
42
Interaction of Proteoglycans
with Receptor Tyrosine Kinases
David K. Moscatello and Renato V. Iozzo
1. Introduction
The control of cell proliferation depends on the interactions between growth factors
and their specific receptor-activated signaling pathways. It is well accepted that the
local extracellular matrix can modulate cellular responses to a given signal in several
ways, such as by modulating the affinity of the ligand for its cognate receptor (1), by
binding and limiting availability of a growth factor (1–3), or by influencing proteolytic
processing and internalization (3). However, it has only recently been shown that
“structural” components of the extracellular matrix can interact directly with, and
activate, receptor tyrosine kinases (RTKs). This was first shown by Vogel et al. and
Shrivastava et al., who demonstrated that the “orphan” receptor tyrosine kinases DDR1
and DDR2 in fact bind fibrillar collagen (4,5). This binding required the native triple-
helical structure of collagen and showed much slower kinetics than observed with
other ligand–receptor interactions (5). Decorin (6–8), a member of a family of small
leucine-rich proteoglycans (3), binds to fibrillar collagen and is an important regulator
of matrix assembly (9–11). Decorin content is elevated in the tumor stroma of colon
cancer (11), and ectopic expression of decorin inhibits cell growth (11–13). The
growth-suppressive properties of decorin are independent of p53 or retinoblastoma
proteins but require functional p21 protein (Waf1/Cip1/Sdi1) (13–16).
We recently reported that decorin activated the epidermal growth factor receptor
(EGFR) in A431 squamous carcinoma cells and other transformed cell lines. This
signaling was mediated by the protein core of decorin and induced MAP kinase
activation and a protracted upregulation of endogenous p21, thereby leading to growth
suppression (17). This activation occurred as a result of a direct interaction of decorin
with the EGF receptor (17,18). However, even “small” proteoglycans such as decorin
(~100 kDa, with a ~42-kDa core protein) are quite large in comparison with most
ligands of receptor tyrosine kinases, which are typically 6–25 kDa, a fact that compli-
cates analysis of crosslinking studies. Thus, in combination with their relatively
