Geckeler K.E., Nishide H. (Eds.) Advanced Nanomaterials
Подождите немного. Документ загружается.


Structural DNA Nanotechnology: Information - Guided
Self - Assembly
Yonggang Ke , Yan Liu , and Hao Yan
869
Advanced Nanomaterials. Edited by Kurt E. Geckeler and Hiroyuki Nishide
Copyright © 2010 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim
ISBN: 978-3-527-31794-3
28
28.1
Introduction
Although the Watson – Crick double helical model [1] of DNA has imparted major
impacts on modern biology for more than 50 years, the signifi cance of this
simple – yet very elegant – model is not limited to one particular area. In 1982 [2] ,
Ned Seeman proposed the building of nanostructures from DNA, an idea which
led to the origination of the fi eld now known as “ structural DNA nanotechnology. ”
During the past decade, this fi eld has witnessed much signifi cant progress, and
in this chapter we will discuss the basic concepts and major research directions
of structural DNA nanotechnology, the important progress that has been made in
recent years, and some future perspectives of the fi eld.
DNA, which serves as the genetic information carrier in most organisms on
Earth, is also an ideal candidate for structural nanotechnology, which targets at
controlling and organizing matter at the nanometer scale. First, DNA is a nanom-
eter - scale object itself, with a diameter of 2 nm and a helical repeat of 10 – 10.5
nucleotide pairs, or ∼ 3.5 nm for the common B - form DNA. Second, DNA hybridi-
zation is highly predictable because of the well - known Watson – Crick base - pairing
that guanine (G) pairs with cytosine (C), and adenine (A) with thymine (T). Third,
whilst single - stranded DNA is quite fl exible, the DNA duplex is more rigid and
has a persistence length of approximately 50 nm. It is this combined fl exibility and
rigidity that permits the design of DNA structures to form different geometric
shapes. Furthermore, as a results of advances in modern chemistry and molecular
biology, DNA molecules with any designed lengths, sequences, and a variety of
functionalities can now be synthesized conveniently, and also manipulated by
using the wide range of enzymes that are available for the cleavage, ligation and
amplifi cation of DNA. All of these facilities have provided scientists with an
extreme degree of control for building DNA nanostructures and maneuvering
DNA nanomachines.
Topologically speaking, DNA duplex is a one - dimensional ( 1 - D ) molecule. In
order to create a two - dimensional ( 2 - D ) or three - dimensional ( 3 - D ) structure, it is
870 28 Structural DNA Nanotechnology: Information-Guided Self-Assembly
necessary to use branched objects. For example, Seeman proposed techniques that
would allow DNA strands to assemble into branched junctions that could further
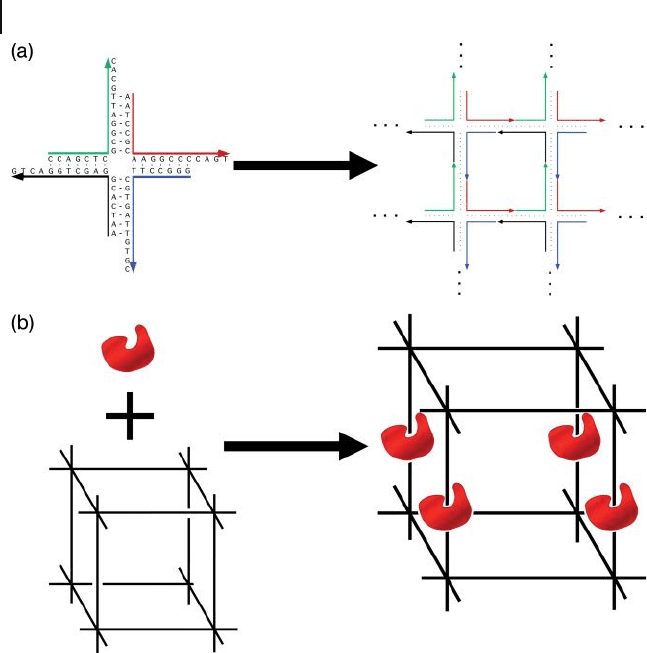
self - assemble into periodic, 2 - D arrays [3] (Figure 28.1 a).
The basic building block here is the branched four - arm junction consisting of
four single - stranded DNA oligonucleotides. To enable DNA junction building
blocks to form higher ordered objects and lattices, single ν stranded DNA over-
hangs called “ sticky ends ” are used to bring DNA junction molecules together.
These sticky ends carries base sequences that are complementary to each other;
for example, the red sticky end is complementary to the black end, and the green
end is complementary to the blue end (Figure 28.1 a). As a result, sticky end cohe-
sion will cause the individual four - arm junctions to be “ glued ” together to form
the 2 - D array. Yet, this simple scheme illustrated a powerful method for building
Figure 28.1 Ned Seeman ’ s original proposal of construction
of a periodic DNA array and its application. (a) Four - arm
junction DNA tiles with sticky ends are connected together to
form a 2 - D periodic array through self - assembly process;
(b) A 3 - D DNA lattice templated protein array could be used
for X - ray crystallography.

28.2 Periodic DNA Nanoarrays 871
DNA nanostructures: fi rst, to design the branched DNA building blocks or “ tiles ” ,
and then to assemble them together using sticky end cohesion.
Although other cohesions have also been used for mortaring DNA nanostruc-
tures – for example, paranemic crossover s ( PX ) cohesion [4] and edge - sharing
[5] – sticky end cohesion is by far the most extensively used in DNA nanostructure
design. Studies of crystal structures have revealed that the duplex formed by com-
plementary sticky ends has an exactly identical structure to B - DNA [6] , a feature
which allows designers to predict and control the relative orientations of the DNA
tiles. It is interesting to note that for sticky ends that are N - bases long, the number
of unique sticky ends can be up to 4
N
, which provides a library of programmable
molecular interactions.
Ideally, the DNA sequences of a DNA nanostructure should be designed to
achieve the highest stability, so that all other less - stable competitive structures
will be less likely to form. On a practical basis, it is necessary to choose a set of
optimized sequences that minimizes sequence symmetry at the branch points [7]
so as to prevent the branch migration of DNA strands.
As originally proposed by Seeman, one potential application of structural
DNA nanotechnology would be to use the highly ordered self - assembling DNA
scaffolds to organize other types of macromolecule into 3 - D crystals. Moreover,
if the macromolecule could be attached to a 3 - D DNA nanostructure (Figure
28.1 b), the self - assembly of DNA would facilitate the organization of macromol-
ecules into a periodic lattice, the periodicity and parameters of which could be
well defi ned. This would in turn facilitate macromolecule structural analysis using
X - ray diffraction.
Besides the above - described potential application, the DNA nanostructure might
also be used as a scaffold to organize different nanomaterials. An example of this
is the organization of nanoelectronic components into an addressable fashion,
leading to the construction of DNA - templated nanoelectronics/nanophotonics
devices. Indeed, recent developments in the use of 1 - D and 2 - D DNA nanostruc-
tures to template nanoparticles into rationally designed patterns have paved the
way towards this goal.
28.2
Periodic DNA Nanoarrays
Seeman ’ s original proposal has inspired many research groups to construct
a variety of DNA tiles with different sizes and geometries, and assemble them
into 2 - D periodic arrays (Figure 28.2 ). For example, Mao et al. designed and con-
structed DNA parallelograms that would grow into micrometer - sized 2 - D arrays
(Figure 28.2 a) [8] .
A series of DNA tile molecules termed double crossover [9] ( DX ) DNA molecules
were originally created by Seeman, and subsequently utilized in many of the later
studies. The DX tile consists of two parallel DNA helices, joined together by two
crossovers through strand exchange. Winfree et al. successfully built DNA 2 - D
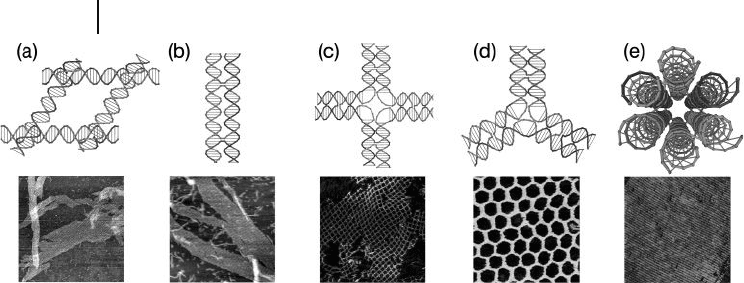
872 28 Structural DNA Nanotechnology: Information-Guided Self-Assembly
arrays through the self - assembly of DX tiles (Figure 28.2 b) [10] . A similar design
strategy, learned from the DX tile construction, was then used by different groups
when designing DNA tiles containing multiple parallel DNA helices; these
included triple crossover tiles [11] , and four - , eight - , and 12 - helix tiles [12, 13] .
Yan et al. used four four - arm DNA junctions to design a cross - shaped tile,
named “ 4 × 4 ” tile [14] . In this design, four four - arm junctions are tethered
together by a long central strand with a dT
4
loop in between (Figure 28.2 c); the
4 × 4 tiles then self - assembled into a 2 - D square lattice. By removing one arm
from, or adding two more arms to the 4 × 4 tile, Mao ’ s group were able to construct
a three - point - star [15] and a six - point star tile [16] that could self - assemble into 2 - D
lattices with hexagonal and triangular cavities (Figure 28.2 d).
Another family of tiles are tube - like tiles, including three - helix [17] , six - helix
[18] (Figure 28.2 e), and eight - helix bundles [19] . With different sticky end
designs, these tubes tile can be assembled to either 1 - D tubes, 2 - D arrays, or even
3 - D lattices.
28.3
Finite - Sized and Addressable DNA Nanoarrays
For the purpose of DNA - directed macromolecule crystallization, the periodic
arrays represent an excellent choice. However, to build a functional DNA nanoelec-
tronic device, it must be possible to control the size of a DNA array and to
attach functional species at particular locations on the array. Such needs
have driven research teams to develop methods for building fi nite - sized and
addressable arrays.
Park et al. [20] reported the construction of a square - shaped, addressable array
consisting of sixteen 4 × 4 tiles (Figure 28.3 a) through an hierarchical assembly
Figure 28.2 DNA tiles and their 2 - D periodic arrays formed
through sticky end cohesion. (a) A DNA parallelogram tile
consisting of four Holliday junction structures; (b) A double
crossover ( DX ) tile; (c) A 4 × 4 cross - shaped tile. Each arm of
the cross is a four - arm DNA junction; (d) A three - point - star
tile; (e) A six - helix - bundle tile.
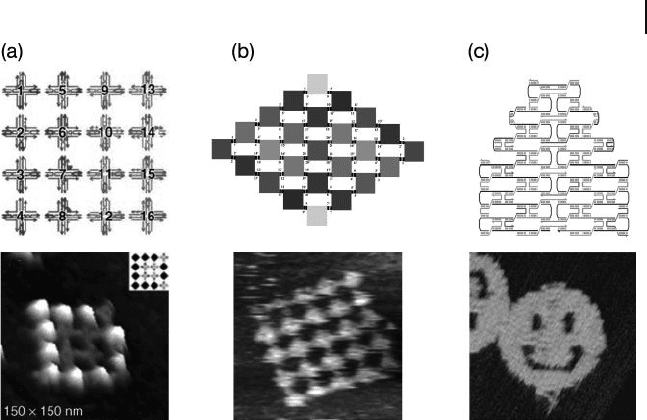
28.3 Finite-Sized and Addressable DNA Nanoarrays 873
Figure 28.3 DNA fi nite - sized and addressable
arrays. (a) A 4 × 4 tile array consisting of
16 distinctive 4 × 4 tiles. This is a fully
addressable array on the level of individual
tiles. Streptavidin protein was attached onto
certain DNA tiles to display a letter “ D ” ; (b) A
5 × 5 tile array consisting of 25 eight - helix
tiles. The number of distinctive tiles was
reduced from 25 to 13 by taking advantage of
the array ’ s C
2
symmetry; (c) Schematic of
Rothemund ’ s scaffolded DNA self - assembly
and atomic force microscopy image of a
nanometer - scale “ smiley face. ” The black
strand is the scaffold DNA that is folded by
other short “ staple ” strands into the
designed shape.
process. In order to demonstrate that the 16 - tile array was fully addressable on the
level of individual tiles, Park and colleagues fi rst functionalized a few tiles on the
array with biotin groups, and then attached streptavidin molecules to the array. In
this way, the protein molecules could be organized to display the letters, “ D ” , “ N ”
and “ A. ”
It is costly to make every tile in an array unique; moreover, the more complex
the system, the greater the chances of self - assembly errors occurring. In order to
build fi nite - sized arrays in a cost - effi cient way, Liu et al. [21] demonstrated a strat-
egy to utilize the geometric symmetry of the array to reduce the number of unique
tiles. For a N - tile fi nite - size array with C
m
symmetry, the number of unique tiles
required is N / m , if N / m is an integral number, or Int( N / m ) + 1, if N / m is a non-
integral number. Consequently, Liu and coworkers demonstrated two 25 - tile array
examples with C
2
and C
4
fold symmetry. The 5 × 5 array with C
2
symmetry required
13 unique tiles instead of 25 (Figure 28.3 b), while the 5 × 5 array with C
4
symmetry
required only seven unique cross - shaped tiles instead of 25.
When comparing these two fi nite - size addressable arrays, it is possible to under-
stand the dilemma that research groups often face when designing a DNA struc-
ture. On one hand, the addressability of an array can be increased by introducing
more unique sequences/tiles into the system, although a low yield and a high error
rate will be expected. On the other hand, the symmetry can be utilized to reduce

874 28 Structural DNA Nanotechnology: Information-Guided Self-Assembly
complexity of the system so as to achieve a high yield and a low assembly error
rate, but the high addressability would be lost. Thus, depending on the purpose
of a DNA nanostructure, it is important for a designer to identify a good balance
between the complexity and addressability of the system.
Another important approach when building an addressable DNA nanoarray is
“ nucleated DNA self - assembly. ” This method uses a long natural or synthetic DNA
strand, which serves as a scaffold, to direct the DNA strands or tiles self - assembly.
Yan et al. demonstrated the use of this technique by effi ciently assembling DX
tiles together into barcode - patterned lattices [22] . In 2006, Paul Rothemund
reported an exciting breakthrough, in which he used more than 200 short “ staple ”
DNA strands to fold 7249 base long, single - stranded M13 viral DNA into 2 - D arrays
( “ DNA Origami ” ) with a variety of shapes [23] . Every staple has its unique sequence,
and can only hybridize with predefi ned parts of M13 according to a predetermined
folding path (Figure 28.3 c). The array resulted is fully addressable at the position
of each individual staple strand. In theory, if the scaffold strand is long enough,
it is possible to design any arbitrarily shaped 2 - D DNA array and to functionalize
staples at any position on that array.
28.4
DNA Polyhedron Cages
One branch of structural DNA nanotechnology focuses on the construction of 3 - D
DNA “ cages. ” Potentially, DNA cages can be used for applications such as target -
specifi c drug delivery or nanoparticle site - specifi c functionalization. For delivery
purposes, the cages can be made to encapsulate cargos during their self - assembly
and to display target recognition tags outside the cages. Because DNA duplex is
linear, it is not surprising that most of the DNA cages built to date are polyhedral,
using DNA duplex(es) as straight edges and branching point(s) at the vertices.
When Chen and Seeman assembled the fi rst DNA polyhedron, a cube consisting
of ten DNA strands [24] , they used endonucleases to cleave specifi c edges of the
cube to prove the tube topology with polyacrylamide gel electrophoresis. Many
years later, several groups have only recently developed a number of methods
to build DNA polyhedral cages and more direct ways to prove their formation
(Figure 28.4 ).
Goodman et al. built a DNA tetrahedron by hybridizing four single - stranded
DNAs together in a one - step annealing process (Figure 28.4 a) [25, 26] . In a later
study, the same group built hairpin loops into the tetrahedron and demonstrated
that the edge of this cage could be opened/closed by the addition of “ fuel DNA ”
strands [27] . This mechanism would allow cargos to diffuse into the structure at
the open stage, and to be captured in the close stage. Shih et al. used a 1.7 kilobase
and fi ve short, single - stranded DNAs to construct an octahedron (Figure 28.4 b)
[28] where the edges of the octahedron were double crossovers (DX) or paranemic
crossovers (PX) DNA motifs. One noteworthy feature of this octahedron was that
the 1.7 kilobase DNA was formed through polymerase chain reaction ( PCR ), which
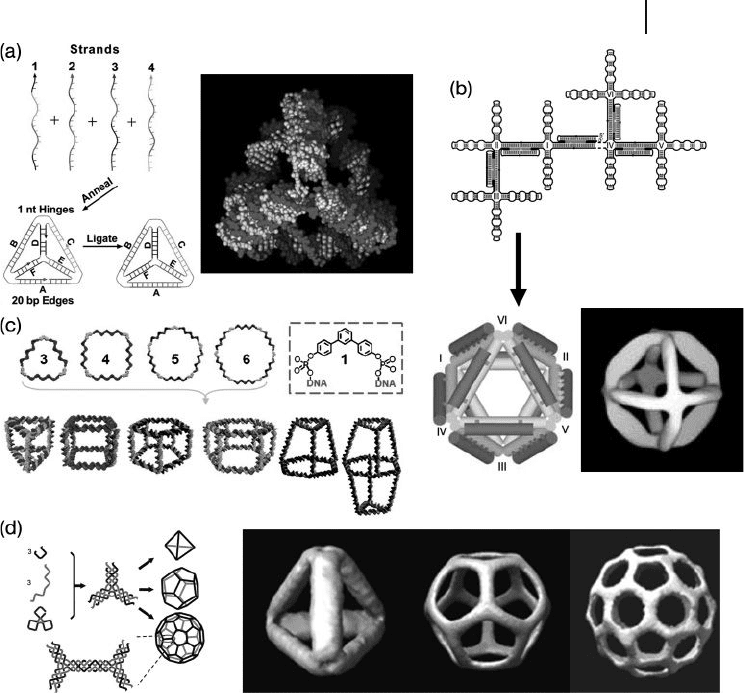
28.4 DNA Polyhedron Cages 875
in turn means that the DNA could be easily amplifi ed to produce large amounts
of the DNA octahedron.
Aldaye et al. recently developed a new approach for building different - shaped
DNA cages [29] . They introduced organic molecules into the circular single -
stranded DNA during the DNA synthesis process (Figure 28.4 c). This allowed the
building of a series of DNA polygons with repeated sequences at each edge and
branched organic molecules at the corners. Linker strands were then used to bring
two or three polygons together to form the DNA cages.
Mao ’ s group reported the design and formation of tetrahedron, dodecahedron,
and buckyballs through hierarchical DNA assembly (Figure 28.4 d) [30] . All three
polyhedra shared the same basic building unit, namely the three - point - star tile
Figure 28.4 DNA polyhedron cages. (a) DNA
tetrahedron: schematic drawing and a physical
model; (b) DNA octahedron model and 3 - D
reconstruction image from cryo - electron
microscopy (EM) experiments; (c) A series
DNA polyhedra. Organic molecules are
incorporated into the DNA circular strands to
help the structures form. (d) DNA
tetrahedron, dodecahedron, and buckyball
assembled from three - point - star tiles. A 3 - D
reconstruction of the cryo - EM images of these
polyhedra is shown on the right.

876 28 Structural DNA Nanotechnology: Information-Guided Self-Assembly
[15] , although at low concentrations (50 – 75 n M ) the tile tended to form discrete
3 - D polyhedra rather than a 2 - D array. Another variable in such a design was the
length of the single - stranded loops at the center part of the tile, which provided a
variable fl exibility to the tile and controlled the angle at which the arms could bend
out of the plane. Longer loops seemed to provide a higher fl exibility at the center
of tiles and allowed them to form polyhedra that required larger bending angles
at the vertexes. For example, fi ve - base loops were found to promote the formation
of tetrahedrons, while three - base loops were suitable for the formation of dodeca-
hedrons. By linking two of the three - point - star tiles, a tile with four arms was
created that could self assemble into a buckyball - shaped DNA structure. In a later
study, a fi ve - point - star tile was shown to self - assemble into an icosahedron, using
a similar design/assembly strategy [31] .
28.5
DNA Nanostructure - Directed Nanomaterial Assembly
As noted at the start of this chapter, one central task of DNA nanotechnology is
to control functional materials at the nanometer scale, and the DNA templated
self - assembly of nanomaterials represents an important direction towards this
goal. Until now, several groups have demonstrated the assembly of nanometer -
scale materials such as metallic nanoparticles, quantum dot s ( QD s) and proteins
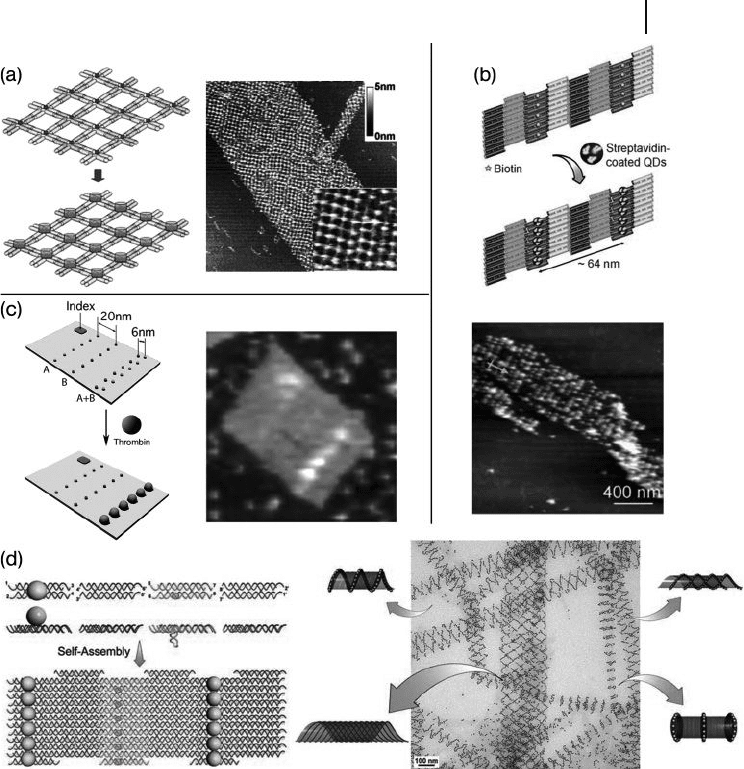
on DNA nanostructures (Figure 28.5 ). The recognition events used to capture
these functional materials included protein – protein/small molecule/aptamer
( in vitro selected short DNA or RNA that can specifi cally bind to certain protein)
interactions, functionalized metallic nanoparticle – DNA interactions, and DNA –
DNA hybridization.
By using the well - known interaction between streptavidin and biotin, streptavi-
din protein molecules can be organized onto 2 - D DNA nanoarrays with a control-
led periodicity and spacing [32] . The streptavidin – biotin interaction has also been
shown to be useful for organizing nanoparticles. For example, Sharma et al. suc-
cessfully built a patterned QD array by reacting streptavidin - coated QDs with
biotin groups on the 2 - D DNA nanoarray [33] . By attaching single/multiple copies
of DNA onto the Au nanoparticle surface [34 – 36] , Au nanoparticles could be
rationally organized onto self - assembled DNA nanoarrays through DNA – DNA
hybridization. Recently, Yan ’ s group demonstrated that different - sized Au nano-
particles could be used to control the conformation of DNA nanotubes (Figure
28.5 d) [37] .
Liu et al. incorporated thrombin - binding aptamers into a linear, three - helix DNA
tile array and used aptamer/protein recognition to capture thrombin proteins into
periodic arrays [38] . Recently, Rinker et al. [39] were able to take one step further
by displaying two different thrombin aptamers, each target at a distinctive site of
the protein, on a DNA array. The distances between the two aptamers were sys-
tematically tuned such that a ∼ 5.7 nm spacing led to an optimal “ multivalent
binding, ” which has much higher binding affi nity than does any of the single
aptamers (Figure 28.5 c).
28.6 Concluding Remarks 877
28.6
Concluding Remarks
The fi eld of structural DNA nanotechnology has witnessed numerous break-
throughs during the past decade, although only a small fraction of the prominent
studies are outlined here. In that time, the complexity of the DNA structures has
grown dramatically, from less than 100 nucleotides (four - arm DNA junction) to
more than 10 000 nucleotides ( “ DNA Origami ” ), with series of 3 - D objects having
Figure 28.5 DNA nanostructure - directed nanomaterial
assembly. (a) Streptavidin assembly on DNA 4 × 4 periodic
2 - D array; (b) Quantum dot assembly on a double crossover
(DX) array; (c) Thrombin line assembled on M13 viral DNA
scaffolded 2 - D array; (d) Gold nanoparticles were assembled
on a DX 2 - D array and forced the array to form tubes with
different spiral patterns.

878 28 Structural DNA Nanotechnology: Information-Guided Self-Assembly
been built and characterized. Today, the capability is available to attach proteins,
metallic and semi - conducting nanoparticles onto DNA arrays so as to create a
variety of patterns of these materials. An increasing knowledge of the rules
to design DNA structures has led to the building of more - complex DNA self -
assemblies, and with fewer errors. With such progress, there is much optimism
that DNA nanotechnology can potentially be applied to build DNA nanostructure -
based nanocircuits and to control chemical/biochemical reactions in an ordered
fashion that mimics enzyme cascade reactions. Nonetheless, many challenges
remain. Although some functional materials have been successfully patterned by
DNA arrays, the key ability is still lacking to create well - controlled, multicompo-
nent nanoarchitectures. Whilst the current success with DNA 3 - D objects has been
limited to the building of polyhedron cages for purposes such as protein encap-
sulation and drug delivery, the task remains to create a universal strategy for
building highly ordered 3 - D structures. The “ designer DNA ” nanostructures pro-
duced, with their controlled geometry and topology, might fi nd their use in biologi-
cal applications such as interfacing cellular components through DNA scaffolds.
Yet, much remains to be done in studying the biocompatibility, delivery, and
stability of DNA nanostructures inside living systems. With the fi eld of nanotech-
nology having successfully “ borrowed ” DNA from biological systems, it will not
be surprising to see DNA nanotechnology contribute to the in vivo applications of
nanotechnology in the future.
Acknowledgments
These studies were supported by grants from the National Science Foundation
( NSF ), the Army Research Offi ce ( ARO ), and the Technology and Research Initia-
tive Fund from Arizona State University to Y.L., and by grants from NSF, ARO,
Air Force Offi ce of Scientifi c Research, Offi ce of Naval Research, and the National
Institute of Health to H.Y.
References
1 Watson , J.D. and Crick , F.H.C. ( 1953 ) A
structure for deoxyribose nucleic acid .
Nature , 171 , 737 – 738 .
2 Seeman , N.C. ( 1982 ) Nucleic acid
junctions and lattices . J. Theor. Biol. , 99 ,
237 – 247 .
3 Holliday , R. ( 1964 ) A mechanism for
gene conversion in fungi . Genet. Res. , 5 ,
282 – 304 .
4 Zhang , X. , Yan , H. , Shen , Z. and
Seeman , N.C. ( 2002 ) Paranemic cohesion
of topologically - closed DNA molecules .
J. Am. Chem. Soc. , 124 , 12940 – 12941 .
5 Yan , H. and Seeman , N.C. ( 2003 )
Edge - sharing motifs in structural DNA
nanotechnology . J. Supramol. Chem. , 1 ,
229 – 237 .
6 Qiu , H. , Dewan , J.C. and Seeman , N.C.
( 1997 ) A DNA decamer with a sticky
end: the crystal structure of
d - CGACGATCGT . J. Mol. Biol. , 267 ,
881 – 898 .
7 Seeman , N.C. ( 1990 ) De novo design of
sequences for nucleic acid structural
engineering . J. Biomol. Struct. Dyn. , 8 ,
573 – 581 .
