Faulon J.L., Bender A. Handbook of Chemoinformatics Algorithms
Подождите немного. Документ загружается.

388 Handbook of Chemoinformatics Algorithms
chemoinformatics uses a much broader set of data types. Bioinformatics applications
use a huge variety of algorithms but are generally restricted to microarray (numerical)
and sequence (string) data. Chemoinformatics applications generally use two to three
clustering algorithms (Jarvis–Patrick, Ward clustering, and k-means) but use very
large datasets and a huge number of chemical descriptors (see Chapters 2–4) as their
input.
Clustering in both bioinformatics and chemoinformatics is fairly mature. Both
fields have standard approaches that work well for their data types and applications.
Cross-fertilization, however, is certain to benefit both areas of study. Bioinformat-
ics, for example, has spurred the development of many new alternative clustering
techniques, some of which work on large datasets. After adaptation to data types
from chemoinformatics, it seems likely that many of these algorithms will be useful.
Conversely, the diversity of data types in chemoinformatics and the general interest
of drug design in bioinformatics have spurred the development of new techniques,
including graph kernels in support vector machines (SVMs) (see Section 14.2.2) and
the inference of protein–chemical interaction networks (Section 14.2.3).
14.2 APPLICATIONS OF CLASSIFICATION AND REGRESSION
In this section, we consider applications of methods in classification and regression to
problems in bioinformatics and chemoinformatics. In the previous section (Equation
14.1), we saw that the choice of clustering algorithm varied widely between the two
fields. This is due to historical reasons, as well as dataset size and type differences
between biology and chemistry. In contrast, the choice of algorithm in classifica-
tion and regression is fairly uniform between bioinformatics and chemoinformatics.
Instead, most of the difference lies in the choice of data description. In bioinformatics,
descriptors are often based on sequence (largely due to the vast amount of sequence
data available), while in chemoinformatics, descriptors are typically tied more directly
to structure.
Classification and regression are both methods for correlating elements in a dataset
with properties of those elements. In classification, elements are correlated with dis-
crete properties, as in the case of the classification of a molecule according to its
activity(e.g., activeor inactive).In regression, elements are correlated with continuous
properties, as in the case of relating molecular structure to boiling point or log P.
There are many available algorithms for use in both classification and regression. In
classification, popular algorithms include k-nearest neighbors [41], neural networks
[42], SVMs [43], and numerous statistical methods [44,45]. In regression, popular
algorithms include linear and multiple regression, principal component regression,
and partial least squares [46,47]. In addition, there are several algorithms that can be
used for both classification and regression, including neural networks and SVMs.
Since techniques in classification and regression are applied fairly uniformly in
both bioinformatics and chemoinformatics, we focus our discussion on SVMs. SVMs
can be used for classification [43,48], regression [49], and even clustering [50]. We
highlight data-type differences between bioinformatics and chemoinformatics and
how these differences can be accommodated using SVMs.

Machine Learning–Based Bioinformatics Algorithms 389
FIGURE 14.3 A linear SVM. The SVM decision boundary is defined by the support vectors,
which are the examples falling on the dotted lines. The distance between the dotted lines is
known as the margin and is maximized to obtain the SVM decision boundary, shown as the
solid line separating the circles (class 1) from the x marks (class −1).
At its core, an SVM is a linear binary classifier. Suppose that we have a dataset
{x
i
}⊆R
n
and that each point x
i
in our dataset has a corresponding class label
y
i
∈{±1}. Our goal is to separate the points in our dataset according to their class
label. Since there are two classes, this is known as binary classification. An SVM
attempts this classification by using a linear hyperplane w
T
x +b, w = 0, as shown
in Figure 14.3.
Assuming that our dataset is in fact linearly separable, there will in general be
many possible hyperplanes that can achieve the separation. An SVM uses an optimal
separating hyperplane known as the maximal margin hyperplane. The hyperplane
margin is twice the distance from the separating hyperplane to the nearest point in
one (or the other) of the two classes. In Figure 14.3, this is the distance between the
two dotted lines. The SVM hyperplane is found by solving the quadratic programming
problem [51,52]:
max
α
1
2
i,j
y
i
y
j
α
i
α
j
x
T
i
x
j
−
i
α
i
s.t.
i
y
i
α
i
= 0, 0 ≤ α
i
≤ C,
(14.2)
where w =
i
y
i
α
i
x
i
is the normal to the SVM hyperplane. Using w we form the
SVM decision function f (x) = sign(w
T
x +b), where b is obtained implicitly [51,52].
We note that α
i
= 0 only when x
i
is a support vector.
The SVM problem given in Equation 14.2 only applies to datasets {x
i
}⊆R
n
. Often,
however, we want to use an SVM on a dataset that is not a subset of R
n
. This occurs
in the case of biology and chemistry problems when we are likely to use amino acid
sequences or chemical structures to describe our data. Fortunately, there is a ready
solution to this problem, formalized in the use of kernel functions.
390 Handbook of Chemoinformatics Algorithms
Suppose that our data {x
i
}⊆S, where S might be the set of all finite length protein
sequences or all finite diameter chemical graphs. We can then define a kernel function
as a map k : S ×S → R such that
k(x
i
, x
j
) = Φ(x
i
)
T
Φ(x
j
), (14.3)
where Φ: S → F is a map from our original data space S into a space F with a defined
dot product such as R
N
.
Once we have defined a kernel function, we simply replace the dot product x
T
i
x
j
in
Equation 14.2 with the kernel k(x
i
, x
j
) to obtain the full SVM quadratic programming
problem. A similar procedure can be used for any method that is written in terms of
dot products, thus giving rise to the moniker kernel methods.
SVMs have a number of advantages over competing methods, including a unique
solution of a fairly straightforward quadratic programming problem (compared to a
nonunique solution for a neural network), ability to employ nonlinearity by choice
of kernel, and widespread availability of software [53,54]. On the other hand, SVMs
can be memory intensive O(n
2
) and the flexibility in choice of kernel can make
SVMs difficult for beginners. Neither of these disadvantages, however, has slowed
the widespread use of SVMs in both bioinformatics and chemoinformatics.
14.2.1 APPLICATIONS IN BIOINFORMATICS
The central applications of classification and regression methods in bioinformatics
are to protein function prediction [55] and protein interaction prediction [56]. These
two topics are not unrelated and are often approached using the same basic methods.
In particular, both protein function and protein interaction prediction have benefited
from the application of classification and regression methods.
The success of function and interaction prediction often depends more on the
formulation of the problem than on the algorithm used for the classification or regres-
sion. For proteins, this means the use of either sequence or structure and the particular
assumptions used when the data are put into the algorithm. For sequence data, pre-
dictions are often made by comparing proteins using sequence similarity (homology)
across organisms [24,57], using subsequence motifs or identifying motifs and using
them to predict either function or interaction [58,59] and by using amino acid residue-
specific features such as van der Waals volume, polarity, hydrophobicity, charge, and
so on to represent a sequence as a vector [60,61]. For structural data, predictions can
be made by comparing proteins based on geometric considerations such as amino
acid proximity and similarity of 3D ball protein models [62].
In terms of SVMs, data types used to predict function include gene expression
data [63], motif-based subsequences [58], and feature vectors based on sequence [61]
and structure [62]. Data used to predict interactions include generalized subsequences
[64], feature vectors [60], and combinations of sequence and structure [65,66].
The success of classification and regression methods has been widespread in bioin-
formatics, due largely to the availability of vast amounts of sequence data. Although
many different algorithms have been applied in the field, SVMs have been a favorite
for a variety of data types. One reason that SVMs are popular is that SVMs encourage
Machine Learning–Based Bioinformatics Algorithms 391
a very useful separation between data and method. A computer scientist interested
in developing algorithms or methods can take as a starting point a kernel of the
type described in Equation 14.3, while a biologist or chemist interested in a particular
problem needs only provide a pairwise kernel similarity in order to apply the methods
developed by the computer scientist.
To illustrate the use of an SVM kernel in bioinformatics, we briefly describe
the subsequence kernel used for predicting protein interaction used by Martin et al.
[64]. Suppose that we have two protein sequences that we would like to compare.
In mathematical terms, a protein sequence is a finite length string over an alphabet
of 20 letters corresponding to the 20 possible amino acid residues. To calculate the
similarity between two proteins we must calculate the similarity between two strings.
Hence we use a string kernel.
To define a string kernel, we first define a map Φ
l
s
: {finite length amino acid
strings} → Z
N
l
, where N
l
is the number of possible amino acid sequences of length
l. If we denote by z
j
the bases of Z
N
l
where each basis vector z
j
corresponds to an
amino acid sequence of length l, then Φ
l
s
is given by
Φ
l
s
P
i
=
j
σ
j
z
j
, (14.4)
where P
i
is a finite length amino acid string and σ
j
is the number of times that the
amino acid string corresponding to z
j
occurs in the string P
i
. We then define the string
kernel k
l
s
(P
i
, P
j
) between two proteins P
i
and P
j
by
k
l
s
(P
i
, P
j
) = Φ
l
s
(P
i
)
T
Φ
l
s
(P
j
). (14.5)
As an example, suppose that we have amino acid strings LVMLVM and LVMTTM.
We want to calculate Φ
3
s
(LVMLVM), Φ
3
s
(LVMMTT), and k
3
s
(LVMLVM, LVMTTM).
There are four substrings of length 3 (also known as trimers) in LVMLVM, namely
LVM, VML, MLV, and LVM. There are also four substrings of length 3 in LVMTTM,
namely LVM, VMT, MTT, and TTM. Suppose that z
1
corresponds to LVM, z
2
corre-
sponds to VML, z
3
corresponds to MLV, z
4
corresponds to VMT, z
5
corresponds to
MTT, and z
6
corresponds to TTM. We see that Φ
3
s
(LVMLVM) = (2, 1, 1, 0, 0, 0)
T
and
Φ
3
s
(LVMMTT) = (1, 0, 0, 1, 1, 1)
T
, so that k
3
s
(LVMLVM, LVMMTT) = 2. A visual
demonstration arriving at Φ
3
s
(LVMLVM) is shown in Figure 14.4.
14.2.2 APPLICATIONS IN CHEMOINFORMATICS
The main application of classification and regression in chemoinformatics is to the
development of QSARs and QSPRs. We refer to both QSARs and QSPRs using the
term “QSAR” only. The use of QSAR is widespread in chemoinformatics, occur-
ring historically in the study of drugs [29] and theoretical organic chemistry (see
Refs. [67,68]). Pioneering work was conducted by Hansch and Fujita [69] and Free
and Wilson [70] and substantial continuing research is ongoing in the area of drug dis-
covery [29,30]. The use of SVMs in the field of QSAR has occurred more recently and
includes applications to various chemical properties [71–73], drug activity [74,75],
and mutagenicity prediction [76,77].
392 Handbook of Chemoinformatics Algorithms
1
0
0
0
0
. . .
0
1
0
0
0
. . .
0
0
1
0
0
. . .
0
0
0
1
0
. . .
+ 1 + 1 + 0 +
.
. .
LVM VML MLV VMT
Φ
S
3
(LVMLVM) = ∑
j
σ
j
z
j
= 2
FIGURE 14.4 Representing an amino acid string as a vector. Here we show how a finite
length amino acid string can be represented as a vector. The string VLMVLM is mapped to a
vector Φ
l
s
(VLMVLM) by counting the number of occurrences of different (trimer) substrings
of length 3.
There are as many different algorithms for QSAR as there are applications of
the QSARs themselves. As in bioinformatics, the success of a given QSAR often
depends more on the formulation of the problem than on the algorithm used for the
classification or regression. In the case of QSAR, this boils down to the choice of the
molecular descriptor.
Molecular descriptors used in QSAR are numerous and include examples discussed
earlier in this book such as topological indices, fragmental descriptors and pharma-
cophores (Chapters 2 through 4). Any descriptor may be used in an SVM-based
QSAR, although SVMs are particularly well suited for complex high-dimensional
descriptors such as fragments of labeled graphs that may not be handled as easily
using other methods (e.g., multiple linear regression).
To illustrate the use of labeled graph fragments in SVM-based QSARs, we describe
a simplified version of the method developed by Mahe et al. [77]. The method we
describe here is based on work done to predict protein–chemical interactions [78,79].
Suppose we have two molecular structures that we would like to compare, both given
as labeled graphs, where vertices correspond to atoms and edges correspond to bonds.
Mathematically, we represent such a molecular graph using a vector counting the
number of subgraphs in our original graph. This representation is known as molecular
signature [80–82].
Formally, we definea map Φ
h
g
: {molecular graphs} → Z
N
h
, where N
h
is the number
of possible atomic signatures of height h. As in Section 14.2.1, we denote by z
j
the
bases of Z
N
h
where each basis vector z
j
corresponds to a height h subgraph of a
molecular graph. If M
i
denotes a molecular graph, then Φ
h
g
is given by
Φ
h
g
(M
i
) =
j
σ
j
z
j
, (14.6)
where σ
j
is the number of times that the molecular subgraph corresponding to z
j
occurs in M
i
. Now we define a graph kernel just like we defined the string kernel.
Namely, the graph kernel k
h
g
(M
i
, M
j
) between two molecules M
i
and M
j
is given by
k
h
g
(M
i
, M
j
) = Φ
h
g
(M
i
)
T
Φ
h
g
(M
j
). (14.7)
Machine Learning–Based Bioinformatics Algorithms 393
z
1
z
7
z
2
z
3
z
8
z
4
z
5
z
6
z
1
z
2
z
3
z
4
z
5
z
6
1 C(OHCC)
2 C(OHHC)
5 H(C)
1 H(O)
4 O( N)
1 O(HC)
2 O(NC)
2 N(O O O)
1 C(OHCC)
2 C(OHHC)
5 H(C)
3 O(NC)
3 N(O O O)
6 O( N)
Nitroglycerine 1,2-Dinitroglycerine
N
O
N
O
O
O
O
O
O
HO
O
O
N
O
O
O
O
O
N
N
O
(3,6,3,5,2,1,0,0)
T
(2,4,2,5,2,1,1,1)
T
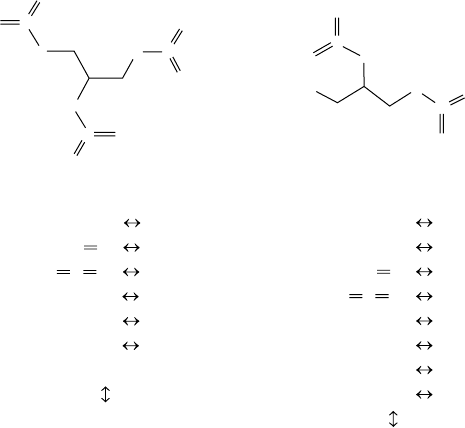
FIGURE 14.5 Height-1 signature representations of nitroglycerine (left) and 1,2-
dinitroglycerine (right). Proceeding from the top to the bottom we show (top row) the molecular
graph representations of the two molecules, (middle row) the number of occurrences of height-
1 atomic signatures and (bottom row) the signature vector representation of the two molecules.
The atomic signature occurrences give the number of times a given chemical fragment occurs in
the molecules. In the case of nitroglycerine, we have three oxygen atoms bonded to a nitrogen
atom and a carbon atom [shown as 3 O(NC)], six oxygen atoms double bonded to a nitrogen
atom [shown as 6 O(
=
N)], three nitrogen atoms bonded to an oxygen atom and double bonded
to two other oxygen atoms [shown as 3 N(O
=
O
=
O)], and so on.
To provide an example, consider the two molecules shown in Figure 14.5. Both
molecules, nitroglycerine and 1,2-dinitroglycerine, are represented as undirected
edge- and vertex-labeled molecular graphs (carbons and hydrogens are implicit). To
obtain vectors from these graphs, we first visit each node in each graph and record the
subgraph formed by that node and its neighbors. This is known as a height 1 atomic
signature. If we wanted to compute height 2 signature, we would have to visit the
neighbors of the neighbors.
The atomic signatures are recorded as strings A
1
(b
2
A
2
b
3
A
3
...), where A
1
is the
vertex type of the root node (the node we are visiting), A
2
is a neighbor of A
1
with bond
type b
2
,A
3
is a neighbor of A
1
with bond type b
3
, and so on. If b
2
, b
3
, ...are single
bonds, then they are omitted. This representation is canonical if we alphabetize the
list (b
2
A
2
, b
3
A
3
, ...) of bonds and atoms [81]. In Figure 14.5, we have written oxygen
bonded to nitrogen and carbon as O(NC) and oxygen double bonded to nitrogen as
O(
=
N).
After visiting each node of each molecular graph, we obtain a list of atomic signa-
ture string representations. This list is identified with a set z
1
, z
2
, ...of basis vectors.
394 Handbook of Chemoinformatics Algorithms
In Figure 14.5, we have identified O(NC) with z
1
and O(
=
N) with z
2
. Using these
basis vectors, we record the number of times a given atomic signature subgraph occurs
in a molecular graph to obtain our molecular signature vector representation. Since
O(NC) occurs three times in nitroglycerine and O(
=
N) occurs six times, the first two
entries of our signature vector for nitroglycerine are 3 and 6. The full vector is (3, 6,
3, 5, 2, 1, 0, 0, 0)
T
. This analysis is also performed on 1,2-dinitroglycerine to get the
signature vector (2, 4, 2, 5, 2, 1, 1, 1)
T
.
Once we have molecular signature vectors for the various molecular graphs
in our dataset, it is a simple matter to compute kernel similarities by taking dot
products of signature vectors. Using the signature vectors for nitroglycerine and 1,2-
dinitroglycerine, we compute a similarity of (3, 6, 3, 5, 2, 1, 0, 0)
T
· (2, 4, 2, 5, 2, 1,
1, 1)
T
= 66.
14.2.3 COMPARISONS
The use of classification and regression are fairly mature in both bioinformatics
and chemoinformatics. The two fields, however, are again benefiting from cross-
fertilization, this time in terms of problem formulation and molecular descrip-
tion. A motivating example is drug design, where we are interested in predicting
protein–chemical interaction [78,83].
One of the standard chemoinformatic approaches to drug design is the use of
virtual screening, where a library of chemicals is examined (e.g., using QSAR)
for a certain activity. In this approach, the corresponding protein is usually fixed
and all attention is focused on the drug. In bioinformatics, it is more com-
mon to consider a network of interactions, as in the case of a protein–protein
interaction network [56]. If we combine both types of approaches, we arrive
at the prospect of predicting a protein–chemical interaction network [78,83].
[Although the general area of large-scale (informatics-based) protein–chemical inter-
action prediction is relatively unexplored, both approaches mentioned here use
SVMs.]
REFERENCES
1. Jain, A., Murthy, M. N., and Flynn, P., Cluster analysis: A review. ACM Comput. Surv.
1999, 31(3), 264–323.
2. Xu, R., and Wunsch, D., Survey of clustering algorithms. IEEE Trans. Neural Netw. 2005,
16(3), 645–678.
3. de Hoon, M. J., Imoto, S., Nolan, J., and Miyano, S., Open source clustering software.
Bioinformatics 2004, 20(9), 1453–1454.
4. Downs, G. M. and Barnard, J. M., Clustering methods and their uses in computational
chemistry. In: Reviews in Computational Chemistry, K. B. Lipkowitz and D. B. Boyd
(eds), Wiley, Hoboken, NJ, 2002; Vol. 18.
5. Krause, A., Stoye, J., and Vingron, M., Large scale hierarchical clustering of protein
sequences. BMC Bioinform. 2005, 6, 15.
6. Loewenstein, Y., Portugaly, E., Fromer, M., and Linial, M., Efficient algorithms for
accurate hierarchical clustering of huge datasets: Tackling the entire protein space.
Bioinformatics 2008, 24(13), i41– i49.
Machine Learning–Based Bioinformatics Algorithms 395
7. Jarvis, R. A. and Patrick, E. A., Clustering using a similarity measure based on shared near
neighbors. IEEE Trans. Comput. 1973, C-22(11), 1025–1034.
8. Rubin, V. and Willett, P., A comparison of some hierarchical agglomerative clustering
algorithms for structure–property correlation. Anal. Chim. Acta 1983, 151, 161–166.
9. Willett, P., A comparison of some hierarchical agglomerative clustering algorithms for
structure–property correlation. Anal. Chim. Acta 1982, 136, 29–37.
10. Willett, P., Evaluation of relocation clustering algorithms for the automatic classification
of chemical structures. J. Chem. Inf. Comput. Sci. 1984, 24(1), 29–33.
11. Willett, P., Barnard, J. M., and Downs, G. M., Chemical similarity searching. J. Chem.
Inf. Comput. Sci. 1998, 38(6), 983–996.
12. Schena, M., Shalon, D., Davis, R. W., and Brown, P. O., Quantitative monitoring of gene
expression patterns with a complementary DNA microarray. Science 1995, 270(5235),
467–470.
13. Shalon, D., Smith, S. J., and Brown, P. O., A DNA microarray system for analyzing
complex DNA samples using two-color fluorescent probe hybridization. Genome Res.
1996, 6(7), 639–645.
14. Eisen, M. B., Spellman, P. T., Brown, P. O., and Botstein, D., Cluster analysis and
display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 1998, 95(25),
14863–14868.
15. Spellman, P. T., Sherlock, G., Zhang, M. Q., Iyer, V. R., Anders, K., Eisen, M. B., Brown,
P. O., Botstein, D., and Futcher, B., Comprehensive identification of cell cycle-regulated
genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell
1998, 9(12), 3273–3297.
16. Wilson, C. S., Davidson, G. S., Martin, S. B., Andries, E., Potter, J., Harvey, R., Ar, K.,
et al., Gene expression profiling of adult acute myeloid leukemia identifies novel biologic
clusters for risk classification and outcome prediction. Blood 2006, 108(2), 685–696.
17. Jiang, D., Tang, C., and Zhang, A., Cluster analysis for gene expression data: A survey.
IEEE Trans. Knowl. Data Eng. 2004, 16(11), 1370–1386.
18. Tavazoie, S., Hughes, J. D., Campbell, M. J., Cho, R. J., and Church, G. M., Systematic
determination of genetic network architecture. Nat. Genet. 1999, 22(3), 281–285.
19. Tamayo, P., Slonim, D., Mesirov, J., Zhu, Q., Kitareewan, S., Dmitrovsky, E., Lander, E.
S., and Golub, T. R., Interpreting patterns of gene expression with self-organizing maps:
Methods and application to hematopoietic differentiation. Proc. Natl. Acad. Sci. USA
1999, 96(6), 2907–2912.
20. Shamir, R. and Sharan, R., Click: A clustering algorithm for gene expression analysis.
In: Proceedings of the 8th International Conference on Intelligent Systems for Molecular
Biology (ISMB), AAAI Press, San Diego, CA, 2000; Vol. 8, pp. 307–316.
21. Ben-Dor, A., Shamir, R., andYakhini, Z., Clustering gene expression patterns. J. Comput.
Biol. 1999, 6(3–4), 281–297.
22. Cheng, Y. and Church, G. M., Biclustering of expression data. In: Proceedings of the 8th
International Conference on Intelligent Systems for Molecular Biology (ISMB), AAAI
Press, San Diego, CA, 2000; Vol. 8, pp. 93–103.
23. Getz, G., Levine, E., and Domany, E., Coupled two-way clustering analysis of gene
microarray data. Proc. Natl. Acad. Sci. USA 2000, 97(22), 12079–12084.
24. Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J., Basic local alignment
search tool. J. Mol. Biol. 1990, 215(3), 403–410.
25. Cohen, A. M. and Hersh, W. R., A survey of current work in biomedical text mining. Brief
Bioinform. 2005, 6(1), 57–71.
26. Janssens, F., Glanzel, W., and DeMoor, B., Dynamic hybrid clustering of bioinformatics by
incorporating text mining and citation analysis. In: Proceedings of the 13th ACM SIGKDD
396 Handbook of Chemoinformatics Algorithms
International Conference on Knowledge Discovery and Data Mining, San Jose, CA, 2007;
pp. 360–369.
27. Rodgers, J. R. and Cebon, D., Materials informatics. Mater. Res. Soc. Bull. 2006, 31.
28. Mayr, L. M. and Fuerst, P., The future of high-throughput screening. J. Biomol. Screen
2008, 13(6), 443–448.
29. Kubinyi, H., From narcosis to hyperspace: The history of QSAR. QSAR 2002, 21(4),
348–356.
30. Selassie, C. D., History of Quantitative Structure–Activity Relationships (6th edition),
Wiley, New York, NY, 2003.
31. Willett, P., Winterman,V., and Bawden, D., Implementation of nonhierarchic cluster anal-
ysis methods in chemical information systems: Selection of compounds for biological
testing and clustering of substructure search output. J. Chem. Inf. Comput. Sci. 1986, 26,
109–118.
32. Doman, T. N., Cibulskis, J. M., Cibulskis, M. J., McCray, P. D., and Spangler, D. P.,
Algorithm5: A technique for fuzzy similarity clustering of chemical inventories. J. Chem.
Inf. Comput. Sci. 1996, 3(6), 1195–1204.
33. McGregor, M. J. and Pallai, P. V., Clustering of large databases of compounds: Using the
MDL “keys” as structural descriptors. J. Chem. Inf. Comput. Sci. 1997, 37(3), 443–448.
34. Menard, P. R., Lewis, R. A., and Mason, J. S., Rational screening set design and compound
selection: Cascaded clustering. J. Chem. Inf. Comput. Sci. 1998, 38(3), 497–505.
35. Shemetulskis, N. E., Dunbar, J. B., Jr., Dunbar, B. W., Moreland, D. W., and Humblet, C.,
Enhancing the diversity of a corporate database using chemical database clustering and
analysis. J. Comput.-Aided Mol. Des. 1995, 9(5), 407–416.
36. Nouwen, J. and Hansen, B., An investigation of clustering as a tool in quantitative
structure–activity relationships (QSARs). SAR QSAR Environ. Res. 1995, 4(1), 1–10.
37. Lawson, R. G. and Jurs, P. C., Cluster analysis of acrylates to guide sampling for toxicity
testing. J. Chem. Inf. Comput. Sci. 1990, 30(2), 137–144.
38. van Geerestein, V. J., Hamersma, H., and van Helden, S. P., Exploiting molecular diversity:
Pharmacophore searching and compound clustering. In: Computer-Assisted Lead Finding
and Optimization, D. H. van de Waterbeemd, P. B. Testa, and P. D. G. Folkers (eds),
Wiley-VCH Basel, Switzerland, 2007; pp. 157–178.
39. Wild, D. J. and Blankley, C. J., VisualiSAR: A web-based application for clustering,
structure browsing, and structure–activity relationship study. J. Mol. Graph. Model 1999,
17(2), 85–89, 120–125.
40. Engels, M. F., Thielemans, T.,Verbinnen, D., Tollenaere, J. P., andVerbeeck, R., CerBeruS:
A system supporting the sequential screening process. J. Chem. Inf. Comput. Sci. 2000,
40(2), 241–245.
41. Dasarathy, B., Nearest Neighbor Pattern Classification Techniques. IEEE Computer
Society, Los Alamitos, CA, 1991.
42. Gurney, K., An Introduction to Neural Networks. CRC Press, London, UK, 1997.
43. Shawe-Taylor, J. and Cristianini, N., Support Vector Machines and other Kernel-Based
Learning Methods. Cambridge University Press: Cambridge, 2000.
44. Fukunaga, K., Introduction to Statistical Pattern Recognition. Academic Press, San
Francisco, CA, 1990.
45. Theodoridis, S. and Koutroumbas, K., Pattern Recognition. Academic Press, San Diego,
CA, 2003.
46. Frank, I. E. and Friedman, J. H., A statistical view of some chemometrics regression tools.
Technometrics 1993, 35(2), 109–135.
47. Montgomery, D. and Peck, E., Introduction to Linear Regression Analysis. Wiley, New
York, 2001.
Machine Learning–Based Bioinformatics Algorithms 397
48. Vapnik, V., Statistical Learning Theory. Wiley: New York, 1998.
49. Smola, A. and Scholkopf, B., A Tutorial on Support Vector Regression; NeuroCOLT NC-
TR-98-030, University of London, UK, 1998.
50. Ben-Hur, A., Horn, D., Siegelmann, H. T., and Vapnik, V., Support vector clustering. J.
Mach. Learn. Res. 2001, 2, 125–137.
51. Bennett, K. and Campbell, C., Support vector machines: Hype or hallelujah? SIGKDD
Explor. 2000, 2(2), 1–14.
52. Burges, C., A tutorial on support vector machines for pattern recognition. Data Min.
Knowl. Discov. 1998, 2, 121–167.
53. Chang, C.-C. and Lin, C.-J., LibSVM: A library for support vector machines,
http://www.csie.ntu.edu.tw/∼cjlin/libsvm, 2001.
54. Joachims, T., Making large-scale SVM learning practical. In: Advances in Kernel
Methods—Support Vector Learning, B. Scholkopf, C. Burges, and A. Smola, (eds). MIT
Press, Cambridge, MA, 1999.
55. Pandey, G., Kumar,V., and Steinbach, M., ComputationalApproaches for ProteinFunction
Prediction. Wiley Book Series on Bioinformatics, Hoboken, NJ, 2008.
56. Shoemaker, B. A. and Panchenko, A. R., Deciphering protein–protein interactions. Part II.
Computational methods to predict protein and domain interaction partners. PLoS Comput.
Biol. 2007, 3(4), e43.
57. Pellegrini, M., Marcotte, E. M., Thompson, M. J., Eisenberg, D., andYeates, T. O.,Assign-
ing protein functions by comparative genome analysis: Protein phylogenetic profiles. Proc.
Natl. Acad. Sci. USA 1999, 96(8), 4285–4288.
58. Ben-Hur, A. and Brutlag, D., Protein Sequence Motifs: Highly Predictive Features of
Protein Function. Springer: Berlin, 2006.
59. Sprinzak, E. and Margalit, H., Correlated sequence-signatures as markers of protein–
protein interaction. J. Mol. Biol. 2001, 311(4), 681–692.
60. Bock, J. R. and Gough, D. A., Predicting protein–protein interactions from primary
structure. Bioinformatics 2001, 17(5), 455–460.
61. Cai, C. Z., Han, L. Y., Ji, Z. L., Chen, X., and Chen, Y. Z., SVM-Prot: Web-based sup-
port vector machine software for functional classification of a protein from its primary
sequence. Nucleic Acids Res. 2003, 31(13), 3692–3697.
62. Wang, C. and Scott, S. D. New kernels for protein structural motif discovery and function
classification, In: Proceedings of the 22nd International Conference on Machine Learning
(ICML), AAAI press, Bonn, Germany, 2005; pp. 940–947.
63. Brown, M. P., Grundy, W. N., Lin, D., Cristianini, N., Sugnet, C. W., Furey, T. S., Ares,
M., Jr., and Haussler, D., Knowledge-based analysis of microarray gene expression data
by using support vector machines. Proc. Natl. Acad. Sci. USA 2000, 97(1), 262–267.
64. Martin, S., Roe, D., and Faulon, J. L., Predicting protein–protein interactions using
signature products. Bioinformatics 2005, 21(2), 218–226.
65. Bradford, J. R. and Westhead, D. R., Improved prediction of protein–protein binding sites
using a support vector machines approach. Bioinformatics 2005, 21(8), 1487–1494.
66. Koike, A. and Takagi, T., Prediction of protein–protein interaction sites using support
vector machines. Protein Eng. Des. Sel. 2004, 17(2), 165–173.
67. Hammett, L. P., The effect of structure upon the reactions of organic compounds. J. Am.
Chem. Soc. 1937, 59(1), 96–103.
68. Taft, R. W., Polar and steric substituent constants for aliphatic and o-benzoate groups from
rates of esterification and hydrolysis of esters. J. Am. Chem. Soc. 1952, 74, 3120–3128.
69. Hansch, C. and Fujita, T., A method for the correlation of biological activity and chemical
structure. J. Am. Chem. Soc. 194, 86, 1616–1626.
