Faulon J.L., Bender A. Handbook of Chemoinformatics Algorithms
Подождите немного. Документ загружается.

368 Handbook of Chemoinformatics Algorithms
Going back to the 1960s, Edmonds and Matula [19] showed that the maximum
common subtree (MCST, in short) between two unordered trees can be computed in
polynomial time, where a tree T is called a maximum common subtree of T
1
and T
2
if T is a subtree of both T
1
and T
2
and T has the maximum number of nodes. Using
efficient computation of bipartite graph matching, their MCST algorithm works in
O(n
2.5
) time [20]. Furthermore, some extensions of the MCST algorithm to more
general graphs have been studied [21,22]. Since it is often useful to regard glycans as
unordered trees, it is reasonable to develop algorithms for the comparison of glycans
based on the MCST algorithm.
However, in order to develop biologically meaningful algorithms, score matrices
such as PAM and BLOSUM and local alignment should also be taken into account
because these two have been used quite successfully in the analysis of the DNA and
protein sequences [23]. Thus, KCaM algorithms were developed by combining the
MCST algorithm, score matrices, and local alignment.
13.3 GLYCAN STRUCTURES
Glycans are special kinds of chemical compounds. The basic component of glycans
is the monosaccharide unit, or sugar, of which a handful are most common in higher
animal oligosaccharides (see Table 13.1). Each unit is linked to one or more other
monosaccharides by various types of linkages, depending on the anomer (i.e., α or β)
and the hydroxyl group numbers to which they are attached on the monosaccharides.
Most glycans have rooted tree structures, where nodes correspond to monosaccha-
rides and edges correspond to linkages between monosaccharides (see Figure 13.4).
Although glycans might be regarded as ordered trees, it is better to regard them as
unordered trees for providing more flexible matching methods.
There are several classes of glycan based on certain basic patterns mainly in the
core structure (i.e., a substructure near to the root), which include N-glycan, O-glycan,
glycoside, and sphingolipid [1]. Parts of these structures are recognized by various
TABLE 13.1
Common Monosaccharide Names, their
Abbreviations, and their Symbols [4]
Sugar Name Abbreviations Symbols
Glucose Glc
Galactose Gla •
Mannose Man *
N-acetyl neuraminic/sialic acid NeuNAc
N-acetylglucosamine GlcNAc
N-acetylgalactosamine GalNAc
Fucose Fuc ,
Xylose Xyl -
Sequence Alignment Algorithms 369
b6
b3
b4a2
b3b4
b6
b3
b4
b4
a3
a2
a3
a2
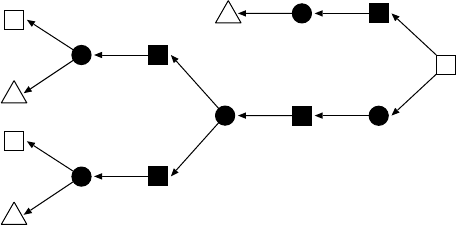
FIGURE 13.4 An example of glycan structure (KEGG Glycan ID: G02178). A label of each
edge denotes a pair of ID numbers of atoms connected by the corresponding chemical bond.
agents such as pathogens and proteins and, thus, are closely related to their functions.
Thus, analysis of tree patterns of glycan structures is important for understanding and
predicting functions of glycans.
13.4 BASIC ALGORITHMS
Before describing KCaM algorithms, we review the MCST algorithm [19,20] and the
global and local sequence alignment algorithms because KCaM algorithms are based
on them.
13.4.1 MCST ALGORITHM
Different from tree edit, we hereafter consider unordered trees. A subtree of tree T
is a tree whose nodes and edges are subsets of those of T. Two trees T
1
and T
2
are
said to be isomorphic if and only if there is a bijection M between nodes in T
1
and T
2
such that the following conditions are satisfied:
• ∀(v
1
, v
2
) ∈ M, label(u) = label(v)
• (u
1
, v
1
) ∈ E(T
1
) if and only if (u
2
, v
2
) ∈ E(T
2
)
Then, the MCST problem is defined as a problem of, given two unordered, labeled,
rooted trees T
1
and T
2
, finding the tree T
c
that is isomorphic to a subtree of both
T
1
and T
2
and whose number of nodes is the maximum among all such possible
trees.
The MCST algorithm is based on dynamic programming. For a node v in tree
T, T(v) denotes the subtree of T induced by v and its descendants. For each pair
(u, v) ∈ V(T
1
) ×V(T
2
), the MCST algorithm computes the size (i.e., the number of
nodes) of the MCST between T
1
(u) and T
2
(v), which is denoted by R[u, v]. M(u, v)
denotes the set of one-to-one mappings between Chd(u) and Chd(v). Then, R[u, v]
can be computed by using the following dynamic programming procedure (see also
370 Handbook of Chemoinformatics Algorithms
u u
u
1
u
2
u
3
u
1
u
2
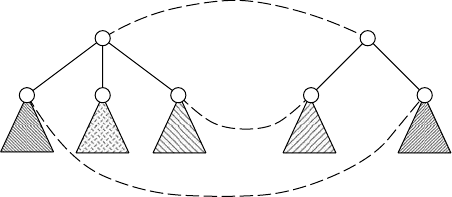
FIGURE 13.5 Explanation of the MCST algorithm. In order to compute score R[u, v], all
possible mappings between {u
1
, u
2
, u
3
} and {v
1
, v
2
} are examined. In this example, M =
{(u
1
, v
2
), (u
3
, v
1
)} is selected as a mapping between Chd(u) and Chd(v) with the maximum
score, where triangles with similar patterns mean similar subtrees.
Figure 13.5):
R[u, v]=
⎧
⎪
⎪
⎨
⎪
⎪
⎩
0, if u = , v = or label(u) = label(v),
1 + max
M∈M(u,v)
⎧
⎨
⎩
(u
i
,v
j
)∈M
R[u
i
, v
j
]
⎫
⎬
⎭
, otherwise,
where denotes an empty tree. It is to be noted that the left-to-right ordering of siblings
is not taken into account in the above (since there is no restriction on mappings M).
Computation of max
M∈M(u,v)
{···}can be carried out in polynomial time by using the
maximum bipartite graph matching [19,20]. The resulting MCST can be retrieved by
using a traceback procedure, where the traceback is a standard technique in dynamic
programming.
13.4.2 GLOBAL AND LOCAL SEQUENCE ALIGNMENT
Sequence alignment is one of the most basic and important algorithms in bioinfor-
matics. Since sequence alignment is explained in detail in another chapter, we briefly
review the algorithms.
Let s = s
1
s
2
...s
m
and t = t
1
t
2
...t
n
be sequences of DNAs or proteins. Global
sequence alignment with linear gap cost is computed by the following dynamic
programming procedure:
S[i,0]=d ·i,
S[0, j]=d ·j,
S[i, j]=max
⎧
⎪
⎪
⎨
⎪
⎪
⎩
S[i, j −1]+d,
S[i − 1, j]+d,
S[i − 1, j −1]+w(s
i
, t
j
),
where d < 0 is a penalty for a gap and w(x, y) is the score between nucleotides or
amino acids x and y. S[i, j] gives the score of an optimal global alignment between
Sequence Alignment Algorithms 371
s
1
s
2
...s
i
and t
1
t
2
...t
j
and thus the score of an optimal global alignment between s
and t is given by S[m, n].
Local sequence alignment is defined as a problem of finding a global sequence
alignment with the maximum score between s
and t
where s
and t
are any consec-
utive subsequences of s and t, respectively. Local sequence alignment with linear gap
cost is computed by the following dynamic programming procedure:
S[i,0]=0,
S[0, j]=0,
S[i, j]=max
⎧
⎪
⎪
⎪
⎪
⎪
⎨
⎪
⎪
⎪
⎪
⎪
⎩
0,
S[i, j −1]+d,
S[i − 1, j]+d,
S[i − 1, j −1]+w(s
i
, t
j
).
In this case, S[i, j]denotes the score of an optimal local alignment ending at (s
i
, t
j
) and
thus the score of an optimal local alignment between s and t is given by max
i,j
S[i, j].
13.5 KCaM ALGORITHMS
KCaM algorithms are combinations of the MCST algorithm and the global/local
sequence alignment algorithms. There are basically two versions, global glycan align-
ment and local glycan alignment, where the former corresponds to global sequence
alignment and the latter corresponds to local sequence alignment. Furthermore, we
have variants of these glycan alignment algorithms in which gaps are not allowed.
The original versions are called approximate matching algorithms and the variants are
called exact matching algorithms. In this section, we begin with global glycan align-
ment, then explain local glycan alignment, and finally briefly explain exact matching
algorithms.
13.5.1 GLOBAL GLYCAN ALIGNMENT
The global glycan alignment algorithm is a simple combination of the MCST algo-
rithm and the global sequence alignment algorithm. More precisely, it is obtained
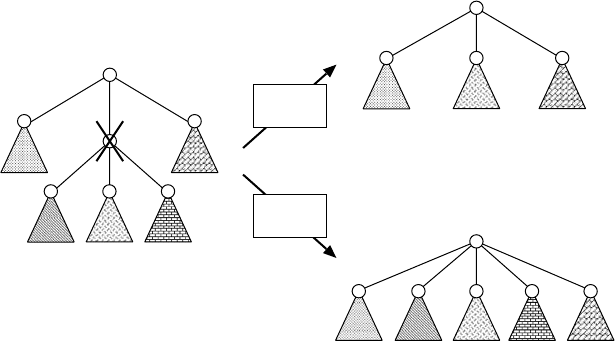
from the MCST algorithm by allowing deletion of a middle node. However, the dele-
tion operation is different from that of tree edit. In tree edit, all children of the deleted
node u become children of the parent of u. However, in glycan alignment, only one
child can be a child of the parent of u and the other children are deleted along with
their descendants (see Figure. 13.6).
Since it would be a bit complicated to define appropriate edit operations, we do
not give edit operations for glycan alignment. Instead, we directly give the dynamic
372 Handbook of Chemoinformatics Algorithms
u
u
1
u
b
u
p
u
p
u
p
u
a
u
2
u
3
u
a
u
2
u
b
u
a
u
2
u
3
u
1
u
b
Glycan
alignment
Tree
edit
FIGURE 13.6 Difference of deletion operations between glycan alignment and tree edit. If u
is deleted, all the children of u become children of the parent of u (denoted by u
p
) in tree edit,
whereas only one child of u can become a child of u
p
in glycan alignment.
programming procedure as follows:
Q[u,0]=
u
i
∈T
1
(u)
d(u
i
),
Q[0, v]=
v
i
∈T
2
(v)
d(v
i
),
Q[u, v]=max
⎧
⎪
⎪
⎪
⎪
⎪
⎪
⎪
⎪
⎨
⎪
⎪
⎪
⎪
⎪
⎪
⎪
⎪
⎩
max
v
i
∈Chd(v)
6
Q[u, v
i
]+d(v) +
v
j
∈Chd(v)−{v
i
}
Q[0, v
j
]
7
,
max
u
i
∈Chd(u)
6
Q[u
i
, v]+d(u) +
u
j
∈Chd(u)−{u
i
}
Q[u
j
,0]
7
,
w(u, v) +max
M∈M(u,v)
⎧
⎪
⎪
⎨
⎪
⎪
⎩
(u
i
,v
j
)∈M
Q[u
i
, v
j
]+
u
k
∈Chd(u)−M
1
Q[u
k
,0]+
v
k
∈Chd(v)−M
2
Q[0, v
k
]
⎫
⎪
⎪
⎬
⎪
⎪
⎭
.
In the above, d(u) denotes the cost for deleting a node u (corresponding to gap penalty
in sequence alignment), w(u, v) represents the similarity between nodes u and v, and
M
i
denotes the set of nodes of T
i
appearing in M. The meanings of three terms (three
terms in outer max) appearing in the right-hand side of the third recursion is as follows.
The first term corresponds to the deletion of v. The second term corresponds to the
deletion of u (see Figure 13.6). The third term corresponds to the matching between
u and v. In order to compute the third term, a maximum score matching between
the children of u and the children of v is computed as in the MCST algorithm (see
Figure. 13.5). However, in this case, the deletion costs for non-matching nodes and
their descendants (represented by Q[u
k
,0] and Q[0, v
k
]) are taken into account.
Sequence Alignment Algorithms 373
As in the case of tree edit, we can relate the score with a kind of mapping or align-
ment. A ⊆ V(T
1
) ×V(T
2
) is called a glycan alignment if the following conditions
are satisfied for any pair (u
1
, v
1
), (u
2
, v
2
) ∈ A:
i. u
1
= u
2
iff. v
1
= v
2
ii. u
1
is an ancestor of u
2
iff. v
1
is an ancestor of v
2
iii. If u
1
∈ V(T
1
) (respectively v
1
∈ V(T
2
)) does not appear in A, at most one
child of u
1
(respectively v
1
) and its descendants can appear in A
It is to be noted that condition (iii) is not included in tree edit mapping. Instead,
the condition on sibling orderings is not included here. We define the score of an
alignment A by
(u,v)∈A
w(u, v) +
u∈V(T
1
)−A
1
d(u) +
v∈V(T
2
)−A
2
d(v),
where A
i
denotes the set of nodes of T
i
appearing in A. Then, the score of an optimal
glycan alignment A is equal to Q[r
1
, r
2
] where r
1
and r
2
are the roots of T
1
and T
2
,
respectively.
In the above, the similarities of edges are not taken into account. However, edges
have important biological meanings. Thus, the similarities of edges should be taken
into account. For that purpose, we define w(u, v) as follows:
w(u, v) = max
⎧
⎪
⎪
⎨
⎪
⎪
⎩
0,
α ·δ(label(u), label(v))
−β ·(1 −δ(ulabel(p(u), u), ulabel(p(v), v)))
−β ·(1 −δ(dlabel(p(u), u), dlabel(p(v), v))),
where p(u) denotes the parent of u, label(u) denotes the name of the monosaccharide
unit or sugar, and ulabel(p(u), u) (resp. dlabel(p(u), u)) indicates
the carbon number
(id) of sugar p(u) (respectively sugar u) to which the edge (p(u), u) is connected. In
the current implementation, we use α = 100.0 and β = 25.0 based on several trials.
13.5.2 LOCAL GLYCAN ALIGNMENT
As in sequence alignment, we can develop a local version of glycan alignment. The
local glycan alignment problem is defined as a problem of finding a global glycan
alignment with the maximum score between T
1
and T
2
where T
1
and T
2
are any
subtrees of T
1
and T
2
, respectively.
The algorithm for local glycan alignment is obtained by combining the local
sequence alignment algorithm and the global glycan alignment algorithm. The
374 Handbook of Chemoinformatics Algorithms
following is its dynamic programming procedure:
Q[u,0]=0,
Q[0, v]=0,
Q[u, v]=max
⎧
⎪
⎪
⎪
⎪
⎪
⎨
⎪
⎪
⎪
⎪
⎪
⎩
0,
max
v
i
∈Chd(v)
6
Q[u, v
i
]+d(v) +
v
j
∈Chd(v) −{v
i
}
Q[0, v
j
]
7
,
max
u
i
∈Chd(u)
6
Q[u
i
, v]+d(u) +
u
j
∈Chd(u) −{u
i
}
Q[u
j
,0]
7
,
w(u, v) +max
M∈M(u,v)
6
(u
i
,v
j
)∈M
Q[u
i
, v
j
]
7
.
13.5.3 EXACT MATCHING ALGORITHMS
There exists another variant of the above mentioned glycan alignment algorithms.
In this variant, gaps are not allowed. Furthermore, the degree of specificity can be
specified by selecting either to match just monosaccharide names (i.e., sugar names)
or both names and linkage information (i.e., sugar names and bond types). As in
approximate matching, there exist two versions: exact global matching and exact
local matching. Since the algorithms are almost the same as the ones for approximate
matching, we do not give algorithms or pseudocodes of the exact algorithms. See
Ref. [3] the details of the exact matching algorithms.
13.6 PSEUDOCODE
Although outlines (i.e., recursions) of the glycan alignment algorithms have been
provided above, some details are unclear. In particular, how to compute maximum
matchings is unclear. Therefore, we here provide pseudocodes of the algorithms. In
these codes, each node is given an ID number, where the numbers are given in the
postorder traversal: the ID number of a parent is larger than the ID numbers of its
children. The node with ID number i (respectively j)inT
1
(respectively T
2
) is denoted
by u
i
(respectively v
j
). For each node with ID number i in T
1
, Chd(i) denotes the set
of ID numbers of the children of u
i
, and ChdID(u
i
, k) denotes the ID number of the
kth child of v
i
. Chd(j) and ChdID(v
j
, k) are defined in the same way for T
2
. For a
tree T and a set of vertices W, T(W) denotes a subtree induced by W.
In both cases, we give codes only for computing the score of an optimal alignment
and do not give codes for retrieving an optimal alignment itself because an optimal
alignment can be obtained by using the standard traceback technique.
13.6.1 CODE FOR GLOBAL GLYCAN ALIGNMENT
We start with the code for global glycan alignment. In this code, GlobalGlycan
Alignment(T
1
, T
2
) is the main procedure. F[i, j] stores the score (i.e., Q[u
i
, v
j
]) for
T
1
(u
i
) and T
2
(v
j
). It is to be noted that F[i,0](respectively F[0, j]) corresponds to the
score for deleting T
1
(u
i
) (respectively T
2
(v
2
)). Match(i, j) is a subroutine to compute
the score of a maximum matching between the children of u
i
and the children of v
j
.
Sequence Alignment Algorithms 375
Since the number of children is small (less than four in most cases), we did not employ
an efficient maximum bipartite matching algorithm. Instead, we employed a simple
exhaustive procedure MatchSub(i
1
, deg0, CurScore). In this procedure, we examine
all complete bipartite matchings between deg0 children of u
i
and deg0 children of v
j
,
where deg0 = max{deg(u
i
), deg(v
j
)} and the score between the i
1
th child of u
i
and
j
1
th child of v
j
is stored in M[i
1
, j
1
]. It is to be noted that if i
1
> deg(u
i
) (respectively
j
1
> deg(v
j
)), the i
1
th (respectively j
1
th) child is regarded as a null child and M[i
1
, j
1
]
denotes the score for deletion of T
2
(v
j
1
) (respectively T
1
(u
i
1
)).
MatachSub(i
1
, deg0, CurScore) computes the scores of all possible matchings in
a recursive manner. It starts with the null matching and extends partial matching
gradually by adding pairs of nodes (including pairs with null nodes) one-by-one.
When MatchSub(i
1
, deg0, CurScore) is called, an assignment for up to the (i
1
−1)th
children of u
i
was already computed and CurScore keeps the score for such a partial
assignment. When i exceeds deg0, a complete matching is obtained and the maximum
score is updated if the current score is greater than the previous maximum score.
Procedure GlobalGlycanAlign(T
1
, T
2
)
begin
Let 1, ···, n
1
be the id numbers of nodes in T
1
in postorder traversal;
Let 1, ···, n
2
be the id numbers of nodes in T
2
in postorder traversal;
for i=1 to n
1
do
F[i, 0]←d(u
i
) +
k∈Chd(i)
F[k, 0];
for j = 1 to n
2
do
F[0, j]←d(v
j
) +
k∈Chd(j)
F[0, k];
for i = 1 to n
1
do
for j = 1 to n
2
do
begin
x
1
← Match(i,j) +w(u
i
, v
j
);
x
2
← min
k∈Chd(j)
{F[i, k]+d(v
j
) +
h=k,h∈Chd(j)
F[0, h]};
x
3
← min
k∈Chd(i)
{F[k, j]+d(u
i
) +
h=k,h∈Chd(i)
F[h, 0]};
F[i, j]←max{x
1
, x
2
, x
3
}
end;
return F[n
1
, n
2
]
end
Procedure Match(i,j)
begin
deg0 ← max{deg(u
i
), deg(v
j
)};
for i
1
= 1 to deg0 do
for j
1
= 1 to deg0 do
if i
1
> deg0 then M[i
1
, j
1
]←F[0, ChdId(v
j
, j
1
)]
else if j
1
> deg0 then M[i
1
, j
1
]←F[ChdId(u
i
, i
1),0
]
else M[i
1
, j
1
]←F[ChdId(u
i
, i
1
), ChdId(v
j
, j
1
)];
for j
1
= 1 to deg0 do ASSIGNED [j
1
]←0;
376 Handbook of Chemoinformatics Algorithms
MatchScore ←−∞;
MatchSub(1,deg0,0.0);
return MatchScore
end
Procedure MatchSub(i
1
, deg0, CurScore)
begin
if i
1
> deg0 then
if CurScore > MatchScore then MatchScore ← CurScore;
else
for j
1
= 1 to deg
0
do
if ASSIGNED[j
1
]=0 then
begin
ASSIGNED[j
1
]←1;
MatchSub(i
1
+1, deg0, CurScore + M[i
1
][j
1
]);
ASSIGNED[j
1
]←0
end
end
Here we briefly analyze the time complexity of this glycan alignment algorithm.
Since the maximum number of children in glycans is bounded by a constant, we can
assume that Match(i, j) works in constant time. Since the main loop of the main pro-
cedure is iterated n
1
n
2
times, the total time complexity of the global glycan alignment
algorithm is O(n
1
n
2
). It is easily seen that the space complexity of the algorithm is
also O(n
1
n
2
).
13.6.2 MODIFICATION FOR LOCAL GLYCAN ALIGNMENT
The code for global glycan alignment can be modified for local glycan alignment.
The required modification is simple and is a direct consequence of the change of the
recursion.
We first modify the initialization part for F[i,0] and F[0, j] as below.
for i = 1 to n
1
do F[i,0]←0;
for j = 1 to n
2
do F[0, j]←0;
Next, we replace “F[i, j]←max{x
1
, x
2
, x
3
}” with “F[i, j]←max{x
1
, x
2
, x
3
,0}.”
Finally, we replace “return F[n
1
, n
2
]” with “return max
i,j
F[i, j].” As in the case of
global glycan alignment, the time and space complexities of the local glycan alignment
algorithm are O(mn).
13.7 ILLUSTRATIVE EXAMPLE
Here we provide an example for illustrating the global glycan alignment algorithm.
We employ two glycan data with KEGG Glycan ID numbers G06886 and G05554
(see also Figure 13.7).
