Blake A.J.(ed.) Crystal Structure Analysis
Подождите немного. Документ загружается.

172 Refinement of crystal structures
13.1.4 Structure factor from a parameterized model
F
hkl
≈
f
j
.
e
2πi(hx
j
+ky
j
+lz
j
)
. (13.4)
The continuous electron density in a unit cell can be replaced by a
parameterized model, that provides a convenient approximate repre-
sentation of the true electron density. It is this model which is normally
called the ‘crystal structure’. Remember that X-rays do not see atoms –
they see average electron density. We replace this by discrete atoms as
a convenience.
13.2 Reasons for performing refinement
Refinement usually means adjustment of the values in the param-
eterized model according to some criteria. Both Fourier and least-
squares methods are regularly applied in crystallography. Refinement
is undertaken for a number of reasons, as follows.
13.2.1 To improve phasing so that computed electron
density maps more closely represent the actual
electron density
Commonly used variations of (13.3) are as follows.
F = F
o
, α = α
c
. The resultant map contains features from both the
observed intensities and the computed phases. It is easily demonstrated
that the phases have a profound influence on the features in the map,
so that their veracity is fundamental. Better phases lead to better maps,
which in their turn can lead to better phases, and so on.
F = F
o
− F
c
, α = α
c
. The ‘difference map’. If the structure amplitudes
generated from the model lead to F
c
values differing only from the
observed amplitudes by their random errors, then this map will be fea-
tureless. This criterion is normally assumed to indicate that the structure
has been properly parameterized, and it is assumed that, if F
c
is the
same as F
o
, then α
c
will be the same as the (unmeasured) α
o
. The prob-
lem is that there are unspecified (and possibly systematic) errors in F
o
.
The Fourier transform of the residual F
o
− F
c
and α
c
may contain fea-
tures that will enable the model to be improved. Isolated positive peaks
indicate atoms missing from the model. Positive peaks adjacent to neg-
ative peaks indicate misplaced atoms, and positive or negative peaks
surrounded by ripples indicate either inappropriate atomic scattering
factors or inappropriateisotropic displacement parameters. Pairs of pos-
itive and negative peaks forming a ‘clover leaf’ indicate inappropriate
anisotropic displacement parameters.
F = 2F
o
− F
c
or F = 3F
o
− 2F
c
, α = α
c
. Hybrid maps having proper-
ties in common with both the normal F
o
map and a difference map. They
reveal features from the current model, and as-yet unparameterized
13.2 Reasons for performing refinement 173
features, and are commonly used in macromolecular crystallography. If
the data are subject to a θ cut-off before the majority of the reflections
are unobservably weak, the map will show series termination ripples
with a wavelength slightly less than the resolution limit.
F = xF
o
.(1 − x)E, α = α
c
. E is the normalized structure amplitude, as
used in direct methods, so that the peaks appearing in the map are
accentuated. The optimal value for x is resolution dependent.
The phases, α
c
, are generally obtained from the current parameter-
ized model, which may itself have been refined by either Fourier or
least-squares methods. Rarely, the computed phase may contain a con-
tribution from the discrete Fourier transform of part of an existing map
(SQUEEZE in PLATON; Spek, 2003). Interpretation of maps is the most
common way of locating items missing from the parameterized model,
though other forms of model building may also be applicable (addition
of hydrogen atoms at their expected positions, geometric completion of
other regular shapes, e.g. PF
−
6
groups).
13.2.2 To try to verify that the structure is ‘correct’
Because there is no way of directly and unambiguously computing the
structure from the data, there is always the possibility that the proposed
structure is ‘wrong’. Thereare broadlytwo waysof assessingthe validity
of a structure.
1. From the X-ray data alone. This is the most fundamental method,
and is the only one applicable to totally novel materials. It is also
the most difficult method to apply. Techniques for parameter opti-
mization that rely on fitting F
c
to F
o
cannot be certain that there
is not another solution giving about the same goodness of fit for
the amplitudes, but a better fit for the (unmeasured) phases. In
addition, in reality we know little about the actual errors in the
observed data, and there is the risk that over-parameterization of
the model will simply result in a model that better fits the uniden-
tified errors. In protein crystallography, where there is a very real
possibility that a structure will be novel (in that there are still no
reliable ways for predicting protein folding), R
free
is used to mon-
itor parameterization. A random subset of the data, often 10%, is
excluded from the parameterization stage of the refinement, but
has its value calculated whenever there is a substantial change in
the modelling. If R
free
fails to fall by a significant amount, it is prob-
able that the change in the modelling is following noise in the data
(the signal represents a common trend throughout all the data, the
noise is individual to each data point, though systematic errors
can be regarded as correlated noise). R
free
is rarely used in small-
molecule work since 10% of a typical data set does not contain
enough reflections to give a reliable estimator.
2. From comparisons with the ‘known’ properties of structures. This
is a Bayesian method, but requires that the analyst correctly
174 Refinement of crystal structures
identify which properties are known, and what their values are.
It cannot reveal if a structure is ‘right’, but may reveal that it is
‘wrong’.
‘Wrongness’ comes in several forms.
(a) Correct local geometry, but the whole structure is misplaced
in the cell. This phenomenon was common with early direct
methods programs, and may also occur for structures solved
by Patterson methods when the ‘heavy’ atom is not all that
heavy. Symptoms include the following.
i. The structure looks generally OK, but gives a high R factor,
often as low as 20%, but failing to fall any lower.
ii. Unusual bond lengths and adps. These rarely fall into a
systematic pattern, as might be the case for refinement in a
space group of too-low symmetry.
iii. Unreasonable intermolecular contacts. Translational mis-
placement of a structure within a cell will generally bring
it too close to a symmetry element, leading to very short
non-bonded contacts.
iv. Noisy difference maps. The difference map may be gener-
ally noisy, show some inexplicable peaks, or occasionally
show ghost structural fragments.
v. Strongly featured DIFABS map
1
or highly anisotropic R-
1
DIFABS fits a Fourier series in polar co-
ordinates in reciprocal space to the ratio
of F
o
:F
c
. The value of this function can be
plotted out to reveal if this ratio deviates
stronglyfrom unity in any partofreciprocal
space (Walker and Stuart, 1983).
tensors.
2
These can also be due to uncorrected systematic
2
The R-factor tensor is a tensor that rep-
resents the variation of local R-factor as
a function of direction in reciprocal space
(Parkin, 2000).
errorsin the data, e.g. absorption, crystal decay, crystal mis-
centring, icing, beam inhomogeneity.
(b) Incorrect but plausible local geometry. This is relatively rare in
small-molecule crystallography. The most common occurrence
is generally due to disorder. Symptoms are as follows.
i. R factor higher than anticipated, though often as decep-
tively low as 6–8%.
ii. Novel molecular features. These must sometimes occur,
otherwise there is little point in much structure analysis.
However, if they cannot be rationalized by accepted chem-
ical or physical reasoning,there remains the possibility that
the structure is false.
iii. Weird adps. Weird may mean unexpectedly large, small or
anisotropic. Usually, if something is simply ‘wrong’, there
is no evident relationship between the adps of adjacent
atoms.
iv. A few particularly large F
o
− F
c
discrepancies, though the
most common cause for this is some kind of failure in the
data collection or pre-processing.
v. Noisy difference maps. The maps are generally rather fea-
turelessexcept for a few substantial peaks, though cases are
known where the maps were quite featureless even though
subsequent events showed that the proposed structure was
in serious error.
13.3 Data quality and limitations 175
13.2.3 To obtain the ‘best’ values for the
parameters in the model
Once the analyst is convinced that the structure is essentially correct,
weighted least-squares refinement is used to optimize the parameter
values.
13.3 Data quality and limitations
The quality of the data should be as high as is economically practical.
Spending a little time choosing a good crystal from a mixed batch is time
well spent, though it also harbours the risk that the chosen crystal is not
truly representative. Spending very long times on data collection is use-
ful only if the crystal is of excellent quality, since the signal-to-noise ratio
only doubles if the counting time is increased by a factor of 4. Factors
arising in data collection that can affect later refinement are as follows.
13.3.1 Resolution
With copper radiation and organic molecules, useful data can usually
be measured to the instrument θ limit. With molybdenum radiation,
the operator needs to use experience. If there are a few heavy ele-
ments present, they will dominate the high-angle data, and they should
be measured to reduce series termination (diffraction ripple) effects.
If there are only light atoms, the mean (Wilson) temperature factor B
will show when reflections become indistinguishable from background.
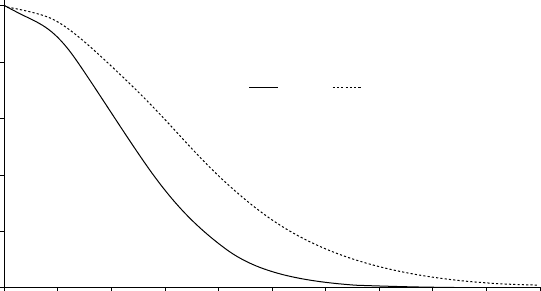
Figure13.3showsthevariation of relativeintensity as a function ofBragg
angle for B values of 4 and 8.
0
1
0.8
Variation of mean intensity (I/I
o
)
as a function of theta
Theta
B = 8 B = 4
Mean intensity, I/I
o
0.6
0.4
0.2
0
5 101520253035404550
Fig. 13.3 The variation of mean intensity as a function of Bragg angle for two different B
values.
176 Refinement of crystal structures
Lack of properly measured high-angle data will restrict the quality
of the analysis. However, if the Wilson temperature factor is unusu-
ally high, this indicates that the crystal itself is of poor quality, with
substantial static or dynamic disorder. Re-collection of the data at a
lower temperature should reduce dynamic disorder. Unless there is
a phase change with conservation of crystallinity, struggling to work
at extremely low temperatures rarely has much influence on this
type of problem. The best strategy is either to try to understand the
nature of the disorder, or to try growing better crystals by a different
method. Simply including high-angle refections that are indistinguish-
able from noise into a refinement in order to achieve some required
observation-to-parameter ratio is neither productive nor desirable.
13.3.2 Completeness
All kinds of refinement are to some extent sensitive to the systematic
omission of substantial sections of the reciprocal lattice. Fourier meth-
ods, depending upon a summation over the whole reciprocal lattice, are
most sensitive to missing reflections. Least-squares methods are more
robust. Loss of data in the ‘cusp’ region of some diffractometers is rarely
important. More serious is not collecting data to similar resolution in all
directions in the asymmetric unit of reciprocal space.
13.3.3 Leverage
While all the reflections should be used to compute the electron density
at any point in the unit cell, in least-squares refinement some reflec-
tions will have a particular influence on certain parameters. This effect is
known as leverage. Once a structure has been approximately solved, it is
possible to determine which reflections are important for which param-
eters. These reflections can be carefully remeasured and introduced into
the refinement with appropriate weights or used as discriminators to
test the effectiveness of new parameterizations.
13.3.4 Weak reflections and systematic absences
As noted above, high-angle weak reflections have very little information
content. Weak lower-angle reflections can be really important, for exam-
ple in deciding between a centrosymmetric or non-centrosymmetric
space group. The problem is to get systematic-error-free determinations
of these reflections. Programs that perform analysis of the systematic
absences generally reveal that, within a given θ range, these reflections
have a net positive value, i.e. are not strictly absent. Reasons for this
include the Renninger effect, thermal diffuse scattering, λ/2 contamina-
tion and deviations from strict space group symmetry. Thesenet positive
values for absences suggest that all weak low-angle reflections may be
systematically in error, a condition that cannot be properly handled by
least-squares refinement.
13.4 Refinement fundamentals 177
13.3.5 Standard uncertainties
Even when most data were collected on serial diffractometers there were
differences of opinion about the correct way to compute the standard
uncertainties, particularly of the very strong and very weak reflections.
With the advent of area detectors, with their very sophisticated numer-
ical data pre-processing, the uncertainty in the uncertainty has grown.
Even the massive redundancy possible with these machines creates a
false sense of security (remember the parable of the Emperor of China’s
new robes
3
). If the errors in the individual observations are large but
3
Discussedinanarticle‘How precise are mea-
surements of unit-cell dimensions from single
crystals?’ (Herbstein, 2000).
more or less randomly distributed about the ‘true’ value, then repeated
measurements will yield a result approaching the true result (central
limit theorem). In this case, even if R
int
is large, the structure can be well
refined to a reasonably trustworthy solution. If the individual observa-
tions are highly reproducible, but systematically wrong, this will lead
to a low R
int
, but an incorrect final structure.
13.3.6 Systematic trends
One might expect that, for a given instrument, all structures of a similar
constitution would refine to about the same R value, and this is often
the case. In those cases where the R value is anomalously low, we might
suspect that the R factor is dominated by an anomalous feature in the
structure, such as the presence of heavy atoms. In those cases where
it is anomalously high, the analyst should seek an explanation. The
following are possible explanations.
i. Weak data, i.e. the intensities disappear into the background at a
relativelylow θ angle.This can be due to static or dynamic disorder,
poor crystallinity, or large solvent content.
ii. Features in the data that cannot be represented by the normal struc-
tural model. Either the experiment should be repeated, taking care
to avoid these unwanted trends in the data, or some kind of ‘cor-
rection’ should be applied to the data to remove them. Statistically,
this kind of tampering with the data is not recommended, and the
statistician would prefer the model to be extended to reflect these
perturbations in the data. In practice, it turns out that the tinker-
ing works reasonably well (e.g. empirical absorption corrections,
SADABS-, DIFABS- and SORTAV-like procedures).
13.4 Refinement fundamentals
For the bulk of small-molecule structures, refinement means some
process related to least squares. The process seeks to minimize some
function
M =
w(Y
1
− Y
2
)
2
. (13.5)
178 Refinement of crystal structures
13.4.1 w, the weight
For a statistically well-understood problem, it can be shown that, for
uncorrelated errors in the observations, the optimal parameter values
and their standard uncertainties are obtained when the weight is equal
to the inverse of the variance of the observation. In crystallography,
we are by no means certain about the distribution of the errors in our
observations, nor can we be certain that our model is capable of fully
explaining the observations; for example, there may be systematic errors
in the data that should be accounted for by additional parameters in the
model. Weights are now usually chosen to satisfy the criterion that there
is no appreciable trend in M for any rational ranking of the data. This
enables the weights to reflect errors in both the data and the model, and
leads to optimal parameter values and s.u.s for that model. Computing
these model-dependent weights when the model is substantially incor-
rect may lead to the error being concealed. It is therefore recommended
that purely statistical or modified unit weights be used in the early
stages. Note that unit weights are robust for F
o
refinement, and should
be replaced by (1/2F) or 1/σ
2
(F
2
o
) for F
2
o
refinement. Note also that the
assumption that the errors in observations are uncorrelated is generally
unfounded, though no widely available single-crystal programs use the
full data variance-covariance matrix.
Omission of data according to some s.u. threshold is a brutal way of
down-weighting (to zero!) the weak reflections, and in principle can-
not be justified. However, there is little justification either in including
hundreds of high-angle data that are indistinguishable from noise. An
appropriate θ cut-off is more acceptable than an I/σ (I) cut-off. The small
numbers of weak reflections remaining in the refinement, even if posi-
tively biased as explained above, probably do little harm, and may even
be required for some kinds of analysis.
Weighting schemes can be modified in various ways to accelerate
convergence, to reduce the influence of outliers, or to enhance some
feature of the structure.
13.4.2 Y
1
, the observations
The community is still divided as to whether Y
1
should be F
o
, F
2
o
or I. The
debate is finely balanced, and in practice perfectly acceptable results are
obtained whichever representation is used. Traditional nomenclature,
in which the structure factor is written |F
o
|, has added to the confusion.
The moduli signs were introduced to remind the reader that one can
measure only the magnitude of the diffracted beam but not the phase. It
has nothing todo with takingthe square root of F
2
. Negative F values are
permitted in least-squares refinement. The application of a non-linear
transformation (taking the square root) to the data raises several issues.
Rollett and Prince have independently shown that, with appropri-
ate weighting, both F and F
2
refinements yield the same parameters
and s.u.s.

13.4 Refinement fundamentals 179
Non-linear transformations of the data lead to a skewing of the error
distribution. For medium and strong reflections, a 1/2F term success-
fully accommodates this, but there is an evident problem if F is ≤ 0.
In the absence of a proper knowledge of the error distribution in F
2
for weak reflections, an approximate value has to be estimated for the
s.u. of F
o
.
Why was refinement against F
o
ever introduced? Transformation of
data or variables is a technique commonly used to improve the numer-
ical stability of a calculation, which would certainly have been an issue
before digital computers. In the case of transformation from F
2
to F,
except for very small reflections, unit weights are close to the optimal
weighting, again an advantage before computers. Neither of these con-
siderations is important now for routine work, though F refinement
remains resistant to the effects of outliers (especially partially occluded
observations), particularly among the strong reflections.
Although the minima for both refinements should be the same, the
pathsfromagiven starting model willbedifferent.Therearesuggestions
that F
2
refinement is less influenced by false minima, but I have only
seen contrived artificial examples of this.
Contrary to common belief, for the software user F
2
refinement has no
advantage over F
o
refinement for either determining the Flack parame-
ter, or treating twinned data. The programmer has a little more work to
do in the case of F
o
refinement.
Note that the F versus F
2
debate is quite independent of the debate
about the inclusion/exclusion of weak reflections.
13.4.3 Y
2
, the calculations
This is generally F
c
or F
2
c
to match Y
1
. In maximum-likelihood refine-
ment, the following function is minimized:
M =
1
σ
2
ML
(F
o
−
F
o
)
2
. (13.6)
F
o
is the expected value of F
o
, and is a modified form of F
c
.Itis
intended to include a contribution to the minimization function due to
the uncertainty in the phase angle, and seems to be particularly useful
in protein crystallography, where the models rarely achieve the same
resolution as those for small molecules. As the model improves, F
o
approaches F
c
, and such is the power of modern direct methods pro-
grams that it is generally believed that initial structures are so good
that maximum-likelihood methods have little to offer small-molecule
crystallographers.1/σ
ML
isa special weighting function. Maximumlike-
lihood may have somerolein cases of pseudo-centred cells, where a very
large percentage of the data is systematically weak. Edwards (1992) has
shown that maximum-likelihood least squares is unaffected by the F
2
to F transformation.
180 Refinement of crystal structures
13.4.4 Issues
During least-squares refinement, (13.7) is solved. There are some
important things to notice.
i. The observations of restraint are handled in the same way as the
X-ray observations.
ii. The matrix of constraint is applied to the whole matrix of deriva-
tives, so will override any conflicting restraints.
iii. The shift-limiting restraints are included as equations in the matrix
work – they influence the terms in the matrix, and hence its
numerical processing.
δ
x
applied
= P.(M
.A
.W.A.M)
−1
.M
.A
.W.Y
A =
⎛
⎝
∂F
o
/∂x
∂R
t
/∂x
1
⎞
⎠
A is the matrix of derivatives
R
t
is a restraint target value
Y =
F
o
−F
c
R
t
−R
c
0.0
Y is the vector of residuals
x
applied
= P.δ
x
leastsquares
P is the matrix of partial shifts
x
physical
= M.x
leastsquares
+ c
M is the matrix of constraint
c is a vector of constants.
(13.7)
iv. The matrix of partial shifts is applied after the matrix inversion,
and so cannot help in the control of singularities.
v. The terms in the design matrix do not involve the values of the
observations, only terms computed from the current model. If the
model is seriously wrong, these terms will also be seriously wrong.
vi. The vector of residuals involves both the actual observations and
values calculated from the model. If the values of F
c
are hopelessly
wrong, these residuals will drive the refinement towards a false
minimum.
13.5 Refinement strategies
If there were a known, single, reproducible refinement strategy, it would
have been programmed long ago. In the 1970s it was hoped that such a
strategy was on the point of being discovered. Now, 30 years later, com-
puterized strategies do exist for the processing of good-quality data
from well-behaved structures, but there is an ever-growing number
of structures requiring some kind of human insight and intervention.
If a structure does not develop and refine in the normal way, some
13.5 Refinement strategies 181
possible actions are listed below. A refinement ‘blows up’ when the
R factor begins to rise uncontrollably, or the displacement parameters
take on nonsensical values, or massive parameter shifts make the struc-
ture unrecognizable. At later stages, it can also mean that the extinction
parameter or some displacement parameters have gone substantially
negative.
The crucial thing to remember about refinement is that both Fourier
and least-squares methods have limited ranges of convergence. Both
(13.3) and (13.7) involve the current model. The better the currentmodel,
the greater the chance of computing reliable estimates of changes to
make to the parameters. In difficult cases, model development should
be undertaken cautiously, with non-crystallographic information used
to hold parameters at sensible values. Many cases are known where the
structure solution was difficult, yet for which the final fully parame-
terized model refined quite stably. Exclusion of valid atoms is generally
less harmful than inclusion of false ones, and in any case the valid atoms
generally reappear in subsequent maps.
i. If the direct methods solution does not appear to reveal the struc-
ture, try changing some of the initial parameters, or try changing
program. If SIRxx initially shows a promising structure, which falls
to pieces during the automatic refinement, turn off the refinement
and process the initial solution manually. Note that very poor data
or data with systematic errors can yield an interpretable E-map that
may never refine well.
ii. If the direct methods give a reasonable figure of merit but the
structure looks wrong, try alternative programs for assembling
molecules. Molecule assembly routines are affected in different
ways by missing or spurious peaks. If this fails, compute a pack-
ing diagram and set the bonding criteria to a little more than their
usual values. Packing diagrams are especially useful if the molecule
straddles symmetry elements. A chemist, knowing what to look
for, may spot molecular fragments amongst a jumble of spurious
peaks.
iii. If the structure is plausible overall, but ‘blows up’ on normal refine-
ment, try Fourier refinement. For Fourier refinement it is best to
omit really dubious atoms from the model used to compute phases.
iv. If the main features are recognizable, use geometric model build-
ing to regularize the molecular parameter values (bonds, angles,
planarity).
v. Least squares can be used to identify potentially spurious atoms.
A few cycles of refinement of U
iso
(with fixed x, y, z) may reveal
spurious atoms as those with very high values.
vi. Least squares has no mechanism for introducing new atoms. This
can be done only by Fourier methods or model building. 2F
o
− F
c
or 3F
o
−2F
c
maps are often the easiest to interpret.
vii. If the model persists in blowing up, try increasing the effect of
shift-limiting restraints. Fix the isotropic displacement parameters
