Vlak J.M., de Gooijer C.D., Tramper J., Miltenburger H.G. (Eds.) Insect Cell Cultures: Fundamental and Applied Aspects
Подождите немного. Документ загружается.


122
Production of libraries of very high complexity will
require further advances in baculovirus recombinant
DNA technology. For instance, it may be possible to
achieve efficient conversion of plasmid into virus and
a high yield of recombinants by combining the in vit-
ro site-specific recombination system with the Ganci-
clovir, p35
,
or ORF1629 selections for recombinant
viruses.
Acknowledgments
I am grateful to Kristen Mayo for reviewing this man-
uscript.
References
Abremski K, Hoess R & Sternberg N (1983) Studies on the proper-
ties of P1 site-specific recombination: evidence for topologically
unlinked products following recombination. Cell 32: 1301–1311.
Ayres MD, Howard SC, Kuzio J, Lopez-Ferber M & Possee RD
(1994) The complete DNA sequence of Autographa californica
nuclear polyhedrosis virus. Virology 202: 586–605.
Bonning BC & Hammock BD (1992) Development and potential
of genetically engineered viral insecticides. In: Tombs MP(ed.)
Biotechnology and genetic engineering reviews, Vol. 10(pp.455–
489) Intercept, Andover.
Clem RJ, Fechheimer M & Miller LK (1991) Prevention of apoptosis
by a baculovirus gene during infection of insect cells. Science
254: 1388–1390.
Copeland WC & Wang TS-F (1993) Mutational analysis of the
human DNA polymerase a. J. Biol. Chem. 268: 11028–11040.
Day A, Wright T, Sewall A, Price-Laface M, Srivastava N & Fin-
layson M (1995) Rapid procedures for the isolation and PCR
analysis of recombinant baculovirus. In: Richardson (ed.) Meth-
ods in Molecular Biology, Vol. 39: Baculovirus Expression Pro-
tocols (pp. 143–159) Humana Press, Totowa, NJ.
Davies AH (1994) Current methods for manipulating baculoviruses.
Bio/Technology 12: 47–50.
Ernst WJ, Grabherr RM & Katinger HWD (1994) Direct cloning
into the Autographa californica nuclear polyhedrosis virus for
generation of recombinant baculoviruses. Nucleic Acids Res. 22:
2855–2856.
Fung M-C, Chiu KYM, Weber T, Chang T-W & Chang NT (1988)
Detection and purification of a recombinant human B lym-
photropic virus hhv–6 in the baculovirus expression system by
limiting dilution and DNA dot-blot hybridization. J. Virol. Meth.
19: 33–42.
Godeau F, Saucier C & Kourilsky P (1992) Replication inhibition
by nucleoside analogues of a recombinant Autographa californi-
ca multicapsid nuclear polyhedrosis virus harboring the herpes
thymidine kinase gene driven by the IE–1(0) promoter: a new
way to select recombinant baculoviruses. Nucleic Acids Res. 20:
6239–6246.
Granados RR & Federici BA (1986) The biology of baculoviruses,
Vol. II. CRC Press, Boca Raton, Florida.
Grosse F & Manns A (1995) Fast isolation of recombinant bac-
ulovirus by antibody screening. In: Richardson (ed.) Methods in
Molecular Biology, Vol. 39: Baculovirus Expression Protocols
(pp. 179–185) Humana Press, Totowa, NJ.
Hartig PC & Cardon MC (1992) Rapid efficient production of bac-
ulovirus expression vectors. J. Virological Methods 38: 61–70.
Hershberger PA, Dickson JA & Friesen PD (1992) Site-specific
mutagenesis of the 35-kilodalton protein gene encoded by Auto-
grapha californica nuclear polyhedrosis virus: cell line-specific
effects on virus replication. J. Virol. 66: 5525–5533.
Hoess R & Abremski K (1984) Interaction of the bacteriophage
P1 recombinase Cre with the recombining site loxP. Proc. Natl.
Acad. Sci. USA 81: 1026–1029.
Kitts PA (1995) Production of recombinant baculoviruses using lin-
earized viral DNA. In: Richardson (ed.) Methods in Molecular
Biology, Vol. 39: Baculovirus Expression Protocols (pp. 129–
142) Humana Press, Totowa, NJ.
Kitts PA, Ayres MD & Possee RD (1990) Linearization of bac-
ulovirus DNA enhances the recovery of recombinant virus expres-
sion vectors. Nucleic Acids Res. 18: 5667–5672.
Kitts PA & Possee RD (1993) A method for producing recombinant
baculovirus expression vectors at high frequency. BioTechniques
14:810–817.
Lalumière M & Richardson CD (1995) Production of recombinant
baculoviruses using rapid screening vectors that contain the gene
for In: Richardson (ed.) Methods in Molecular
Biology, Vol. 39: Baculovirus Expression Protocols (pp. 161-
177) Humana Press, Totowa, NJ.
Lerch RA & Friesen PD (1993) The 35-kilodalton protein gene (p35)
of Autographa californica nuclear polyhedrosis virus and the
neomycin resistance gene provide dominant selection of recom-
binant baculoviruses. Nucleic Acids Res. 21: 1753–1760.
López-Ferber M, Sisk WP & Possee RD (1995) Baculovirus trans-
fer vectors. In: Richardson (ed.) Methods in Molecular Biology,
Vol. 39: Baculovirus Expression Protocols (pp. 25–63) Humana
Press, Totowa, NJ.
Luckow VA, Lee SC, Barry GF & Olins PO (1993) Efficient gen-
eration of infectious recombinant baculoviruses by site-specific
transposon-mediated insertion of foreign genes into a baculovirus
genome propagated in Escherichia coli. J. Virol. 67: 4566–4579.
Manns A & Grosse F (1991) Fast isolation of recombinant bac-
ulovirus by antibody screening. BioTechniques 10: 154–158.
Martens JWM, van Oers MM, van de Bilt BD, Oudshoorn P & Vlak
JM (1995) Development of a baculovirus vector that facilitates
the generation of pl0-based recombinants. J. Virological Meths.
52: 15–19.
Patel G, Nasmyth K & Jones N (1992) A new method for the isolation
of recombinant baculovirus. Nucleic Acids Res. 20: 97–104.
Peakman T, Page M & Gewert D (1989) Increased recombinational
efficiency in insect cells irradiated with short wavelength ultra-
violet light. Nucleic Acids Res. 17: 5403.
Peakman TC, Harris RA & Gewert DR (1992) Highly efficient gen-
eration of recombinant baculoviruses by enzymatically mediated
site-specific in vitro recombination. Nucleic Acids Res. 20: 495–
500.
Pen J, Welling GW & Welling-Wester S (1989) An efficient proce-
dure for the isolation of recombinant baculovirus. Nucleic Acids
Res. 17:451.
Peng S, Sommerfelt MA, Berta G, Berry AK, Kirk KL, Hunter E &
Sorscher EJ (1993) Rapid purification of recombinant baculovirus
using fluorescence-activated cell sorting. BioTechniques 14: 274–
277.
Pennock GD, Shoemaker C & Miller LK (1984) Strong and regulated
expression of Escherichia coli in insect cells with
a baculovirus vector. Mol. Cell. Biol. 4: 399–406.

123
Pham DQ-D, Hice RH, Sivasubramanian & Federici BA (1993)
The 1629-bp open reading frame of the Autographa californica
multinucleocapsid nuclear polyhedrosis virus encodes a virion
structural protein. Gene 137: 275–280.
Possee RD, Sun T-P, Howard SC, Ayres MD, Hill-Perkins M &
Gearing KL (1991) Nucleotide sequence
o
f the Autographa cali-
fornica nuclear polyhedrosis 9.4 kbp EcoRI-I and -R (polyhedrin
gene) region. Virology 185: 229–241.
Richardson C, Attia J, Dunn R, Gupta S, O’Connor M, Semeniuk D,
Tam J, Hamel M, Lambert G, Dennis M, Jacobs F, Martin L, Iorio
C & Vialard J (1992a) Engineering glycoproteins for secretion
using the baculovirus expression system. In: Vlak JM, Schlaeger
E-J & Bernard AR (eds) Baculoviruses and recombinant protein
production processes (pp. 67–74) Roche, Basel.
Richardson CD, Banville M, Lalumière M, Vialard J & Meighen
EA (1992b) Bacterial luciferase produced with rapid screening
baculovirus vectors is a sensitive reporter for infection of insect
cells and larvae. Intervirology 34: 213–227.
Smith GE, Fraser MJ & Summers MD (1983) Molecular engineering
of the Autographa californica nuclear polyhedrosis virus genome:
deletion mutations within the polyhedrin gene. J. Virol. 46: 584–
593.
Sternberg N & Hoess R (1983) The molecular genetics of bacterio-
phage P1. Ann. Rev. Genet. 17: 123–154.
Summers MD & Smith GE (1987) A manual of methods for bac-
ulovirus vectors and insect cell culture procedures. Texas Agri-
cultural Experiment Station Bulletin No. 1555.
Vialard J, Lalumière M, Vernet T, Briedis D, Alkhatib G, Henning
D, Levin D & Richardson C (1990) Synthesis of the membrane
fusion and hemagglutinin proteins of measles virus, using a novel
baculovirus vector containing the gene. J. Virol.
64: 37–50.
Vialard JE & Richardson CD (1993) The 1,629-nucleotide open
reading frame located downstream of the Autographa cali-
fornica nuclear polyhedrosis virus polyhedrin gene encodes a
nucleocapsid-associated phosphoprotein. J. Virol. 67: 5859–
5866.
Vlak JM, Schouten A, Usmany M, Belsham GJ, Klinge-Roode EC,
Maule AJ, van Lent JWM & Zuidema D (1990) Expression of
cauliflower mosaic virus gene I using a baculovirus vector based
upon the p10 gene and a novel selection method. Virology 179:
312-320.
Weyer U, Knight S & Possee RD (1990) Analysis of very late gene
expression by Autographa californica nuclear polyhedrosis virus
and the further development of multiple expression vectors. J.
Gen. Virol. 71: 1525–1534.
Wood HA & Granados RR (1991) Genetically engineered bac-
uloviruses as agents for pest control. Ann. Rev. Microbiol. 45:
69–87.
Zuidema D, Schouten A, Usmany M, Maule AJ, Belsham GJ,
Roosien J, Klinge-Roode EC, van Lent JWM & Vlak JM (1990)
Expression of cauliflower mosaic virus gene I in insect cells using
a novel polyhedrin-based baculovirus expression vector. J. gen.
Virol. 71:2201–2209.
Address for correspondence: Paul A. Kitts CLONTECH Laborato-
ries, Inc., 1020 East Meadow Circle, Palo Alto, CA 94303–4230,
U.S.A.
This page intentionally left blank.

Cytotechnology 20
:
125–137, 1996.
125
© 1996 Kluwer Academic Publishers. Printed in the Netherlands.
Passage effect of virus infection in insect cells
Peter J. Krell
Department of Microbiology, University of Guelph, Guelph, Ontario, N1G 2W1 Canada
Key words: baculovirus, genomic alteration, passage, bioreactors, viral persistence, viral fitness
Introduction
Genomic plasticity is the hallmark of all biological
processes and for viruses is more apparent because
of their short generation times and high number of
“progeny” produced during each replication cycle or
passage. Despite the inherent fidelity of DNA replica-
tion, DNA genomes, including those of baculovirus-
es, continuously undergo variations in their nucleotide
sequences. Due to recombination and errors in DNA
replication there is a certain probability of error at
each passage which for the baculovirus genome leads
to point mutations, deletions, rearrangements, inver-
sions, reiterations and acquisition of host cell DNA.
It is in this background that baculoviruses replicate
either in cells of insect larvae or of insect tissue cul-
tures. Whether the baculovirus replicates in its insect
host as a “natural” pathogen, or in cultured cells for the
production of microbial insecticides and foreign pro-
teins of economic or scientific interest, the potential
for such genomic alterations is omnipresent.
Faulkner (1981) first suggested that the production
of variants, atypical morphogenesis and lower viru-
lence of baculovirus preparations occurred as a con-
sequence of repeated serial passage, herein referred to
as the “passage effect”. The result of prolonged (undi-
luted) passage of baculovirus in vitro
,
was first shown
by MacKinnon et al. (1974) for Trichoplusia ni NPV
(TnNPV) derived from “late passage” (up to 50) in
T. ni TN–368 cells. At late passage fewer polyhedra
per cell and less virulent polyhedra were produced.
Moreover polyhedral morphology mutants and aber-
rant and shorter nucleocapsids resulted and the number
of normal occluded virions per polyhedral inclusion
body (PIB; the form of baculoviruses responsible for
insect to insect horizontal transmission) also decreased.
Payne (1988) noted the importance of passage effect
in the production of effective viral pathogens for insect
control. Tramper & Vlak (1986) first recognized the
importance of the passage effect in bioreactor produc-
tion of baculoviruses and foreign protein in the context
of decreased productivity.
The accumulation of genomic alterations through
several replication cycles either through individual
insects or tissue culture flasks or through the sever-
al cycles of replication in bioreactors can be referred
to as the passage effect. Some of these new genotypic
variants could replicate at the expense of the origi-
nal parental virus and could, with sufficient numbers
of passages become dominant. The major problems
with the passage effect on baculovirus replication and
expression is the loss of virulence of PIB for virus
grown in tissue culture and, for viruses used as expres-
sion vectors, a reduced ability to produce the foreign
protein of interest. For growth of PIB in tissue culture
there is no selection pressure for virus capable of pro-
duction of infectious polyhedra. Similarly for foreign
genes in recombinant baculoviruses there is often no
selection to maintain and express those genes.
Baculovirus genomic alterations
That genomic alterations occur frequently among bac-
uloviruses was evident from the many genotypic vari-
ants (McIntosh et al., 1987) initially detected by
restriction fragment length polymorphism of many nat-
ural baculovirus isolates including Autographa cali-
fornica nuclear polyhedrosis virus (AcMNPV; Lee &
Miller, 1978; Miller, 1984), Spodoptera frugiperda
126
NPV (SfNPV; Knell & Summers, 1981), S. exempta
MNPV (Brown et al., 1985), S. litura NPV (Maeda et
al., 1990) and Panolis flammea MNPV (Weitzman et
al., 1992). That baculovirus genomes can recombine
was initially demonstrated by Summers et al. (1980)
for AcMNPV and Rachiplusia ou NPV and also by
recombination between mutant strains of AcMNPV
(Carstens et al., 1987). Moreover the development of
baculoviruses as expression vectors is predicated on
homologous recombination between baculovirus and
transfer vector DNAs.
Of the many genomic changes that can occur some
are silent and provide no selective advantage, some
are lethal and, except under unusual circumstances,
would be eliminated with passage. Certain changes
could enhance replicative advantage so that the mutant
virus could outcompete the original genotype during
successive passages. A single nucleotide change which
alters a codon but not the corresponding amino acid
could, except for factors such as codon usage (Ayres
et al., 1994), have no effect. An alteration in a codon
leading to substitution of a different amino acid could
lead to proteins such as polyhedrin with altered struc-
ture (e.g., mutant M29 and M934, Carstens et al., 1987;
1992) or influence the activity of viral enzymes criti-
cal for viral replication such as the AcMNPV helicase
(Lu & Carstens, 1991). Other changes such as dele-
tions or insertions of DNA in an essential gene, could
alter the reading frame. If deletions are large enough,
entire genes could be eliminated. Another alteration
which could influence gene expression is reiteration of
viral DNA such as reported by Lu & Carstens (1990).
Those genotypic changes which enhance the replica-
tion ability in host cells, or insects will be amplified at
each passage and will eventually dominate the original
genotype. The outcome depends on the virus species
and genotype, the cell (species and tissue source of
cells in culture or in insects), the nature of the genotypic
changes (genes affected, source and location of DNA
insertions or deletions), the method of virus culture
(passage in insects, cells in suspension, monolayers or
bioreactors, frequency of transfer) and multiplicity of
infection (moi).
For AcMNPV and other baculoviruses there are
numerous isolates, strains, variants or mutants which
have been extensively analyzed to determine the nature
of the genotypic variations (Eraser, 1987; Kool et al.,
1991; Kool&Vlak, 1993; Xiong et al., 1991; Table 1).
Genotypic variation in the AcMNPV and other bac-
ulovirus genomes, include point mutations, both small
and large deletions and insertions, recombination and
reiterations. There may be hot spots for certain genom-
ic alterations such as insertions due to transposable ele-
ments or the deletions in the hypervariable DA26 gene
region (O’Reilly et al., 1990), but all regions of the
genome appear capable of variation. Some variations
such as transposon-mediated insertions occur readily
even under low passage number and low moi (Fraser
et al., 1985). Other variations occur under conditions
of genomic stress during prolonged passage (Kumar
& Miller, 1987; O’Reilly & Miller, 1990; Miller &
Miller, 1982; Friesen et al., 1986) or under high mul-
tiplicity passage (HMP) in flasks or bioreactors (Kool
et al., 1991).
Deletions of viral DNA occurs readily. For
example, the genomes of the majority of the 667
AcMNPVLl-X plaques analyzed by Kumar & Miller
(1987), after only 10 passages at an moi of 0.1, had
deletions of 0.7 kb in Pst1 G (335) or 0.05 kb in Pst1
I (321). Many other deletions ranging in size from 0.1
to 2.3 kb have been reported and some are summarized
in Table 1. Some deletions inactivate viral genes such
as the UDPecdysteroid glucosyltransferase gene (egt;
O’Reilly & Miller, 1990) and the DA26 gene (O’Reilly
et al., 1990). Since these mutants survive, neither gene
is essential for replication in cell culture. The DA26
deletion occurred after passage of AcMNPV through
Manduca sexta larvae suggesting that deletions can
also occur by passage in insects. One consequence of
the passage effect is that some of the genotypic changes
which arise through passage involve sequential and
cumulative deletions of viral DNA (Kool et al., 1991;
Cusack & McCarthy, 1989; Wickham et al., 1991; Lee
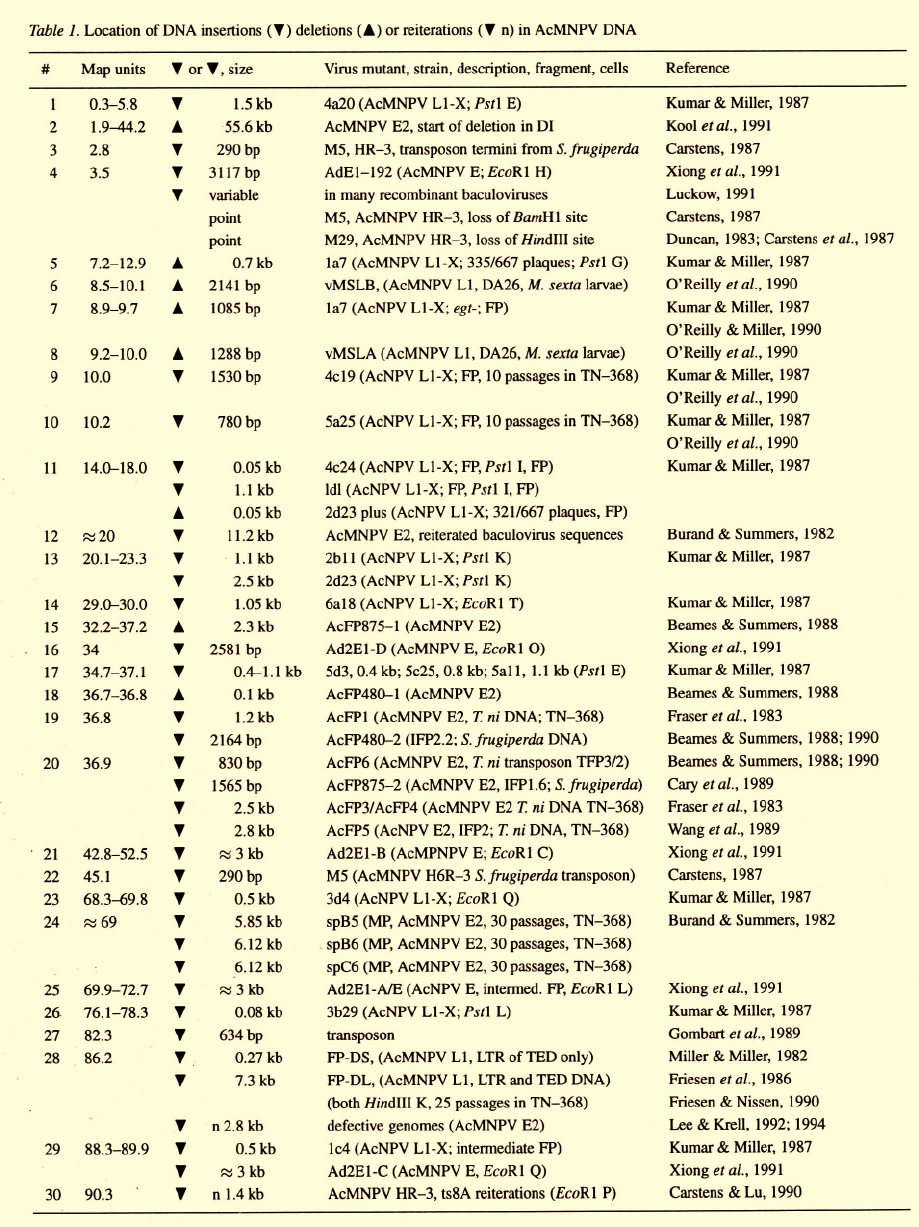
& Krell, 1992; 1994; Croizier et al., 1985; Figure 1)
resulting in major losses of the standard genome. Such
highly deleted genomes are released from the infect-
ed cells as particles referred to as defective interfering
particles (DIP; Huang & Baltimore, 1977). A deletion
of 55 kb occurs during replication of M5, an AcM-
NPV PIB morphology mutant (Carstens, 1982; 1987)
and in the generation of AcMNPV DIP in a bioreac-
tor (Kool et al., 1991). Since DIP lack many essential
genes they cannot independently infect cells. Howev-
er, at HMP both DIP and helper virus can co-infect the
same cell allowing the DIP genome to replicate and
be expressed at the expense of the helper virus (Roux
et al., 1991; Bangham & Kirkwood, 1990). Moreover
the DIP concentration increases relative to the original
parental strain (wild type virus or recombinant virus)
and the virus titre and foreign gene expression would
decline with each passage (MacKinnon et al., 1974; De
Grooijer et al., 1992; Kompier et al., 1988; van Lier et
127
128
129
al., 1992). DIP can also mediate persistent infections
as observed for Heliothis zea NPV (Hz–1) DIP in IMC
Hz–1 and T. ni TN–368 cells (Wood & Burand, 1986).
Insertions of DNA into baculovirus genomes can
also occur at high frequency. Kumar & Miller (1987)
found 100 of 667 viral plaques with insertions of
0.08 to 2.5 kb of DNA, the majority of which (88)
were in Pst1
E
. The sizes of other insertions in bac-
ulovirus genomes range from 0.3 to 3.1 kb and up to
11.2 (Table 1) in both standard and recombinant bac-
uloviruses (Luckow, 1991). Insertion of DNA is usu-
ally mediated by homologous recombination, as for
the generation of recombinant baculoviruses (Luckow,
1991) and as demonstrated by the efficiency of marker
rescue (Carstens et al., 1987). However DNA inser-
tions can also be mediated by non-homologous recom-
bination as shown by Xiong et al. (1991) using aden-
ovirus DNA and, as is becoming increasingly obvious,
by transposons (Miller & Miller, 1982; Fraser et al.,
1983, 1985; Beames & Summers, 1990; Gombart et
al., 1989; Cary et al., 1989; Wang et al., 1989; O’Reil-
ly et al., 1990; Blissard & Rohrmann, 1990). DNA
insertions can be from the cell (e.g., transposons), co-
infecting viruses (Summers et al., 1980; Croizier et al.,
1994) or from transfected plasmids (Luckow, 1991;
Xiong et al., 1991). Other changes include reiterations
of viral DNA (Burand & Summers, 1982; Carstens &
Lu, 1990; Lee &Krell, 1994). These genetic exchanges
or rearrangements could dramatically change the virus
by altering its virulence or even host range as demon-
strated for an AcMNPV which exchanged as little as
79 bp of the Bombyx mori NPV (BmNPV) helicase
gene, allowing AcMNPV to replicate in B. mori cells
(Croizier et al., 1994).
Passage, genotypic changes and the few polyhedra
(FP) phenotype
The serial passage of any virus results in the accumu-
lation of genotypic variations among any virus popu-
lation, including that of baculoviruses. One of the first
variations which arose during serial passage of bac-
uloviruses described at the genetic level, was a change
from the parental, many polyhedra per cell (MP) phe-
notype, to the few polyhedra per cell (FP) phenotype
reported for both AcMNPV and Galleria mellonella
NPV (GmNPV; Fraser, 1987; Fraser et al., 1983, 1985;
Wang et al., 1989) and TnNPV (Potter et al., 1976).
That this was the earliest and most common pheno-
typic change during serial passage (Kumar & Miller,
1987) reflects the high frequency of genetic alterations
in the FP-locus and the non lethal nature of this change
for virus grown in tissue culture.
Potter et al. (1976) showed for TnNPV that FP
plaques could be recovered as early as passage 9 of an
undiluted serial passage in TN–368 cells. The propor-
tion of FP plaques recovered increased during passage
so that by passage 14, no MP plaques were observed.
Also non occluded virus (NOV; i.e., extracellular virus)
from the FP isolates were released at a faster rate than
MP PIB. Although FP NOV were infectious to TN–368
cells and to insects when injected into the haemocoel,
the corresponding FP polyhedra were much less infec-
tious per os than the MP NOV. When TnNPV MP NOV
was passaged by injection into T. ni larvae, FP NOV
was observed by 4 passages and by passage 14 rep-
resented about 95% of plaques isolated from infected
haemolymph (Potter et al., 1978). FP virus was detect-
ed only sporadically for virus passaged as PIB and
per os infection of larvae, but was not maintained nor
was there a change in virulence in per os transmitted
virus. Thus one way to maintain the MP character of an
inoculum is to ensure that it is passaged as PIB through
insect larvae.
The generation of some AcMNPV FP mutants
occurs even during a single passage at low moi and
most are due to transposon insertion in the FP-locus
(Miller & Miller, 1982; Fraser, 1987). The very high
frequency of FP mutants from several insect cells and
insect species suggested that transposon mutagenesis
was common. From the first description of the FP
plaques by Hink & Vail (1973), the incidence of FP
and their molecular basis have been of major interest.
Miller & Miller (1982) in their analysis of a 7.3 kb
insertion in the genome of an FP mutant (FP-DL) later
established the role of transposon-mediated mutagen-
esis in the generation of some FP mutations. For AcM-
NPV, FP mutants were detected within only one or
two passages in TN–368 cells (Hink & Strauss, 1976).
FP virus was also detected in vivo from per os (PIB)
infected T. ni
,
Heliothis virescens
,
Helisidota caryae
,
and
S
. frugiperda larvae. For these, the LD
50
of the
parental MP PIB was 38 times higher than for FP PIB.
MP virus could be “rescued” (from 21 to 67%) if a
predominantly FP virus preparation was passed in vivo
(
per os
)
through T. ni larvae. Host DNA insertions of
0.8 to 2.8 kb were detected in the genomes of AcMN-
PV and GmNPV FP mutants (Fraser, 1987; Fraser et
al.,
1983)
Fraser & Hink (1982) first demonstrated how fast
the passage effect can manifest itself in the generation
130
of FP virus. The FP phenotype was detected within
two passages of GmNPV in TN–368 cells or in G. mel-
lonella larvae. The proportion of FP virus was virtually
100% by the 10th passage in TN–368 cells or by 5 ser-
ial injection passages in G. mellonella larvae. GmNPV
FP virus is a stable genotypic variant which was main-
tained through at least 10 serial injection passages in
G. mellonella larvae. The MP virus always generated
both MP and FP virus whether passaged through TN–
368 cells or by injection through G. mellonella larvae.
Thus the FP virus resulted from frequent spontaneous
mutations of a single MP virus rather than the FP virus
being present in the original MP virus preparation. For
GmNPV, the FP virus was up to 350 fold less infectious
than the parental MP virus.
Genomic changes and serial passage
Wickham et al. (1991) followed the effect of serial
passage and moi on genomic changes in a recombi-
nant, AcMNPV in Sf21 cells. Virus production
decreased from 814 pfu/cell for the original isolate to
35 pfu/cell for a late passage isolate. DIP at late pas-
sage were detected by electron microscopy showing
up to 60% short (200 nm) particles. The DIP genome
had a deletion of contiguous Eco
R
1 A, I, J, K, N, and
O fragments which includes the gene and corre-
sponded to the region that Kool et al. (1991) reported
was missing in their DIP (Figure 1E). The DIP prepa-
ration also interfered with replication of the original
AcMNPV. When DIP-containing virus (moi of
100) was added, the amount of AcMNPV pro-
duced per cell decreased from 814 pfu/cell (without
DIP) to 40 pfu/cell. However the percent of standard
virus in the DIP preparation increased from 15 to 65%
after a single passage at a much reduced moi of 0.001.
While the production of per cell decreases due
to the DIP, this effect is less obvious at low moi. It is
unfortunate that Wickham et al. (1991) did not give
sufficient details (e.g., initial moi, frequency of pas-
saging, passage number analyzed) so that their results
could be compared to those of Kool et al. (1991) and
others.
In a more systematic study, Kool et al. (1993) fol-
lowed HMP of AcMNPV in Sf21 cells through 40
passages in culture flasks. As for studies of virus in
bioreactors (Kompier et al., 1988) most cells infected
with P40 virus no longer produced polyhedra and the
yield of NOV was about 100 fold lower than that of the
inoculum. Much of the genome was deleted in late pas-
sage DIP and seven novel and hypermolar AcMNPV-
specific Eco
R
1 fragments were detected within cells
infected with P40 virus and in P41 NOV preparations.
Since these fragments supported infection-dependent
DNA replication, Kool et al. (1993) suggested that
they contain cis sequences important for DNA repli-
cation and would have a selective advantage over the
replication of the larger standard virus genome.
In a more extended study, Lee & Krell (1992, 1994)
followed the generation of defective genomes during
HMP up to passage 81 (Figure 1). Like Kool et al.
(1993) the region from 1.7 to 45.0 m.u. was deleted
within 40 passages in the defective genomes. The size
of the defective genomes became smaller during pas-
sage down to an average size of about 50 kb. Up to
passage 81 even more of the genomic DNA genome
was deleted although the overall size of the genome
was not reduced any further. By passage 80 only about
2.8 kb of the standard genome (m.u. 85.0 to 87.2; Fig-
ure 1) was maintained in the defective genomes isolated
from P80 infected cells and from P81 NOV. This short
sequence was reiterated and originated from the AcM-
NPV Hin
d
III K region which Lee & Krell (1994) sug-
gest contains an origin of DNA replication. Because of
the smaller size and presence of multiple origins such
genomes could compete for essential trans-acting fac-
tors such as DNA polymerase and helicase and would
replicate at the expense of the standard genome.
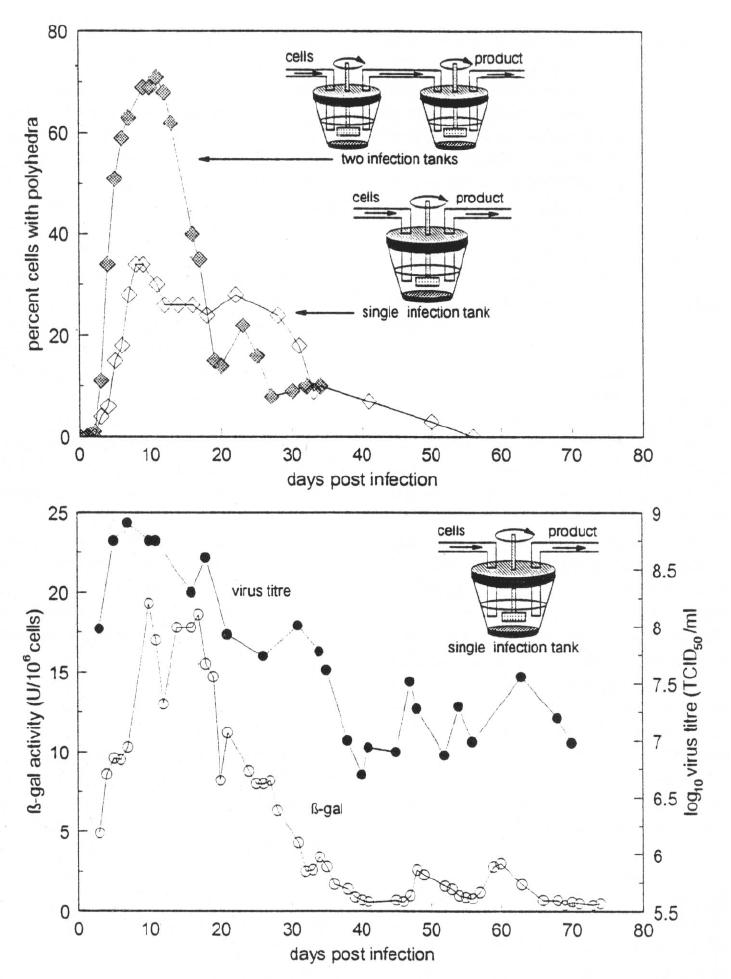
Passage effect and bioreactors
For large scale production of foreign proteins at eco-
nomical levels cells have to be grown and infected in
bioreactors where cell growth, virus infection and for-
eign protein synthesis occur simultaneously within the
same chamber. Under a continuous system the virus
undergoes many passages over a short period of time.
Virus grown in flasks undergoes only a single replica-
tion cycle and virus is normally passaged about every
3 or more days. However, in a bioreactor where unin-
fected cells are being continuously supplied, virus is
“passaged” after each replication cycle time (e.g., 8–
24 hr). In the bioreactor setting, since the virus is being
passaged at a high frequency and at high moi the con-
sequences of the passage effect are noted within about
two weeks (Figure 2).
In one of the first systematic evaluations of the
passage effect on wild type AcMNPV in a bioreac-
tor, Kompier et al. (1988) followed the production
of a standard virus (AcMNPV-E2 in Sf21 cells) in a
131
