Polaina J., MacCabe A.P. (ed.). Industrial Enzymes. Structure, Function and Applications
Подождите немного. Документ загружается.

534 KAUL ET AL.
mixture) for acid productioncan be developed into a high throughput format for the
screen. A simple, colorimetric, pH responsive method was developed for the rapid
enantioselective screening of nitrile hydrolysing enzyme libraries (Banerjee et al.,
2003b). pH sensitive indicators have been used to monitor various enzyme catalysed
reactions such as carbonic anhydrase (Gibbons and Edsall, 1963), cholinesterase
(Lowry et al., 1954), lipase (Baumann et al., 2000) etc. The method is based on the
drop in pH that occurs as the reaction proceeds due to the formation of acid. The pH
drop is reflected by the colour change of the indicator, provided the colour profile
of the indicator falls within the pH range of the enzyme activity. For the colour
change to be proportional to the number of protons released, both the indicator and
the buffer must have similar affinities for protons (pK
a
s within 0.1 unit of each
other), such that the relative amounts of protonated buffer to protonated indicator
remain constant during the course of the reaction. This method was utilized to screen
nitrilase producing micro-organisms for the production of R-−-mandelic acid,
a versatile chiral building block (Kaul et al., 2004b). Since only the R-isomer
of mandelonitrile is commercially available, enantioselectivity was determined by
comparing the rate of colour turnover of the R-isomer with that of the racemate.
Conclusive statements regarding the enantioselectivity of the micro-organisms could
not be made in cases where both changed colour simultaneously, reflecting the
importance of using pure enantiomers. The proposed method is simple and, most
importantly, less time consuming. Using this pH responsive strategy a large number
of micro-organisms can be analysed simultaneously in a short time, thus reducing
the number of samples to be analysed in greater quantitative detail (HPLC analysis).
The assay requires very little substrate thus allowing the use of pure enantiomers
which are not always available in large quantities. The disadvantage of the method
is its qualitative nature. The presence of cells interferes with the spectrophotometric
reading. The use of cell free extracts may solve the problem but will also add an
extra step to the screening procedure. Moreover, this may require special instrumen-
tation (e.g. a microplate reader) which can be avoided by visualising the enzyme
catalysed reaction using a suitable indicator. Hence, by compromising quantitative
aspects screen throughput was increased. The goal was to use the method not for
precise quantitation but for screening large numbers of micro-organisms to facilitate
the identification of those having the desired enantioselectivity acceptable for the
development of a biocatalytic resolution process.
2.1.1. Distribution and physiological role of nitrilases
Nitrilases are found relatively infrequently in nature. The existence of the enzyme
activity in 3 out of 21 plant families (Gramineae, Cruciferae and Musaceae)
(Thimann and Mahadevan, 1964) and in a limited number of fungal genera
(Fusarium, Aspergillus, Penicillium) (Harper, 1977) indicates the relative scarcity
of this activity. Nitrile-degrading activity is more frequent in bacteria, though
without extensive screening it is almost impossible to assess the actual distribution
frequency. A number of bacteria (Acinetobacter, Corynebacterium, Arthrobacter,
Pseudomonas, Klebsiella, Nocardia, Rhodococcus etc.) are known to utilize nitriles
NITRILE HYDROLASES 535
as sole sources of carbon and nitrogen. The physiological role of nitrile hydrolysing
enzymes in micro-organisms is not clear. In plants such activities are implicated
in nutrient metabolism, particularly in the degradation of glucosinolates (Bestwick
et al., 1993) and in the synthesis of indole acetic acid (Bartel and Fink, 1994).The
genome sequence of Arabidopsis thaliana revealed four nitrilase-related sequences
(AtNIT 1, 2, 3 and 4) (Bartling et al., 1992). AtNIT1, 2 and 3 are isoenzymes which
are found only in Brassicaceae (Hillebrand et al., 1998). AtNIT4 on the other hand
is not related to the other three nitrilases and its homologues are found in many plant
species such as tobacco (Tsunoda and Yamaguchi, 1995) and rice (Piotrowski et al.,
2001). The major physiological role for these enzymes appears to be glucosinolate
metabolism. AtNIT4 is a -cyano alanine hydratase and plays a role in cyanide
detoxification (Piotrowski et al., 2001). In microbial systems nitrilases probably
form components of a complex pathway controlling the production and degradation
of aldoximes. Nitriles, which are formed by enzyme activities upstream of aldoxime
dehydratases, may further undergo hydrolysis, oxidation, reduction etc. by different
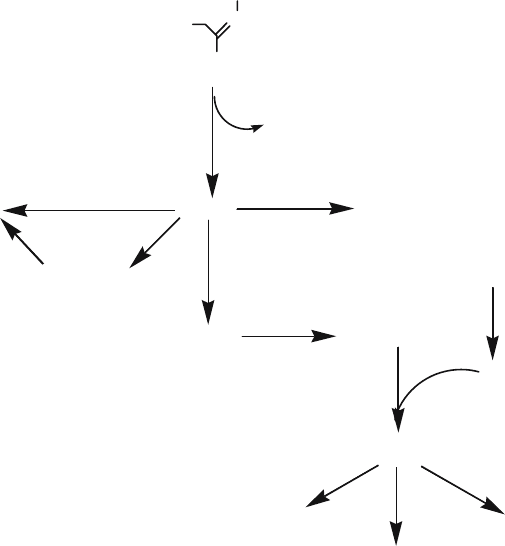
enzymes including nitrilases (Fig. 1).
RCHO + HCN
RCH
2
CH
3
+
NH
3
Oxygenase
Oxynitrilase
Nitrogenase
Aldoxime
Aldoxime
dehydratase
Nitrilase
NHase
Amidase
Metal cyanides
Abiotic
conversion
Accumulated
CO
2
+ NH
3
Cyanase
Cyanide
hydratase
Cyanide
dihydratase
HCOOH + N
3
H
RCH
2
CN
RCH
2
COOH
RCH(OH)CN
R
N
OH
H
H
2
O
RCH
2
CONH
2
HCN
HCONH
2
HCN
Figure 1. Different pathways of nitrile metabolism
536 KAUL ET AL.
2.1.2. Molecular structure and genetic analysis
To date many nitrilases have been purified and their subunit molecular mass
and primary structures have been determined. Most nitrilases consist of a single
polypeptide having a molecular mass in the range 30 to 45 kDa, which aggregate
to form the active holoenzyme under different conditions. The preferred form of
the enzyme seems to be a large aggregate of 6 to 26 subunits. Most of the enzymes
show substrate dependent activation, though the presence of elevated concentrations
of salt, organic solvents, pH, temperature or even the enzyme itself may also
trigger subunit association and therefore activation (Nagasawa et al., 2000). The
hydrophobic effect resulting from the above mentioned conditions might change
the conformation of the enzyme thereby exhibiting hydrophobic sites and enabling
subunit assembly and enzyme activation. The nitrilases of Pseudomonas fluorescens
DSM 7155 and Bacillus pallidus Dac 521 were co-purified with chaperonins
(Almatawah et al., 1999; Layh et al., 1998). It was suggested that chaperonins might
play a role in the assembly of the subunits into high molecular weight complexes
and also their stabilisation. When the nitrilase gene of Comamonas testosteroni
was cloned and over-expressed in E. coli, as much as 30% of the total cellular
protein was found to be nitrilase but only 10% of this was soluble in nature. The
solubility of nitrilase was enhanced by co-overexpression of GroEL (chaperonin)
in the same cell and the activity increased five-fold (Schill et al., 1995). This
further confirms the crucial role of chaperonins in the correct folding and subunit
association of nitrilase.
The genes encoding the nitrilases of Klebsiella pneumoniae ssp. ozaenae (Stalker
et al., 1988), R. rhodochrous strain K22 (Kobayashi et al., 1992), A. faecalis JM3
(Kobayashi et al., 1993), C. testosteroni (Schill et al., 1995) and Bacillus sp. strain
OxB-1 (Kato et al., 2000) have been sequenced and over-expressed. The four
nitrilases of A. thaliana have also been over-expressed in E. coli. AtNIT1, 2 and 3 are
clustered in a 13.8 kb region on chromosome 3 and share 80% identity in sequences
(Hillebrand et al., 1998). AtNIT4, not linked to these genes, shows 63% identity
with AtNIT1-3, which reflects their different functionality in A. thaliana (Piotrowski
et al., 2001). A Cys residue has been shown to be essential for the catalytic activity
of the nitrilases (Kobayashi et al., 1992, 1993). This Cys is proposed to be a
part of the catalytic triad Cys-Glu-Lys which has been identified from the crystal
structures of the NitFhit and N-carbamyl-D-amino acid amidohydrolases, members
of nitrilase superfamily (Nakai et al., 2000). The genetic regulation of nitrilases is
largely unknown. A gene (nitR) was identified downstream of the R. rhodochrous
J1 nitrilase gene and encodes a protein that has significant homology to AraC type
transcriptional regulators (Martin and Rosner, 2001). NitR appears to be a positive
regulator of nitrilase gene (nitA) expression (Komeda et al., 1996a). Unlike most
AraC type regulators whose genes are transcribed divergently from the genes they
are regulating, transcriptional analysis of nitR indicates that it is probably expressed
by read through from the nitA promoter (Komeda et al., 1996b). A putative nitrilase
regulator has also been found in Bacillus sp. strain OxB-1 (Kato et al., 2000).
NITRILE HYDROLASES 537
2.1.3. Reaction mechanism
Hydrolysis of a nitrile of the form R -C ≡ N produces the corresponding amide,
R-C =ONH
2
, with the addition of one molecule of water and the corresponding
acid, R-CO
2
H, with the addition of second molecule of water. Nitrilases are
interesting enzymes in the sense that the substrates are nitriles but the reaction
does not involve the release of or the reaction with a substantial amount of the
corresponding amide. However, it has been shown that the purified nitrilases of
F. oxysporum ssp. melonis (Goldhust and Bohak, 1989), Pseudomonas fluorescens
DSM7155 (Layh et al., 1998) and the ricinine nitrilase of Pseuodomonas sp. (Hook
and Robinson, 1964) produce a small amount of amide product and therefore
have nitrile hydratase activity. In these cases the amide product is usually < 5%
of the total reaction products. AtNIT1 and AtNIT4 also have NHase activity,
especially AtNIT4 which produces 1.5 times more asparagine than aspartic acid
from -cyano alanine (Piotrowski et al., 2001). Some of the nitrilases use amides,
albeit at a very low rate compared to the nitrile substrate, and therefore have
amidase activity (Kobayashi et al., 1998). A reaction mechanism was proposed
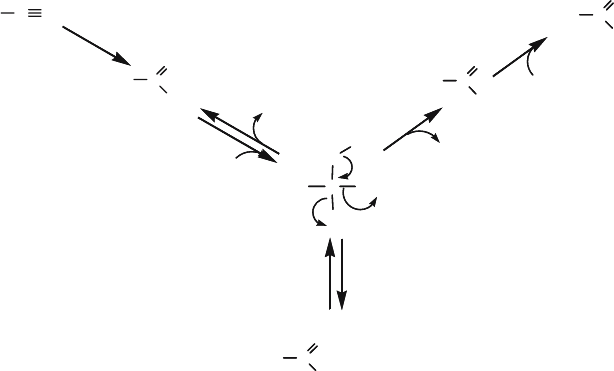
which accounts for all these activities (Hook and Robinson, 1964; Thimann and
Mahadevan, 1964). This mechanism involves a nucleophilic attack on the nitrile
carbon by the sulfhydryl group of the nitrilase, leading to a tetrahedral inter-
mediate via an enzyme thioimidate route (Fig. 2). The tetrahedral intermediate
can frequently break down anomalously to produce an amide instead of the
normal acid product. Nitrilase interaction with its substrates and intermediates
has some geometric constraints: linear substrate (approximately 180
C), planar
R
CN
+
ESH
RC
NH
SE
RC
O
SE
RC
O
OH
R
C
O
R
SE
H
+
+
+
-
+
H
2
O
H
2
O
+
H
2
O
R
C
O
NH
2
NH
2
–NH
3
+
ESH
+
ESH
+
-
Figure 2. Reaction mechanism of nitrilase catalysis
538 KAUL ET AL.
thioimidate and acylenzyme intermediates (approximately 120
C), and tetrahedral
water-bonded intermediates (approximately 1095
C). Since ammonia is a better
leaving group than the enzyme, the first enzyme-dependent water addition does not
produce an acid amide but rather an acylenzyme complex the hydrolysis of which
produces the acid product. Most nitrilases bind strongly to a bulky substrate R group
in a conformation that places the second carbon closer to 120
than to 180
from the
cyano nitrogen. Hence, fitting a distorted nitrile substrate would drive the reaction
towards thioimidation rather than tetrahedral intermediate. In support of this view,
most nitrilases are noted to prefer bulky substrates to non-substituted acetonitrile
(Pace and Brenner, 2001).
2.2. Nitrile Hydratase
Nitrile hydratase (NHase) is a key enzyme in the bienzymatic pathway of the
conversion of nitriles to amides, which are further converted to the corresponding
acid by amidases. A number of micro-organisms having NHase activity have been
isolated and the enzymes have been purified and characterized. These revealed the
wide ranging physiochemical properties and substrate specificities of the NHases,
which are composed of two types of subunits ( and ) complexed in varying
numbers. They are metalloenzymes containing either cobalt or iron. On the basis of
the metal ion present, the NHases can be classified into two broad groups, namely
ferric NHases and cobalt NHases.
2.2.1. Ferric NHases
The NHases from Rhodococcus R312 (formerly known as Brevibacterium R312)
and Pseudomonas chlororaphis B23 are the first examples of non-heme iron
enzymes containing a low spin Fe (III) ion (Sugiura et al., 1987). These have
been well characterized by ESR, extended X-ray absorption (EXFAS), electron
nuclear double resonance (ENDOR) and Raman resonance spectroscopies. These
studies revealed that the NHase from Rhodococcus sp. R312 is a
2
tetramer
containing two low spin non-heme ferric ions which exist in a tetragonally distorted
octahedral ligand field of three histidine imidazoles, two cysteine thiolates and a
hydroxide. The activity of NHase has unique features when exposed to light (Endo
et al., 1999). During aerobic incubation in the dark NHase activity in Rhodococcus
sp. N-771 decreases, but this activity is almost completely recovered upon irradi-
ation with visible light. This photo-reactivity of NHase is intrinsic to the enzyme,
and the mechanism has been biochemically elucidated (Endo et al., 1999). The
chromophore involved in the photo-activation is an iron complex in the subunit,
and light irradiation of the complex induces a conformational change of the subunit.
Moreover, activity is regulated by nitric oxide (NO). An endogenous NO molecule
is bound to the non-heme iron (III) centre in the inactive NHase and is released
upon photo-activation, resulting in recovery of the original NHase activity. Three-
dimensional analysis of Rhodococcus R312 NHase showed that it contains a novel
iron centre (Huang et al., 1997). All the metal ion protein ligands are contained
NITRILE HYDROLASES 539
within the subunit. Three cysteine thilotes and two main chain nitrogen atoms are
ligands. These five iron ligands (Cys 110, Cys113, Cys115, Ser 114 and Cys 115)
are located on five vertices of an octahedron, the sixth position being unoccupied.
Considering the experimental data for hydrogen and oxygen ENDOR resonances,
a hydroxide ion is likely to occupy this position. Furthermore, Cys112 and Cys114
(corresponding to Cys113 and Cys115 in the Rhodococus R312 NHase) are post-
translationaly oxidised to cysteine-sulfinic and sulfenic acids, respectively, in the
Rhodococcus N-771 NHase.
2.2.2. Cobalt NHase
In the presence of cobalt ions the actinomycete R. rhodochrous J1 (Komeda et al.,
1996b) produces two Nhases, depending on the inducer. When cultured in a medium
containing urea and cyclohexane carboxamide, high and low molecular weight
NHases (H- and L-NHases) are selectively induced (Yamada and Kobayashi, 1996).
H-NHases (Nagasawa et al., 1991) act preferentially on aliphatic nitriles, whereas
L-NHases (Komeda et al., 1996b) exhibit higher affinity for aromatic nitriles. H- and
L-NHases have been used for the industrial production of acrylamide and nicoti-
namide from acrylonitrile and 3-cyanopyridine, respectively. Both purified NHases
contain cobalt as co-factor. The cobalt in H-NHase exists as a low-spin Co ion in
a tetragonally distorted octahedral ligand field. The similarities of the pre-edge and
extended X-ray absorption fine structure (EXAFS) spectra suggests that the ligand
environments of the metal ions in the cobalt and iron containing NHases are similar.
Recently, a NHase from Pseudomonas putida with stereoselectivity toward 2-(S)-
(4-chlorophenyl)-3-methylobutyronitrile was found to contain noncorrin low-spin
Co (Payne et al., 1997). The three dimensional structures of both the Rhodococcus
rhodochrous J1 and Pseudomonas putida NHase are probably similar to that of
the Rhodococcus R 312 NHase because of their sequence similarities. The cobalt
NHases have threonine in the –V-C-(T/S)-L-C-S-C-sequence as the active site,
(Payne et al., 1997) whereas the ferric NHase has serine. The difference in the metal
co-factors may be ascribed to the different amino acid residues at this position.
Spectroscopic and structural analyses of novel NHases in Agrobacterium tumifa-
ciens which contain cobalt and iron should improve our understanding of the
functions of these metals. There may be two main reasons for having Co in the
active site: 1) Cobalt is a very good catalyst for CN-triple-bond hydration, and 2)
it is required for the folding of the enzyme. In addition to the function of the active
centre, cobalt ions may play a role in enhancing the folding or stabilization of the
subunit polypeptides of the enzyme.
2.2.3. Reaction mechanism
One possible reaction model is that catalysis proceeds without direct coordination
of the substrate to the metal ion, where the metal ion plays a role as a lewis
acid activating a water molecule. The next step could be either of the following:
(1) the nitrile substrate approaches a metal-bound hydroxide ion which can act as a
nucleophile attacking the nitrile carbon atom (mechanism I), or (2) a metal bound
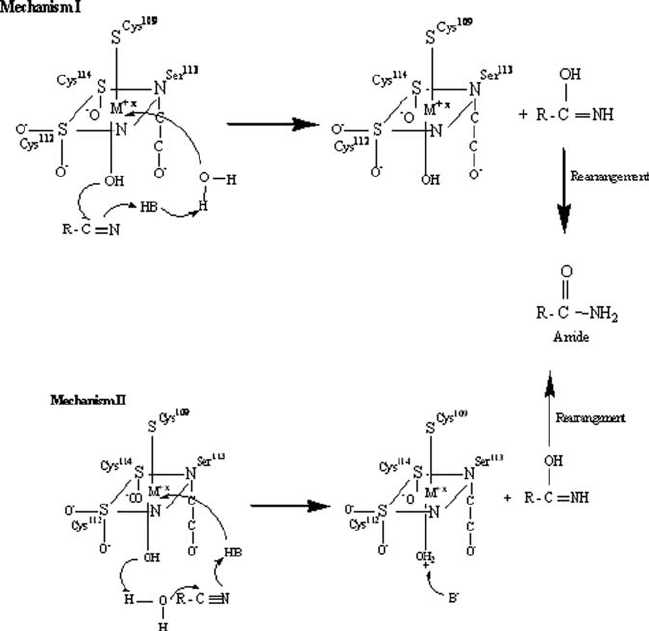
540 KAUL ET AL.
Figure 3. Reaction mechanism of nitrile hydratase catalysed hydrolysis of nitriles
hydroxide ion as a general base activates a water molecule which then attacks the
nitrile carbon (mechanism II) resulting in the formation of an imidate. The imidate
finally tautomerizes to form an amide (Fig. 3).
2.3. Amidase
Amidases catalyse the hydrolysis of carboxylic acid amides to free carboxylic acids
and ammonia. These enzymes are involved in nitrogen metabolism in cells and
are widely distributed in nature. They have been found both in prokaryotic and
eukaryotic cells. Unlike nitrile hydratases, the association of amidases with metals
such as cobalt or iron is reported only in case of K. pneumoniae (Nawaz et al., 1996).
Using site-directed mutagenesis it was confirmed that these are sulfhydryl enzymes.
The other amidases from Pseudomonas chlororaphis B23, R. erythropolis MP50, R.
rhodochrous J1 etc. belong to a group of amidases containing the Gly-Gly-Ser-Ser
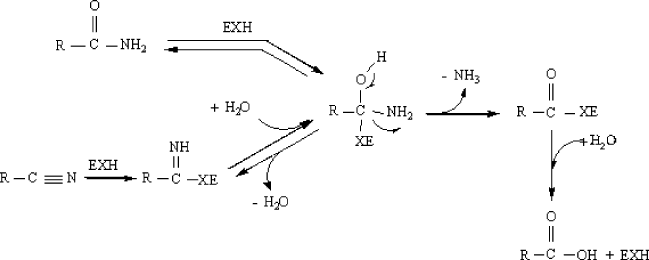
NITRILE HYDROLASES 541
Figure 4. Reaction mechanism of amidase
signature (Chebrou et al., 1996). Surprisingly the amidase of R. rhodochrous J1
is found to catalyse the hydrolytic cleavage of the nitrile functional group to an
acid and ammonia stoichiometrically. The amidase exhibited a K
m
of 3.26 mM for
benzonitrile in contrast to that of 0.15 mM for benzamide as the original substrate
(Kobayashi et al., 1998).Thus the reaction mechanisms of both the nitrilase- and
amidase-catalysed reactions are analogous, but the active nucleophile present in
both enzymes differs. Fig. 4 shows the reaction mechanism of amidase, which
also involves nitrile hydrolysis. The carbonyl group of the amide undergoes a
nucleophilic attack resulting in the formation of a tetrahedral intermediate which
is converted to an acyl-enzyme with the removal of ammonia and subsequently
hydrolysed to acid. Although there is no homology between the amidase and nitrilase
families, comparison of their reaction mechanisms can provide novel insights for
the construction of catalysts required for the hydrolysis of nitriles as well as amides.
All of the different amidases also exhibit acyl transfer activity in the presence
of hydroxylamine (Fournand et al., 1998). Hydroxamic acids thus produced are
known to possess good chelating properties and can act as potent inhibitors of
matrix metalloproteases (Cawston, 1996). They also have the potential to be used
as anti-HIV agents or antimalarial agents (Gao et al., 1995).
3. APPLICATIONS
Biocatalysis offers a new dimension, innovative approaches and enormous
opportunity to prepare useful chiral compounds (Patel, 2001). Research over the
past decade has shown that there are very few barriers to the use of enzymes
and/or whole cells as biocatalysts in organic synthesis (Faber, 1997). Enzymes
are remarkable catalysts capable of accepting a wide array of complex molecules
as substrates and are exquisitely selective, catalysing reactions with unparallel
enantio- and regio-selectivities (Schmid et al., 2001; Kaul et al., 2004a). Enzymatic
transformation of nitriles in particular provides great potential for synthetic chemists.
The ability of the enzyme system to convert cyano functionality to either an acid
542 KAUL ET AL.
or an amide is, in itself, of great use. Traditional chemical methods for conversion
of nitriles to acids or amides have several drawbacks: (1) reactions need to be
carried out either in strongly acidic (6 M HCl) or basic (2 M NaOH) reflux condi-
tions, (2) a requirement for high reaction temperatures, (3) the formation of by-
products such as toxic HCN or large amounts of salts, etc. Biocatalytic hydrolysis
of nitriles is attractive due to its ability to effect reactions in a more ‘green’ manner
and because of the potential for carrying out chemo-, regio-, and enantio-selectiv
transformations.
3.1. Synthetic Biocatalysis
Industrial scale processes for the production of acrylamide (Mitsubishi Rayon
Co. Japan) (Nagasawa, 1993; Yamada and Nagasawa, 1994) and nicotinamide
(Lonza AG, Switzerland) (Chassin, 1996) involving nitrile converting enzymes
are the two most prominent examples of biocatalytic reactions being implemented
at larger scale and pave the way for the development of other processes
involving this class of enzymes. Nitrilases can also selectively convert a single
cyano group of a polynitrile which is virtually impossible using conventional
chemical methods. R. rhodochrous K22 catalyses the conversion of adiponitrile
to 5-cyanovaleric acid, which is an intermediate for the synthesis of nylon-6
(Godtfredsen et al., 1985) and Tranexamic acid, a homeostatic drug, is obtained
by selective mono-hydrolysis of trans 1,4-dicyano cyclohexane by Acremonium
sp. (Nishise et al., 1987). Nitrilases also offer the possibility of stereoselective
transformation which includes the production of several important fine chemicals
and pharmaceutical intermediates, for example S-phenyllactic acid (Hashimoto
et al., 1996), L--amino acids (Bhalla et al., 1992), R −-hydroxy acids (Wu
and Li, 2003), S-ibuprofen (Yamamoto et al., 1990) etc. Although in recent
years several applications of nitrilases have been recognized (Kobayashi and
Shimizu, 1994; Sugai et al., 1997) to date there exist only two industrial scale
processes for the production of 1,5-dimethyl 2-piperidone (DuPont, USA) and
R-−-mandelic acid (BASF, Germany; Mitsubishi Rayon, Japan) employing
these enzymes as biocatalysts. 1,5-dimethyl 2-piperidone (Xolvone
™
) which has
desirable properties for electronics, coatings and solvent applications, is produced
from 4-cyanopentanoic acid, which in turn is generated from regio-selective
nitrilase-mediated transformation of 2-methyl glutaronitrile (MGN) employing
Acidovorax facilis 72W cells (DiCosimo et al., 2000). When compared to existing
chemical process in which MGN is directly converted via hydrogenation to a mixture
of 1,3-DMPD and 1,5-DMPD, the chemoenzymatic method generates less waste and
produces a single lactam isomer at significantly higher yield. BASF and Mitsubishi
Rayon use nitrilase-mediated processes for the production of R-−-mandelic
acid and its derivatives at multiton scale (Endo and Tamura, 1993; Ress-Loschke
et al., 1998). The advantage of this process over its chemical counterpart is that
no organic solvent is required and a high degree of stereoselectivity is achieved
because of the presence of the biocatalyst.
NITRILE HYDROLASES 543
3.2. Bioremediation
Synthetic nitrile compounds are widespread in the environment in the form of
industrial wastewaters. Most of these are toxic, carcinogenic and mutagenic in
nature (Pollak et al., 1991), thus there is a need to control their release into the
environment. A mixed culture of bacteria containing different nitrile hydrolyzing
enzymes (including NHase, amidase and nitrilase) able to metabolise effluents
containing acrylonitrile, fumaronitrile, succinonitrile, etc. were grown in batch
and continuous cultures on these components of industrial waste. A reduction
in COD (75%) and significant removal (99%) of detectable toxic components
was achieved by biodegradation of the effluent from acrylonitrile manufacturing
industries using mixed cultures of bacteria (Wyatt and Knowles, 1995). The use of
specialized consortia of micro-organisms to degrade toxic wastes therefore could
be a viable alternative approach to the classical activated sludge system. Prolonged
exposure to nitrile herbicides [dichlobenil (2,6-dichlorobenzonitrile), bromoxynil
(3,5-dibromo-4-hydroxybenzonitrile) etc] results in symptoms of weight loss,
fever, vomiting, headache and urinary problems (Freyssinet et al., 1996). Nitrile-
metabolising enzymes efficiently degrade these cyano group-containing herbi-
cides and prevent them from entering the food chain. Agrobacterium radiobacter,
a bromoxynil-degrading soil bacterium, was used for the degradation of the
herbicide under non-sterile batch and continuous conditions yielding a 65%
reduction in the bromoxynil concentration in a column reactor after 5 days. The
efficacy of degradation is enhanced by the addition of ferrous, cobaltous or cupric
ions (Muller and Gabriel, 1999). A gene encoding the nitrilase has been cloned
from Klebsiella pneumoniae ssp. ozaenae (McBride et al., 1986) and was used
to raise herbicide resistant plants (Stalker et al., 1988). Bromoxynil resistant
transgenic plants resulting from the introduction of microbial bromoxynil-specific
nitrilase genes into tomato or tobacco are already approved for commercial use
(Freyssinet et al., 1996). Similarly, other nitrile-degrading enzymes could also be
potential candidates for molecular manipulation of bio-degradative systems in plant
biotechnology.
4. CONCLUSIONS
The versatile biocatalytic nature and applications of nitrile converting enzymes
are now increasingly recognized for the production of several pharmaceutically
important compounds and fine chemicals. This relatively new biocatalytic technique
also shows promise for use as a method for the preparation of enantio-, regio-,
chemo-selective amides or acids, the synthesis of which is not feasible by chemical
routes. Commercial scale processes involving kiloton syntheses of acrylamide,
nicotinamide and nicotinic acid using these enzymes are excellent examples of this
methodology. By virtue of their ability to eliminate highly toxic nitriles, these nitrile-
degrading enzymes can also play a significant role in protecting the environment.
Advances in our understanding of biosynthetic regulation, genetics and the structure
