Polaina J., MacCabe A.P. (ed.). Industrial Enzymes. Structure, Function and Applications
Подождите немного. Документ загружается.

422 PARE AND HOBMAN
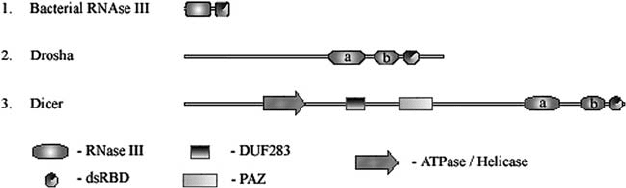
Figure 1. Domain organization of RNase III superfamily members
The members of the E. coli RNase III subfamily are relatively small (150–250
amino acid residues) and are structurally the simplest members of the superfamily.
They are composed of a single RNase III domain together with a double stranded
RNA binding domain (dsRBD) (Fig. 1). By comparison, Drosha subfamily members
are structurally more complex than the bacterial RNase III isoforms. Members of
this group contain two RNase III domains (a and b), a dsRBD and relatively long
amino-termini (Fig. 1). The latter segment contains a proline-rich region and an
arginine/serine-rich (RS-rich) region, both of which are thought to participate in
protein-protein interactions: (Fortin et al., 2002; Wu et al., 2000).
The third class of RNase III enzymes is represented by Dicer family members.
Similar to Drosha, Dicer enzymes contain two RNase III domains and a dsRBD, but
lack proline-rich and RS-rich domains (Fig. 1). They are the only members of the
RNase III superfamily that contain PAZ and RNA helicase domains. In addition, most
Dicer isoforms possess a centrally located domain of unknown f unction (DUF283).
2.2. RNase III
True to its function as a ribonuclease, RNase III was originally isolated from E.
coli as a dsRNA degrading activity (Robertson et al., 1968). Subsequently, it was
determined that members of this subfamily are important for the maturation of
ribosomal RNA, transfers RNAs and mRNAs (Bram et al., 1980; Nicholson, 1996;
Regnier and Grunberg-Manago, 1990; Steege, 2000; Venema and Tollervey, 1999).
In some organisms, RNase III enzymes are also involved in rRNA fragmentation
(Bram et al., 1980; Gegenheimer and Apirion, 1980; Gegenheimer and Apirion,
1981). This process entails the removal of short nucleotide stretches from the mature
domain of rRNA molecules without subsequent re-ligation of the cleaved fragments
(Gerbi, 1995; Gray, 1995). For example, the 16S and 23S rRNA subunits are initially
transcribed as a single primary transcript, which folds into a hairpin structure. The
hairpin is cleaved by RNase III to generate the mature 16S and 23S rRNA subunits.
In addition to their roles in rRNA processing, RNase III enzymes may also have
regulatory functions that are not dependent upon catalytic activity. Specifically, the
RNase III homolog, Rnt1, is required for normal cell cycle progression in budding
yeast (Catala et al., 2004). In this case, the RNase activity of RntI is not required for
DICER: STRUCTURE, FUNCTION AND ROLE 423
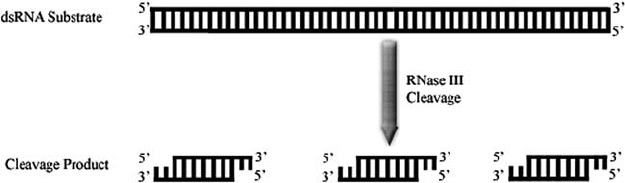
Figure 2. RNase III cleavage. Cleavage of long dsRNA substrates by RNase III enzymes yields uniformly
sized dsRNA cleavage products with 2 nt 3’ overhangs and 5’ phosphate groups
its role in cell cycle regulation. Finally, it has been reported that RNase III enzymes
can bind to certain mRNA substrates, a scenario which is at least consistent with the
potential for these enzymes to affect translation directly or indirectly by decreasing
the stability of the mRNAs (Dasgupta et al., 1998).
Activity of bacterial RNase III enzymes requires the formation of homodimers
(Dunn, 1976; Robertson et al., 1968). Dimerization of two RNase III molecules
results in the creation of a catalytic center comprised of two opposing RNase III
domains. As mentioned above, this arrangement results in the positioning of the
two catalytic sites in such a way that both strands of the dsRNA are cleaved. All
members of the RNase III superfamily generate characteristic cleavage products
that contain 5’ phosphate groups and 2 nt 3’ overhangs (Fig. 2) that terminate
with hydroxyl groups (Nicholson, 1996). Although the lengths of cleavage products
vary depending upon the particular enzyme, the products are of uniform sizes. For
example, bacterial RNase III enzymes generate RNA duplexes that are approxi-
mately 11 nt long (Nicholson, 1996). In contrast, depending upon the isoform, Dicer
cleavage products are duplexes which vary in length from 20 to 25 nt.
2.3. Drosha
Drosha family members localize predominantly to the nucleus and like their Class
I relatives, are important for processing of pre-ribosomal precursors. Originally
termed human RNase III, Drosha was recognized as an essential enzyme from
experiments which showed that inhibiting the expression of this enzyme resulted
in cell death (Wu et al., 2000). Subsequent work by the Kim laboratory revealed
that Drosha activity is required upstream of Dicer in the microRNA (miRNA)
biogenesis pathway (Lee et al., 2003). Specifically, this enzyme is responsible for
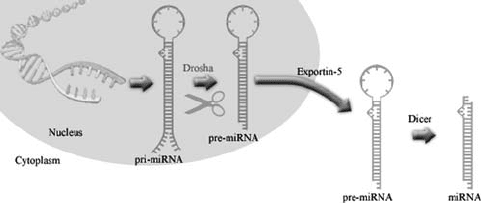
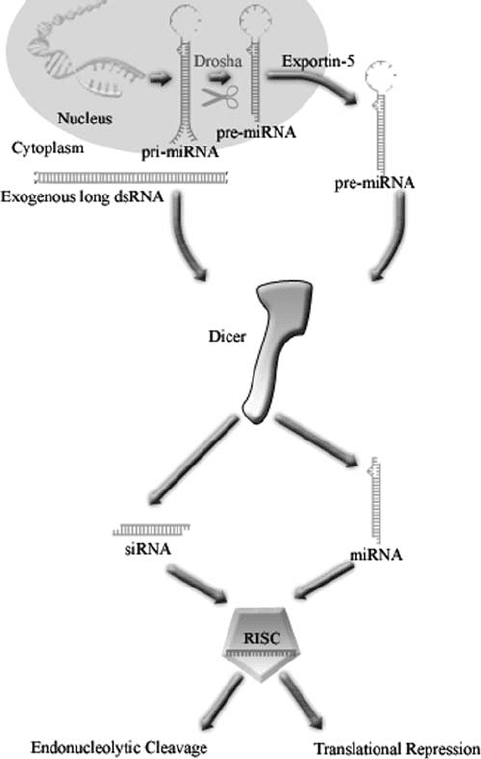
cleaving the long primary-miRNAs (pri-miRNAs) into shorter pre-miRNAs (Lee
et al., 2003) (Fig. 3). Pri-miRNAs are either transcribed as independent transcripts
or are derived from the intronic regions of protein-encoding genes. After they are
processed by Drosha, pre-miRNAs are exported from the nucleus by an Exportin-
5-dependent pathway (Bohnsack et al., 2004; Lund et al., 2004; Yi et al., 2003).
This export factor recognizes the 2 nt 3’ overhangs on Drosha cleavage products.
424 PARE AND HOBMAN
Figure 3. Maturation of miRNAs by Drosha and Dicer. Cleavage of pri-miRNA in the nucleus by
Drosha leads to the formation of pre-miRNAs. The pre-miRNAs are exported to the cytoplasm by
Exportin-5, where they are subsequently processed by Dicer into mature miRNAs
The pre-miRNAs which are exported to the cytoplasm, contain double stranded
stem regions that are cleaved by Dicer to generate mature miRNAs (Fig. 3).
Drosha does not function in isolation in vivo, but rather is part of a multiprotein
complex that has been termed Microprocessor (Denli et al., 2004). The Micropro-
cessor complex, in addition to containing Drosha, includes a dsRNA-binding protein
Pasha (partner of Drosha). Pasha is important for efficient processing of miRNA
precursors as evidenced by the observation that biochemical or genetic depletion
of Pasha activity, interferes with pri-miRNA processing (Denli et al., 2004; Han
et al., 2004; Landthaler et al., 2004).
2.4. Dicer
Combined biochemical and candidate gene selection approaches were used to
identify Dicer as the bidentate ribonuclease activity that generates 22 nt siRNAs
from long dsRNA precursors (Bernstein et al., 2001). Dicer also functions
downstream of Drosha (Fig. 3) to produce mature miRNAs which like siRNAs,
are incorporated into gene-silencing complexes called RNA-induced silencing
complexes (RISCs) (Bernstein et al., 2001; Grishok et al., 2001; Hutvagner et al.,
2001; Ketting et al., 2001).
Dicer enzymes are modular proteins that are composed of up to five different
domains (Fig. 1). Of critical importance to Dicer function is the PAZ domain,
a 130 amino acid residue domain located in the center of the Dicer sequence. This
domain is also present in members of the Argonaute superfamily of proteins (Cerutti
et al., 2000). Together, the Dicer and Argonaute family members comprise the core
components of the RNAi machinery. Originally thought to mediate protein-protein
interactions between Dicer and Argonaute proteins, structural studies revealed that
PAZ domains are in fact oligonucleotide-binding modules (Lingel et al., 2003;
Song et al., 2003; Yan et al., 2003). Dicer appears to have specific requirements for
substrate binding and there is considerable evidence indicating that PAZ domains
are involved in binding the ends of small dsRNA molecules (Lingel et al., 2003;
Song et al., 2003; Yan et al., 2003). In this respect, the characteristic 5’ phosphates
and 2 nt 3’ overhangs that result from Drosha-mediated cleavage of pri-miRNAs,
DICER: STRUCTURE, FUNCTION AND ROLE 425
are important for recognition by Dicer (Lee et al., 2003; Zhang et al., 2004).
Through binding to the ends of dsRNA molecules, PAZ domains help to position
the substrates for cleavage by the RNase III domains (Fig. 4). It is interesting to
note that Dicer PAZ domains contain a large extended loop that is absent from
Argonaute PAZ domains (MacRae et al., 2006). This difference between Dicer
and Argonaute PAZ domains may be an important for differential binding and
potentially transferring RNA substrates between these proteins.
The amino-terminal regions of most Dicer enzymes contain a DExH-box RNA
helicase domain. DExH type helicases contain eight conserved motifs, and are
named so because of the conservation of the amino acid sequence Asp-Glu-X-His
in the fourth motif (Gorbalenya et al., 1989; Linder et al., 1989). Interestingly,
while the sequences of the helicase domains are highly conserved in Dicer family
members (Nicholson and Nicholson, 2002), ATP is not required for the RNase
activity of recombinant mammalian Dicer in vitro (Provost et al., 2002; Zhang
et al., 2002). In fact, some Dicer enzymes in lower eukaryotes lack helicase domains
altogether, yet these enzymes are fully capable of “dicing” dsRNA into uniformly
sized products (MacRae et al., 2006).
While Dicer cleavage products are dsRNA molecules, one strand of the duplex,
the “guide strand” is used to target RISCs to homologous mRNAs (Martinez
et al., 2002). The remaining strand, the “passenger strand”, does not function in
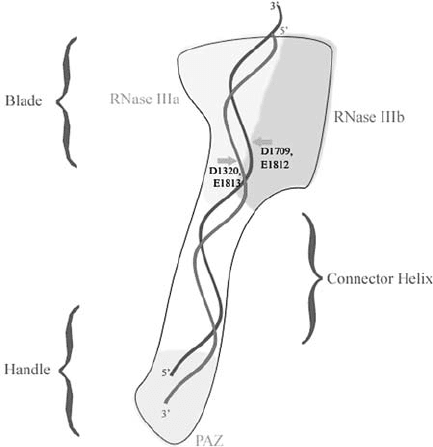
Figure 4. Model of Dicer cleavage. Giardia Dicer forms a hatchet-like structure with the two RNase III
domains forming the blade and the PAZ domain forming the handle. One end of the dsRNA substrate
is held by the PAZ domain and positioned approxiamtely 20–25 nt away from the active center in the
blade region. The catalytic amino acids in the processing center are indicated
426 PARE AND HOBMAN
RISC targeting. A long-standing issue in RNAi biology has been to understand
how unwinding of the siRNA duplex occurs following Dicer cleavage. Up until
very recently, the prevailing theory held that ATP-dependent helicase activity was
required to unwind siRNA duplexes after which the guide strand was loaded onto
RISC (Bartel, 2004; Meister and Tuschl, 2004; Sontheimer, 2005; Tomari and
Zamore, 2005). Certainly, the helicase of Dicer was considered as a likely entity to
perform this function. Studies in Drosophila melanogaster revealed that the RNA
helicase activity of Dicer-2 is necessary for siRNA production in vivo, however,
it is not required for the downstream function of Dicer-2 (Lee et al., 2003). In
mammalian cells, incorporation of miRNA guide strands into RISC was found to
occur in the absence of ATP (Maniataki and Mourelatos, 2005). Indeed, current
evidence suggests that the helicase domain of Dicer is not required for unwinding
dsRNAs before loading onto RISCs. Rather it seems that both the guide strand and
passenger strands of Dicer cleavage products are loaded onto RISC. The passenger
strand is then subsequently cleaved by the RISC leaving the guide strand available
for targeting the mature complex to homologous mRNAs (Leuschner et al., 2006;
Matranga et al., 2005; Rand et al., 2005).
The DUF283 domain is approximately 100 amino acid residues in length and is
also very well conserved in Dicer family members. It is located downstream of the
helicase domain in the primary sequence of Dicer. The structure of this domain has
not been solved yet and as the name implies, its role in Dicer function is not known.
The RNase III domains form the catalytic centers of Dicer enzymes. In all RNase
III family members, dimerization of these domains is required for RNase activity
(Dunn, 1976; Robertson et al., 1968). Dimerization results in alignment of the
active sites from each RNase III domain thereby creating a catalytic center. Such
an arrangement allows these enzymes to cleave two nearby phosphodiester bonds
on opposite strands of the duplex. This type of cleavage gives rise to products with
2 nt 3’ protruding ends (Fig. 2). Based on the crystal structure of Aquifex aeolicus
RNase III, Blaszcyzk et al. suggested that the intersubunit cleft between two RNase
III molecules, contains two compound catalytic centers positioned at the ends of the
cleft. Two clusters of acid amino acid residues within the catalytic centers are in turn
required for cleavage of the phosphodiester bonds (E40, D44, D107 and E110) and
coordinating single Mg
2+
or Mn
2+
ions (E37 and E64). While spatially conserved
acidic amino acid residues corresponding to E37, D44, D107 and E110 are present
in the RNase IIIa domain of Dicer enzymes, amino acid residues equivalent to E37
and E64 are not conserved in RNase IIIb. These observations led to the idea that
only one of the two “active” sites in the RNase IIIb domain is functional (Blaszczyk
et al., 2001; Hannon, 2002; Nicholson, 2003; Zamore, 2001).
The carboxyl termini of most Dicer enzymes include a double strand RNA-
binding domain known as a dsRBD. These domains are relatively small in size
(100 amino acid residues) and are conserved among eukaryotes and prokaryotes
(Fierro-Monti and Mathews, 2000; Green and Mathews, 1992; St Johnston et al.,
1992). The structures of dsRBDs have been studied by X-ray crystallography and by
nuclear magnetic resonance. They adopt an fold, a structure that is common
DICER: STRUCTURE, FUNCTION AND ROLE 427
among oligonucleotide binding pockets (Carlson et al., 2003; Saunders, 2003). The
protein/RNA interface that results from contact between a dsRBD and a dsRNA,
spans two minor and one major groove of the RNA helix (Ryter and Schultz,
1998). It is likely that dsRBDs differentiate dsRNA from DNA by recognition of
the 2’ hydroxyl groups on RNA molecules. Specifically, the amino terminal helix
of the dsRBD binds the 2’ hydroxyl groups that line the minor groove of dsRNA
(Bevilacqua and Cech, 1996; Ryter and Schultz, 1998). In addition, recent studies
have revealed that some dsRBDs contain a carboxyl terminal helix that binds to
hairpin structures (Leulliot et al., 2004). In theory, the dsRBD represents a means
whereby Dicer binds indiscriminately to dsRNA substrates, however, dsRBDs are
not strictly required for the function of Dicer in vivo. For example, the Giardia
intestinalis Dicer homolog does not contain a dsRBD, but is capable of “dicing”
dsRNA substrates into 25 nt products (MacRae et al., 2006). Accordingly, the role,
if any, of this domain in selectively retaining the dsRNA substrates for Dicer
cleavage (i.e. dsRNA containing 2 nt 3’ overhangs, 5’ phosphate groups and 3’
hydroxyl groups) is unknown.
2.5. Mechanism of Dicer Cleavage
As stated above, the RNase III domains of Dicer and other RNase III superfamily
enzymes are only active when they are in dimeric form. Unlike bacterial RNase
III enzymes whose cleavage products are typically 11 nt in length, Dicer cleavage
products are 21–25 nt. Accordingly, models of Dicer cleavage must account for the
differences in product lengths. Two early models proposed that Dicer functions as a
homodimer that is organized in antiparallel or head to tail orientations (reviewed in
Carmell and Hannon, 2004). A major concern regarding these models is that they
both require tight packing of the two Dicer molecules along the dsRNA substrate.
Because of the potential for steric hindrance between the RNase III domains and the
large amino-terminal region that contains the helicase, DUF283 and PAZ domains,
it is uncertain as to whether the RNase III domains of two Dicer molecules can
fit into a small enough space to generate 22 nt products. In fact, if the steric
relationship between the active domains is similar to the E. coli RNase III active
dimer, the products of cleavage are expected to be approximately 30 nt long, rather
than 21–25 nt (Carmell and Hannon, 2004). Finally, and perhaps more importantly,
current evidence suggests that Dicer exists as a monomer (Zhang et al., 2004).
Although the structure of a mammalian or invertebrate Dicer molecule has yet to
be solved, MacRae et al. (2006) recently determined the 3.3 Å resolution structure
of a Dicer homolog from Giardia. Interestingly, this enzyme is considerably smaller
than most Dicer proteins and lacks helicase, DUF 283 and dsRBD domains.
However, it is highly active and is able to generate 25 nt products from long dsRNA
substrates. The results obtained from the functional analyses of the Giardia Dicer
protein, are consistent with data from the Filipowicz group that support the idea
of a single processing center model (Zhang et al., 2004). Central to this model
is the observation that Dicer cleavage occurs from the ends of dsRNA molecules
428 PARE AND HOBMAN
(Zhang et al., 2002). The Giardia Dicer is an elongated molecule that resembles a
hatchet (Fig. 4). The two RNase III domains are arranged to form an intramolecular
dimer with a single processing center resembling a “blade” (Fig. 4). The RNase
blade region is linked to the PAZ-containing handle region by a connector helix.
The distance from the RNase III blade region and the PAZ domain is thought to
govern the length of the RNA cleavage products. In this respect, Dicer has been
compared to a molecular ruler (Carmell and Hannon, 2004; MacRae et al., 2006;
Zhang et al., 2004). In the Giardia Dicer molecule, the active site of the RNase III
dimer is positioned 65 Å or approximately 25 nt away from the substrate terminus
which is anchored by the PAZ domain (Fig. 4). Presumably, in mammalian Dicer
molecules, the distance between the PAZ domain and RNase III processing center
is shorter.
One potential flaw in this model is that not all Dicer molecules appear to contain
PAZ domains. For example, Dicer from the fission yeast Schizosaccharomyces
pombe does not contain a recognized PAZ domain, yet it is capable of generating
siRNAs of the expected size (Reinhart and Bartel, 2002). However, due to the
relatively poor primary sequence conservation among PAZ domains it is possible
that the S. pombe Dicer contains a divergent form of the PAZ domain that is simply
not recognized by sequence analyses algorithms.
3. DICER-BINDING PROTEINS
Given the enormous amount of interest in RNAi mechanisms, it is perhaps
surprising that comprehensive genetic and proteomic screens for Dicer-interacting
proteins in mammalian systems have not been reported yet. Recently, however,
Mello and colleagues used a whole organism approach for identification of Dicer-
binding proteins in C. elegans (Duchaine et al., 2006). The DCR-1 enzyme
appears to interact with at least 20 different proteins. As expected, many of these
proteins are known to be involved in RNAi pathways and include members of the
Argonaute family, RNA helicases, dsRNA binding proteins and RNA-directed RNA
polymerase. Three of the newly discovered DCR-1 binding proteins were studied in
more detail and of these, the PIR-1 phosphatase is perhaps most intriguing because
up to this point, not much thought had been given to the potential roles of kinases
and phosphatases in RNAi. PIR-1 is not required for processing of the trigger
dsRNAs by DCR-1, but amplified dsRNA intermediates are not cleaved by DCR-1
unless PIR-1 is present. Based on how mammalian homologs of PIR-1 function
(Deshpande et al., 1999; Yuan et al., 1998), it is thought that PIR-1 processes the
termini of amplified dsRNAs to generate optimal 5’ ends that are recognized by
DCR-1 and possibly Argonaute proteins.
3.1. RISC
Argonaute proteins are the most well characterized group of Dicer-binding proteins.
Togther with Dicer, they form the core components of RISCs, which are the effectors
DICER: STRUCTURE, FUNCTION AND ROLE 429
of gene-silencing. Biochemical and genetic assays revealed that Dicer/Argonaute
interactions are mediated by the RNase III and PIWI domains of Dicer and
Argonaute respectively (Tahbaz et al., 2004). Interestingly, binding of Argonaute
proteins inhibits the RNase activity of Dicer in vitro. One interpretation of these
data, is that the interaction serves to maintain the specificity of a given RISC for
a single mRNA species by preventing Dicer from engaging other RNA substates.
Activity of the heat-shock protein 90 (Hsp90) was found to be required for stable
binding of Dicer to Argonaute (Tahbaz et al., 2004). The involvement of Hsp90 in
assembly of RISC components suggests that formation of gene-silencing complexes
is highly regulated.
Loading of the guide strand onto Argonaute proteins is required for targeting
mature RISCs to specific mRNAs. This process is discussed in further detail in the
next section. The fate of the RISC-targeted mRNA depends on two major factors.
First, in order for RNA-directed cleavage to occur, siRNAs must be bound to a
cleavage competent Argonaute protein. Of the four mammalian Argonaute proteins,
only Ago2 exhibits cleavage activity (Liu et al., 2004). The second factor is the
level of sequence identity between the guide strand RNA and the mRNA (Elbashir
et al., 2001a; Elbashir et al., 2001b; Paddison et al., 2002). If the guide strand
is 100% identical to the mRNA and base pairing is complete, the mRNA will be
cleaved by the endonuclease activity of Ago2 (Fig. 5). If, however, the base pairing
is incomplete, and the RISC binds to the 3’ untranslated region of the mRNA,
translation is prevented without concomitant degradation (Elbashir et al., 2001a;
Elbashir et al., 2001b; Paddison et al., 2002). Recent evidence suggests that in this
context, Argonaute proteins prevent translation by interfering with recognition of
the cap structure on the mRNA (Pillai et al., 2005).
3.2. The RISC Loading Complex (RLC)
Dicer cleavage products (siRNAs and miRNAs) require the action of a multi-
protein complex, the RISC loading complex (RLC) for functional interaction with
Argonaute proteins. The RLC is best studied in D. melanogaster, whose genome
encodes two Dicer paralogs. Dicer-2 is required for siRNA-mediated RISC activity,
while Dicer-1 is required for miRNA-mediated RISC activity (Lee et al., 2004).
The D. melanogaster RLC includes Dicer-2 and its cognate binding protein R2D2
(Liu et al., 2004). R2D2 is a RNA-binding protein that contains tandem dsRBDs.
It is required for sensing the thermodynamic asymmetry of RNA duplexes (Tomari
et al., 2004a; Tomari et al., 2004b). The ends of asymmetric RNA duplexes have
inherently different thermodynamic properties, and it is hypothesized that the guide
strand is chosen based on these differences. Within the RLC, R2D2 is thought to
bind to the more thermodynamically stable end of the siRNA duplex, and in doing
so, displaces Dicer-2, thereby ensuring that it binds to the less thermodynamically
stable end of the duplex.
Loquacious is a dsRBD-containing protein that binds to D. melanogaster Dicer-1.
The functional relevance of Loquacious is illustrated by the observation that
430 PARE AND HOBMAN
Figure 5. Overview of RNA-dependent post-transcriptional gene-silencing. Drosha generated pre-
miRNAs or long dsRNA are processed by Dicer in the cytoplasm to produce miRNAs or siRNAs
respectively. The guide strands of the Dicer products target RISCs to homologous mRNAs that are
either cleaved or translationally repressed by the Argonaute subunit
Dicer-1-dependent maturation of pre-miRNAs requires interaction with this protein
(Forstemann et al., 2005; Saito et al., 2005). Not surprisingly, binding partners of
mammalian Dicer enzymes, are now being identified. For example, the Loquacious
homolog, trans-activating response RNA-binding protein (TRBP) plays a role in
siRNA- and miRNA- mediated gene silencing (Chendrimada et al., 2005; Haase
et al., 2005). In this case, interaction between TRBP and mammalian Dicer appears
DICER: STRUCTURE, FUNCTION AND ROLE 431
to be required for the assembly of the RISC complex (Chendrimada et al., 2005). In
addition, PACT, a multiple dsRBD-containing protein, binds both Dicer and Ago2
in a 500 kDa protein complex (Lee et al., 2006). Interactions between the helicase-
containing region of Dicer and the most carboxyl terminal dsRBDs of PACT and
TRBP are required for complex formation. Because TRBP and PACT bind to the
same region of Dicer, it has been proposed that RISC assembly in mammalian cells
is promoted by different Dicer-binding proteins (Lee et al., 2006).
Instead of helicase-mediated unwinding of dsRNA before loading onto RISC, it
is now believed that RISC maturation requires Argonaute-mediated cleavage of the
passenger strand (Matranga et al., 2005; Rand et al., 2005). Together, these studies
indicate that passenger strand cleavage plays a significant role in the formation of
mature RISC; however, the existence of a slower, cleavage-independent “bypass”
mechanism that facilitates RISC assembly has also been proposed (Matranga et al.,
2005).
4. DICER AND THE APPLICATION OF GENE-SILENCING
METHODS
Shortly after the discovery of RNAi as a naturally occurring phenomenon,
scientists soon realized that it could be used as an efficient gene-silencing tool
in the laboratory. During this period of time, the development of RNAi, as a
technique, rapidly outpaced the understanding of RNAi biology. For example, the
introduction of in vitro transcribed long dsRNAs (ranging from approximately 500
to 3000 bp) into invertebrates by microinjection, was found to efficiently suppress
the expression of homologous genes in invertebrates (Dzitoyeva et al., 2001). Later,
it was discovered that simply “soaking” organisms such as nematodes in dsRNA-
containing solutions or allowing them to feed on bacteria transformed with plasmids
that encode long dsRNAs, resulted in robust and specific RNAi (Timmons and
Fire, 1998). All of these methods of course, required the in vivo action of Dicer
to cleave the long dsRNAs into siRNAs. By exploiting the RNAi pathway, it is
theoretically possible to “knock down” the expression of any known gene. Indeed,
the application of RNAi has been hailed as the new somatic cell genetics. As our
understanding of gene-silencing pathways advanced, there has been a coincidental
evolution of RNAi reagents that are used to “knock down” expression of genes.
Below, we discuss the evolution of RNAi technology for use in post-transcriptional
gene-silencing. For the sake of brevity, much of the discussion is focused on the
use of RNAi in mammalian cells.
4.1. Long dsRNAs
The first RNAi reagents were simply long dsRNA molecules, which, when
introduced into cells or organisms, resulted in silencing of homologous mRNAs.
Wide-spread use of these reagents followed the discovery that dsRNA molecules
were the triggers for RNAi. In this seminal work, Fire and colleagues observed
