Оберемок В.В. Справочник по ДНК-модифицирующим энзимам и связанным с ними методам молекулярной генетики (на русском и английском языках)
Подождите немного. Документ загружается.

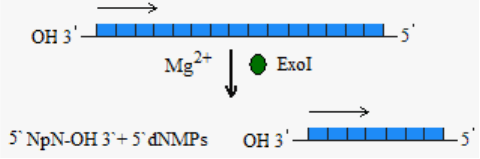
EXONUCLEASES
Exonuclease I (Exo I)
Exonuclease I (ExoI) degrades single-stranded DNA in
3`→5` direction, releasing deoxyribonucleoside 5`-monophosphates
in a stepwise manner and leaving 5`-terminal dinucleotides intact.
Features:
1) it does not cleave DNA strands with terminal 3`-OH groups
blocked by phosphoryl and acetyl groups;
2) active in buffers for PCR;
3) is not suitable for removing 3`-overhangs of dsDNA.
Applications:
1) primer removal from PCR mixtures;
2) removal of single stranded DNA containing a 3`-hydroxyl terminus
from nucleic acid mixtures;
3) assay for the presence of single-stranded DNA with a 3`-hydroxyl
terminus.
10X Reaction buffer
670 mM glycine-KOH (pH 9.5 at 25
o
C), 67 mM MgCl
2
, 10 mM DTT
..
Inhibition and inactivation
Inhibitors: 20 %(w/v) PEG 8000; inactivated by heating at 80
o
C for 15 min.
Fig. 8. Exonuclease I activity
Exonuclease III
Exonuclease III (Exo III) exhibits four different catalytic
activities:
1) 3`→5` exodeoxyribonuclease activity specific for double-
stranded DNA: Exo III degrades dsDNA from blunt ends, 5`-
overhangs or nicks, releasing 5`-mononucleotides from the
51
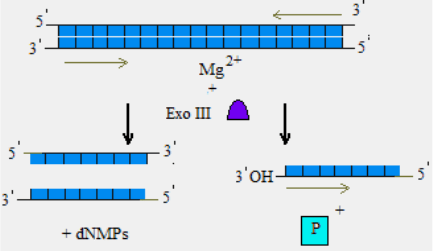
3`-ends of DNA strands and producing stretches of single
stranded DNA. It si not active on 3`-overhang ends of DNA
that are at least four bases long and do not carry a 3`-
terminal C-residue; on single-stranded DNA;
2) phosphatase activity: Exo III removes the 3`-terminal
phosphate and generates a 3`-OH group;
3) RNase H activity: Exo III exonucleolytically degrades the
RNA strand in RNA/DNA hybrids;
4) Apurinic/apirimidinic-endonuclease activity: Exo III cleaves
phosphodiester bonds at apurinic or apyrimidinic sites to
produce 5`-termini that are base-free deoxyribose 5`-
phosphate residues.
Features:
1) the rate of DNA digestion by ExoIII depends upon temperature,
salt concentration and the molar ratio of DNA to enzyme in the
reaction mixture;
2) optimal reaction conditions should be determined experimentally.
Applications:
1) creation of unidirectional deletions in DNA fragments with
conjunction with S1 Nuclease;
2) generation of a single stranded template for dideoxy-sequencing of
DNA;
3) site-directed mutagenesis;
4) cloning of PCR ptoducts;
5) preparation of strand-specific probes.
10X Reaction buffer
660 mM Tris-HCl (pH 8.0 at 30
o
C), 6.6 mM MgCl
2
.
Inhibition and inactivation
Inhibitors: metal chelators, p-chloromercuri benzoate (50-90% inhibitory at
0.1 mM); inactivated by heating at 70
o
C for 10 min.
52
Fig. 9. Exonuclease III activity
DNA LIGASES
T4 DNA ligase
Catalyzes the formation of a phosphodiester between
juxtaposed 5`-phosphate and 3`-hydroxyl termini in duplex DNA or
RNA. The enxyme repairs single-strand nicks in duplex DNA, RNA
or DNA/RNA hybrids, joins DNA fragments with either cohesive or
blunt termini. The T4 DNA Ligase requires ATP as cofactor.
Features:
1) fast – sticky-end ligation is completed in 10 min at room
temperature;
2) active in buffers for restriction enzymes, PCR and RT (when
supplemented with ATP);
3) PEG solution provides efficient blunt-end ligation.
Applications:
1) cloning of restriction enzyme generated DNA fragments;
2) cloning of PCR products;
3) joining of double-stranded oligonucleotide linkers or adaptors to
DNA;
4) site-directed mutagenesis;
5) amplified fragment length polymorphism (AFLP);
6) ligase-mediated RNA detection;
7) nick repair in duplex DNA, RNA or DNA/RNA hybrids;
8) self-circularization of linear DNA.
10X T4 DNA ligase buffer
400 mM Tris-HCl), 100 mM MgCl
2
, 100 mM DTT, 5 mM ATP (pH 7.8 at
25
o
С).
Inhibition and inactivation
Inhibitors: is strongly inhibited by NaCl or KCl at>200 mM; inactivated by
heating at 65
o
С for 10 min or at 70
o
С for 5 min.
53
* Interesting details. Polyethylene glycol (PEG) greatly increases
the rate of ligation of blunt-end DNA; molecular weight – 55.3 kDa.
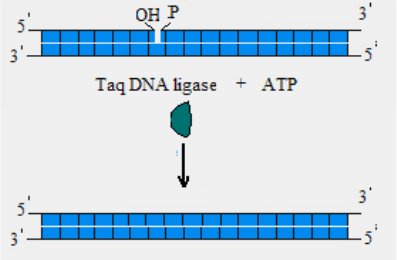
Taq DNA ligase
Taq DNA ligase catalyzes the formation of a phosphodiester bond
between juxtaposed 5´ phosphate and 3´ hydroxyl termini of two adjacent
oligonucleotides which are hybridized to a complementary target DNA. The
ligation will occur only if the oligonucleotides are perfectly paired to the
complementary target DNA and have no gaps between them; therefore, a
single-base substitution can be detected. TaqœDNA ligase is active at
elevated temperatures (45°C-65°C) .
Features:
1) Taq DNA ligase is not a substitute for T4 DNA ligase;
2) incorporates phosphorylated oligonucleotides duringœPCRœand
ligase chain reaction.
Applications:
1) allele-specific gene detection using ligase detection reaction and
ligase chain reaction;
2) mutagenesis by incorporation of a phosphorylated oligonucleotide
during primer extension amplification.
1X Taq DNA ligase buffer
20œmMœTris-HCl; 25œmMœpotassium acetate; 10œmMœmagnesium acetate;
1œmMœNAD; 10œmMœdithiothreitol; 0.1œ%œTriton X-100 (pH 7.6œat 25°C).
Inhibition and inactivation
Stability of Taq DNA ligase at 95°C:
0 min: 64,000 units/ml; 15 min: 32,000 units/ml; 60 min: 16,000 units/ml.
Fig. 10.
Taq DNA ligase
activity
Ligase
chain reaction
A DNA
amplification technique based upon the ligation of oligonucleotide
54
probes. The probes are designed to exactly match two adjacent
sequences of a specific target DNA. The chain reaction is repeated in
three steps in the presence of excess probe: (1) heat denaturation of
double-stranded DNA, (2) annealing of probes to target DNA, and
(3) joining of the probes by thermostable DNA ligase. After the
reaction is repeated for 20-30 cycles the production of ligated probe
is measured. o
Allele-specific LCR employs four oligonucleotides, two
adjacent oligonucleotides which uniquely hybridize to one strand of
target DNA and a complementary set of adjacent oligonucleotides,
hybridize to the opposite strand. Olugonucleotides from one cycle
serve as a matrix during next cycles.
55
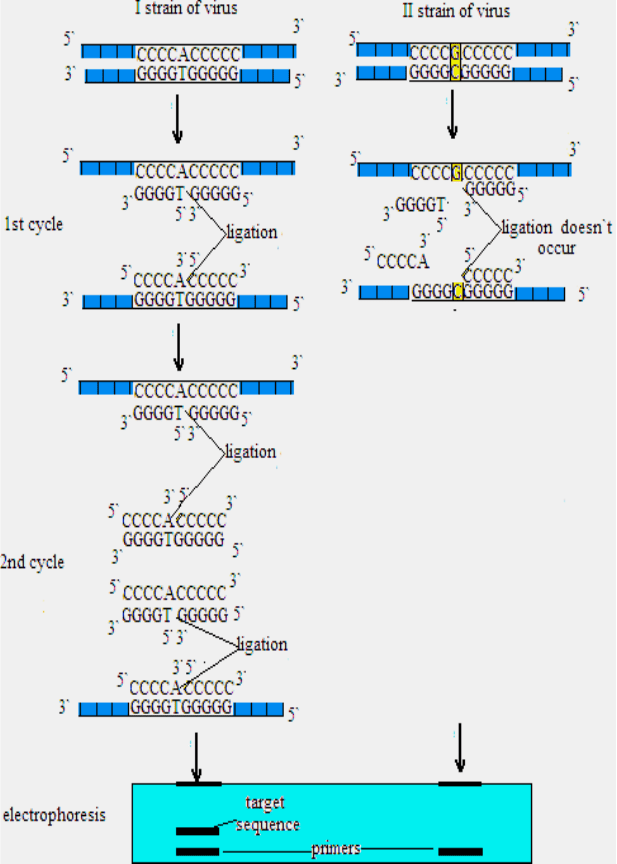
Fig. 11. Scheme of ligase chain reaction
Technical data
56
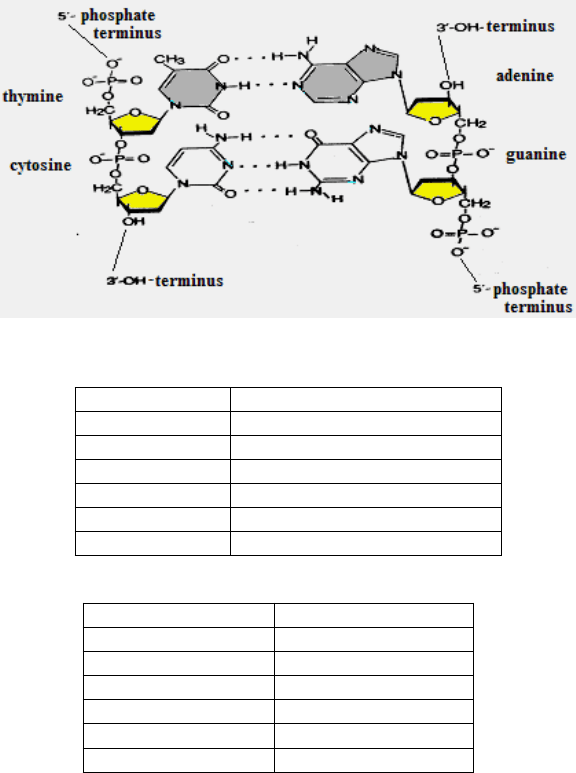
Fig. I. DNA base pairs
Table I. Range of effective separation of DNA fragments in agarose gel
Agarose gel, % Length of DNA fragments, bp
0.5 2000-50000
0.6 1000-20000
1.0 400-8000
1.5 200-3000
3.0 25-1000
5.0 10-300
Table II. General physical constants
Name Constant
Avogadro`s number 6.022x10
23
mol
-1
Planck`s constant 6.626x10
-34
J·s
Boltzmann constant 1.38x10
-23
J·K
-1
Ideal gas constant 8.314 J/mol·K
Speed of light 2.9979x10
8
m/s
Atomic mass unit 1.66x10
-27
kg
*Common conversions of oligonucleotides
Molecular weight:
57
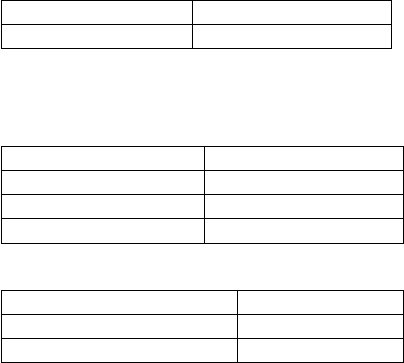
MW =333xN
Concentration of oligonucleotides:
C (µM/ or pmol/µl) = A
260
/0.01xN;
C (ng/ml) = A
260
xMW/0.01xN;
MW – molecular weight, A
260
– absorbance at 260 nm, N – number of bases
*DNA/protein conversion
1 kb of DNA = 333 amino acid = 37 kDa
* Melting temperature of duplex DNA and oligonucleotides
For duplex oligonucleotides shorter than 25 bp, “The Wallace Rule”
T
m
(in
o
C) = 2(A+T) + 4(G+C), where
(A+T) – the sum of the A and T residues in the oligonucleotide and (G+C) –
the sum of G and C residues in the oligonucleotide.
For duplex DNA, <100 bp long
T
m
(in
o
C) = 81.5
o
C+16.6 (log
10
[Na
+
])+0.41(%[G+C]) –675/n –1.0m, where
n – thr number of bases in the oligonucleotide;
m – the percentage of base-pair mismathes
*Commonly used buffers
1 M Tris-HCl [tris(hydroxymethyl)aminomethane]:
Tris base 121.1 g
H
2
O to 800 ml
Adjust to desired pH with concentrated HCl.
Mix and add H
2
O to liter.
1X TBE (Tris-borate-EDTA) electropgoresis buffer:
Adjust to desired pH with concentrated HCl.
Tris base 108 g
Boric acid 55 g
0.5 M EDTA (pH 8.0) 40 ml
H
2
O to 1 liter
1X TE (Tris-EDTA) buffer, pH 7.4, 7.6 or 8.0, per liter:
1 M Tris, pH 7.4, 7.8, 8.0 108 g
0.5 M EDTA (pH 8.0) 2 ml
H
2
O to 1 liter
*Expressing concentrations of reagents
Molarity
58
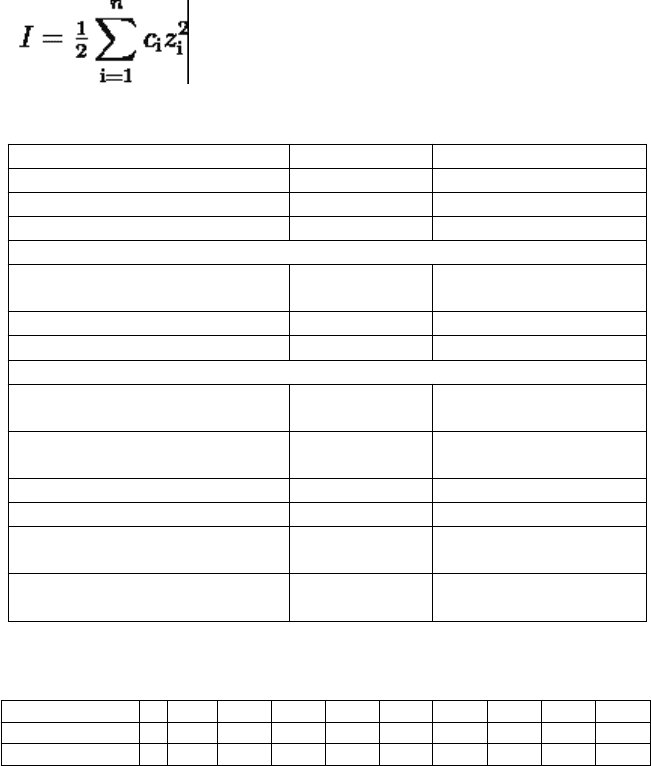
M (mol/l) = number of moles solute/number of liters of solution
Percent composition
Concentration (%) = weight of solute (g)x100%/total weight of solution (g)
Ionic strength
The ionic strength, I, of a solution is a function of
the concentration of all ions present in that
solution.where c
i
is the molar concentration of ion
i (mol·dm
-3
), z
i
is the charge number of that ion,
and the sum is taken over all ions in the solution.
Table III. Genome sizes
Virus Length, nt Approx. MW, Da
Bacteriophage φX174 5,380 3.5x10
6
HIV 9,181 3.1x10
6
SARS Coronavirus 29,751 1x10
7
Bacteria
Agrobacterium tumefaciens
C58-Cereon
4.91x10
6
3.19x10
9
Staphylococcus aureus MW2 4.86x10
6
3.02x10
9
Streptococcus pneumoniae R6 2.04x10
6
1.33x10
9
Eukaryotes
Anopheles gambiae
PEST (malaria mosquito)
2.78x10
8
1.80x10
11
Caenorhabditis elegans
(round worm)
1.21x10
7
7.86x10
9
Drosophila melanogaster 1.37x10
8
8.9x10
10
Homo sapiens 3.15x10
9
2.07x10
12
Saccharomyces cerevisiae
S288C (budding yeast)
1.21x10
7
7.86x10
9
Plasmodium falciparum 3D7
(human malaria parasite)
2.29x10
7
1.49x10
10
Table IV. Atomic numbers and molecular masses of some chemical
elements
H C N O Na Mg P S Cl Ca
Atomic number 1 6 7 8 11 12 15 16 17 20
Atomic mass 1 12 14 16 23 24.3 31 32 35.4 40
Atomic mass – g/mol
Список использованной литературы (References)
59

1. Жимулёв И.Ф. Общая и молекулярная генетика. –
Новосибирск: Сиб. унив. изд-во, 2007. – 479 с.
2. Оберемок В.В. Методические рекомендации к
применению ПЦР-метода. – Симферополь: Таврический
национальный университет им. В.И. Вернадского, 2006. – 35 с.
3. Оберемок В.В. Генетики, молекулярные биологи и их
открытия. – Симферополь: Таврический национальный
университет им. В.И. Вернадского, 2008. – 35 с.
4. www. fermentas.com
5. www. invitrogen.com
6. www. synthesisgene.com
7. www. wikipedia.org
Автор посвящает данную брошюру всем своим учителям в
науке.
По вопросам теоретической и практической
молекулярной генетики можно обращаться на e-mail автора:
genepcr @ mail . ru
60
