Оберемок В.В. Справочник по ДНК-модифицирующим энзимам и связанным с ними методам молекулярной генетики (на русском и английском языках)
Подождите немного. Документ загружается.

simplicity of PCR allowed this method to be used in different
branches of genetic investigations. Natural process of cell replication
lies in the base of PCR.
Every cycle consists of three steps: denaturation, annealing of
primers, synthesis of DNA strand. For every step there is special
temperature and time interval:
1) denaturation (94
o
C, 0.5-1 min);
2) annealing of primers (35-65
o
C, 0.5-1 min);
3) synthesis (72
o
C, 0.5-1 min).
Accurate extraction of DNA, amplification of DNA, and detection
of amplification products (electropheresis) provide high quality polymerase
chain reaciton. Amount of amplicons of DNA fragment from one cell
created during PCR can be foumd with following formula: F = 2
n
, n-number
of cycles.
41
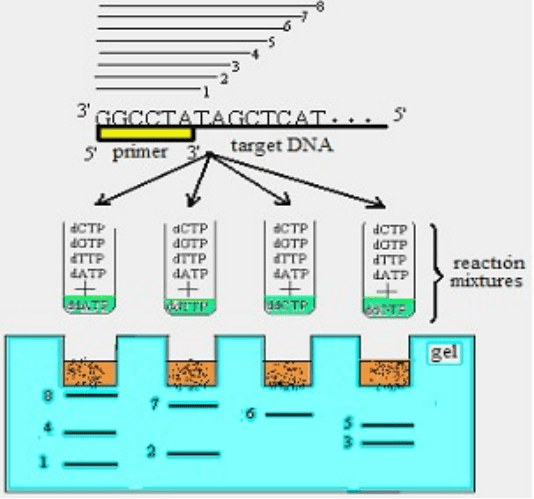
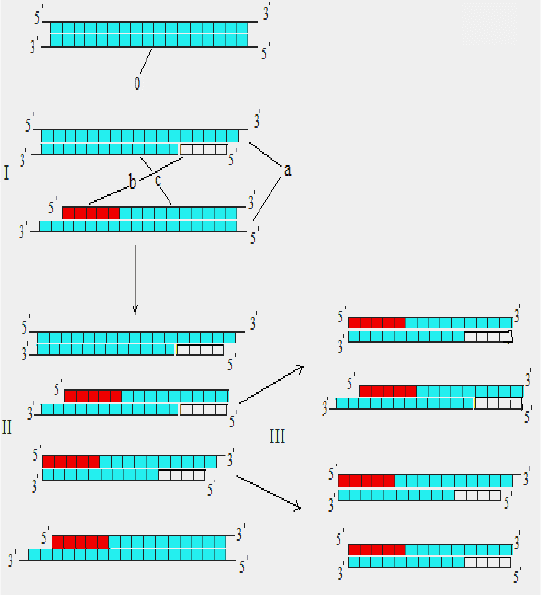
Fig. 3. Scheme of PCR: I, II, III, – 1
st
, 2
nd
and 3
rd
cycles; 0 –
fragment of target DNA; a – denaturation; b – annealing of primers; c –
synthesis
DNA sequencing
Is the method for determining the order of the nucleotide
bases in target DNA. As a matrix in DNA sequencing single strand
DNA is used. To initiate DNA sequencing with DNA polymerase
synthetic primers or natural subfragments got with restriction
endonucleases are used. 3’-end of primer must end before DNA part
that wil be sequenced. This method was invented by UK scientist
Frideric Sanger in 1975 (dideoxy termination method).
Fig. 4. Scheme of DNA sequencing
To reveal sequence of nucleotides in target part of DNA, perform 4
polymerase reations. In each of four reaction mixtures is obligatory to add
deoxynucleotide triphosphates (dCTP, dGTP, dTTP, dATP) and DNA
42
polymerase. Beside this, in each of tubes one of dideoxynucleotide
triphosphates (ddCTP, ddGTP, ddTTP и ddATP) is added. ddNTP in 3’-
position instead of hydroxyl group (OH) contains hydrogen (Н). ddNTP
takes part in DNA polymerisation reaction and ends it. In each reaction
mixture proportion between dCTP, dGTP, dTTP, dATP and one of ddNTP
is 100:1. Because of low concentration of ddNTP polymerase reaction does
not end in the beginning of DNA sequencing. At the same time reaction
mixture contains big amount of DNA fragments of different length (because
of ddNTP connection at different points of growing strand).
Using electrophoresis products of DNA polymerisation are
separated and get sequence of target DNA.
ENDONUCLEASES
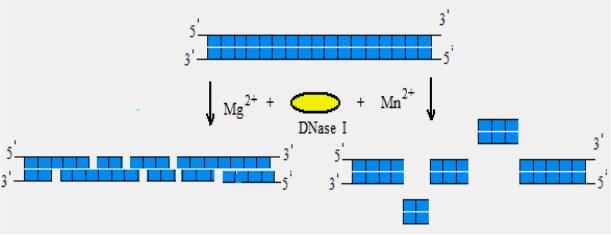
DNase I
Is an endonuclease that digests single- and double-stranded
DNA. It hydrolyzes phosphodiester bonds producing mono- and
oligodeoxyribonucleotides with 5`-phosphate and 3`-OH groups.
Features:
1) the enzyme activity is strictly dependent on Ca
2+
and is
activated by Mg
2+
or Mn
2+
ions;
2) in the presence of Mg
2+
, DNase I cleaves each strand of
dsDNA independently, in a statistically random fashion;
3) in the presence of Mn
2+
ions, the enzyme cleaves both DNA
strands at approximately the same site, producing DNA
fragments with blunt-ends or with overhang termini of only
one or two nucleotides;
4) DNase is sensitive to physical denaturation, that is why mix
gently by inverting the tube, do not vortex.
Applications:
1) preparation of DNA-free RNA;
2) removal of template DNA following in vitro transcription;
3) preparation of DNA-free RNA prior to RT-PCR;
4) DNA labeling by nick translation in cojunction with DNA
Polymerase;
5) studies of DNA-protein interactions by DNase I;
6) generation of a library of randomly overlapping DNA inserts.
Reaction buffer containing Mn
2+
is used.
43
10X Reaction buffer with MgCl
2
100 mM Tris-HCl (pH 7.5 at 25
o
C), 25 mM MgCl
2,
1 mM CaCl
2.
10X Reaction buffer with MnCl
2
100 mM Tris-HCl (pH 7.5 at 25
o
C, 1 mM CaCl
2.
Recommended
concentration of MnCl
2
in 1X reaction buffer is 10 mM.
Inhibition and inactivation
Inhibitors: metal chelators, transition metals, (e.g., Zn) in milimolar
concentrations, SDS (even in concentrations less than 0.1 %), reducing
agents (DTT), ionic strength above 50-100 mM; inactivated by by heating
at 65
o
C for 10 min in the presence of EDTA (use at least 1 mol of EDTA
per 1 mol of Mg
2+
/Mn
2+
).
Fig. 5. DNase I activity in the presence of Mg
2+
or Mn
2+
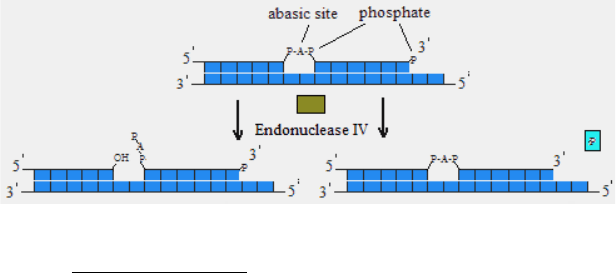
Endonuclease IV, E. coli (Endo 4)
Endonuclease IV recognizes apurinic/apirimidinic (AP) sites
of dsDNA and cleaves phosphodiester bond 5` to the lesion
generating a hydroxyl group at the 3`-terminus. The enzyme can also
act as esterase releasing a 3`-phosphoglycolate or 3`-phosphate from
the damaged ends of dsDNA. Endo IV possesses also a 3`→5`
exonuclease activity.
Features:
1) its progression on substrates is sensitive to ionic strength, metal
ions, EDTA, and reducing conditions;
2) substrates with 3`-recessed ends are preferred substrates for the
3`→5` exonuclease activity;
3) the enzyme has no requirement for Mg
2+
but is more active in the
presence of Mg
2+
.
44
Applications:
1) studies of DNA damage and repair;
2) single cell electrophoresis;
3) antitumour drug research;
4) DNA structure research;
5) SNP analysis.
10X Reaction buffer
100 mM Tris-HCl (pH 8.5 at 25
o
C), 25 mM MgCl
2.
Inhibition and inactivation
Inhibitors: the enzyme is fairly resistant to EDTA during the reaction, but
becomes sensitive to even sub milimolar quantities of chelators when no
DNA substrate is present; inactivated by heating at 80
o
C for 15 min.
Fig. 6. Endonuclease IV activity
Restriction enzymes
Restriction enzymes recognize specific nucleotide sequences
and cleave DNA molecules either within or outside their recognition
site. Digestion results in the degeneration of DNA with “sticky” (5`-
or 3`-overhang or “blunt” ends).
Common properties
The phenomenon of host specificity was first observed by Luria
and Human in the early 1950s. Nearly a decade later, Arber and and Dussoix
predicted its molecular basis. They proposed that host specificity in two-
enzyme system: a restriction enzyme, which recognizes specific DNA
sequences and cleaves foreign DNA upon its entrance into the bacterial cell,
and a modification enzyme (metyltransferase), which protects the host DNA
from degradation by its own restriction enzyme. Both restriction and
modification enzymes recognize the same nucleotide sequence and together
they form a restriction-modification (R-M) system.
45
Restriction-modification systems are wide-spread among bacteria
and have also been isolated from phage, Archaea and eukaryotic algae
viruses. R-M systems have been classified into four types (I, II, III and IV)
depending on the complexity of their structure, cofactor requirements, mode
of action and sequence cleaved. The best characterized and the most
frequently used are type II restriction endonucleases. These enzymes
recognize specific 4-8 bp DNA sequences and cleave a DNA sequence
either within the recognition site, or at a specified position up to 20 base
pairs outside the site.
Often there is more than one enzyme that recognizes a particular
nucleotide sequence. According to restriction enzyme nomenclature, an
enzyme with a unique specificity which has been discovered first is called
prototype. Subsequently discovered enzymes with the same specificity are
termed isoschisomers. An isoschisomer may differ from the prototype in site
preferences, reaction conditions, as well as in sensitivity to methylation and
exhibition of star activity. Restriction enzymes that recognize the same
nucleotide sequence, but cleave DNA at different positions are called
neoschisomers.
Site preferemces by restriction endonucleases
In 1975, Thomas and Davis discovered that EcoRI cleaves its five
recognition sites on phage λ DNA at rates that differ by an order of
magnitude. Similar examples have been documented for other restriction
enzymes. Factors such as flanking sequences, methylation of DNA and the
number of cleavage sites appear to influence cleavage efficiency. Cleavage
of resistant sites can be significantly enhanced by the addition of cleavable
DNA and oligodeoxyribonucleotide containing the recognition site, or
spermidine.
Types of restriction enzymes:
1) type I enzymes cleave at sites remote from recognition site; require
both ATP and S-adenosyl-L-methionine to function;
multifunctional protein with both restriction and methylase
activities;
2) type II enzymes cleave within or at short specific distances from
recognition site; most require magnesium; single function
(restriction) enzymes independent of methylase;
3) type III enzymes cleave at sites a short distance from recognition
site; require ATP (but doesn't hydrolyse it); S-adenosyl-L-
methionine stimulates reaction but is not required; exist as part of a
complex with a modification methylase;
4) type IV enzymes target methylated DNA.
Star activity
46
Restriction enzymes recognize specific nucleotide sequences within
DNA molecules. However their recognitionsite specificity can be reduced in
vitro. Under certain conditions, enzymes are able to recognize and cleave
nucleotide sequences which differ from the canonical site. At low ionic
strength, for example, BamHI (with the recognitionsequence GGATCC) is
able to cleave the following sequences: NGATCC, GPuATCC and
GGNTCC. This phenomenon is called “relaxed” or “star” activity. For most
restriction enzyme applications, star activity is not desirable.
Star activity is the result of:
1) prolonged incubation (over digestion);
2) high enzyme concentration in the reaction mixture;
3) high glycerol concentration in the reaction mixture;
4) presence of organic solvents, such as ethanol, dimethyl sulfoxide
(DMSO) or dimethyl formamide (DMFA), in the reaction mixture;
5) low ionic strength of the reaction buffer;
6) suboptimal pH values of the reaction buffer;
7) substitution of Mg
2+
for other divalent cations, such as Mn
2+
or
Co
2+
.
In some cases, the termini generated by DNA cleavage with a
restriction enzyme at the canonical site have been shown to stimulate
enzyme star activity.
Both star activity and incomplete DNA digestion result in atypical
electrophoresis patterns that can be distinguished by careful examination of
gel images.
Differences between star activity and incomplete digestion
Star activity results in additional DNA bands below the expected
bands below the expected bands and no additional bands above the largest
expected fragment. These additional bands become more intense, while the
expected bands become less intense, when either the incubation time or the
amount of enzyme is increased.
Incomplete DNA digestion results in additional bands above the
expected DNA bands on the gel. Additional bands disappear when the
incubation time or amount of enzyme is increased. No additional bands
below the smallest expected fragment are observed.
Certain restriction enzymes (for example, Alol, Taul, EcoRII)
remain associated with the substrate DNA after cleavage and cause a change
in the mobility of the digestion products during electrophoresis. In these
cases SDS is added, solution is heated for 10 min at 65
o
C and chill on ice
prior to loading on the gel.
Digestion of methylated DNA
47
DNA methylation is the process of transferring a methyl group
from a donor molecule to either cytosine or an adenine by DNA
methyltransferases. Such methylation is the most common and abundant
DNA modification process in living organisms. Thre types of methylated
bases are predominately found in DNA: 1) 5`-methylcytosine (m5C); 2) N4-
methylcytosine (m4C); 3) N6-methyladenine (m6A).
In prokaryotes, DNA cleavage by a cognate restriction enzyme is
prevented by the methylation of DNA by a sequnce-specific
methyltransferase which is an integral component of every restriction-
modification (R-M) system. The majority of E. coli strains used for
propagation of plasmid DNA contain two-site specific DNA
methyltranferases – Dam and Dcm. The methylase encjded by the dam gene
methylates the N6-position of an adenine residue with the GATC sequence.
The methylase encoded by the dcm gene methylates the C5-position of the
internal cytosine residue within the CCWGG sequence.
DNA from higher eukaryotic organisms possesses modified 5-
methylcytosine residues within CpG or CpNpG contexts. These tissue-
specific methylation patterns are heritable. They participate in regulation of
gene expression and cellular differentiation.
In cases where a restriction enzyme target site overlaps a
methylation site, the following digestion results are possible: 1) no effect; 2)
partial inhibition; 3) complete block.
To achieve effective DNA digestion, it is necessary to take into
account both the type of DNA methylation and the sensitivity of the
restriction enzyme to the type of methylation.
Sigle letter code (for describing sites of restriction enzymes)
R = G or A; Y = C or T; W = A or T; M = A or C; K = G or T; S = C or G;
H = A, C or T; V = A, C or G; B = C,G, or T; D = A, G or T; N = G, A, T or
C (A, T, G, C – nitrogen bases).
Examples of restriction enzymes
Alul
Dam, Dcm, CpG – no effect. Becomes inactivated
if heated 20 min at 65
o
C. 4 bp from end of DNA is
required for complete digestion. The optimal
incubation temperature is 37
o
C.
*Interesting details. Generally restriction enzymes are active in
PCR buffer.
XceI (NspI)
48

Dam, Dcm, CpG – no effect. Becomes inactivated
if heated 20 min at 65
o
C. The optimal incubation
temperature is 37
o
C.
Tail (MaeII)
To ensure efficiency of digestion, perform the
cleavage reaction under paraffin oil. Optimal
temperature – 65
o
C. If methylation occurs in CpG
(in cytosine) site the cleavage is blocked ("CpG" is
shorthand for "-C-phosphate-G-", that is, cytosine and guanine separated by
a phosphate, which links the two nucleosides together in DNA). Can not be
inactivated if heated . 2 bp from end of DNA is required for complete
digestion. The optimal incubation temperature is 65
o
C.
*Interesting details. H.O. Smith, K.W. Wilcox, and T.J. Kelley,
working at Johns Hopkins University in 1968, isolated and characterized the
first restriction nuclease whose functioning depended on a specific DNA
nucleotide sequence. Working with Haemophilus influenzae bacteria, this
group isolated an enzyme, called HindII, that always cut DNA molecules at
a particular point within a specific sequence of six base pairs. This sequence
is: 5' G T (T or C) ( A or G) A C 3'
3' C A (A or G) (T or C) T G 5'
They found that the HindII enzyme always cuts directly in the
center of this sequence. Wherever this particular sequence of six base pairs
occurs unmodified in a DNA molecule, HindII will cleave both DNA
strands between the 3rd and 4th base pairs of the sequence.
Restriction
analysis
This method is
based on the ability of
restriction enzymes to
find and cleave
specific sites of DNA.
49
Fig. 7. Scheme of restriction analysis
If change in site sequence occurs (mutatation, recombination,
methylation of DNA, etc.) a restriction enzyme loses its ability to
carry its function. The basic idea is: different sequences of DNA lead
to cutting DNA with restriction enzymes in different sites. Got with
restriction enzymes DNA fragments serve as markers of different
processes that take place inside the cells of investigated organism.
*Lab Shelf
PEG – polyethylene glycol. PEG is used in a
number of toothpastes as a dispersant. PEG is a
commonly used as precipitant for plasmid DNA
isolation and protein
crystallization. When
attached to various
protein medications,
polyethylene glycol
allows a slowed clearance of the carried protein
from the blood. It replaces water in wooden
objects, which makes the wood dimensionally
stable and prevents warping or shrinking of the
wood when it dries. In the field of microbiology,
PEG precipitation is used to concentrate
viruses.
Water - no tea in the evening without it.
50
