Moss Tom. DNA-protein interactions: principles and protocols
Подождите немного. Документ загружается.

Ethylation Interference 243
14. Bushman, F. D., Anderson, J. E., Harrison, S. C., and Ptashne, M. (1985)
Ethylation interference and X-ray crystallography identify similar interactions
between 434 repressor and operator. Nature 316, 651–653.
15. Ptashne, M. (1992) A genetic switch: Phage [lambda] and higher organisms. Cell
and Blackwell Scientific, Cambridge, MA; 2nd edition.
16. Summers, M. F., Powell, C., Egan, W., Byrd, R. A., Wilson, W. D., and Zon, G.
(1986) Alkyl phosphotriester modified oligodeoxyribonucleotides. VI. NMR and
UV spectroscopic studies of ethyl phospotriester (Et) modified Rp–Rp and Sp–Sp
duplexes, {d[GGAA(Et)TTCC]}2. Nucleic Acids Res. 14, 7421–7436.
17. Noble, S. A., Fisher, E. F., and Caruthers, M. H. (1984) Methylphosphonates as
probes of protein–nucleic acid interactions. Nucleic Acids Res. 12, 3387–3304.

Hydroxyl Radical Interference 245
245
From:
Methods in Molecular Biology, vol. 148: DNA–Protein Interactions: Principles and Protocols, 2nd ed.
Edited by: T. Moss © Humana Press Inc., Totowa, NJ
16
Hydroxyl Radical Interference
Peter Schickor, Evgeny Zaychikov, and Hermann Heumann
1. Introduction
Interference studies are just the inverse approach of “footprinting” experi-
ments. In one type of experiment, the effect of a chemical modification of a
single base on the subsequent binding of a sequence-specific protein is deter-
mined, whereas in the other, it is the accessibility of the protein-bound DNA to
modification that is determined. Thus, the experiments necessarily differ in
the order in which the protein binding and DNA modification steps occur. The
“interference” approach is characterized first by chemical modification of the
DNA and by subsequent protein binding. Such studies provide information on
the change of the binding strength following single-base modification. This
change can either be positive or negative and can be quantified by the gel shift
assay (1) (see Chapters 2 and 5).
Here, we describe the use of hydroxyl radicals as the modifying reagent.
This probe has a number of advantages compared to the most commonly used
probes, such as dimethylsulfate (DMS) or ethylnitrosourea. Hydroxyl radicals
cleave the backbone of DNA with almost no sequence dependence, whereas
most other reagents react in a highly sequence-dependent manner with the
bases of the DNA. Furthermore, hydroxyl radicals modify the DNA by elimi-
nation of a nucleoside, producing a “gap” in one DNA strand (for details of the
chemical reaction, see Chapter 5). This allows the study of two kinds of effect:
1. The effect on protein binding caused by missing contacts. Other reagents prevent
binding by introducing bulky groups into a base of the DNA. Whether this reflects
the importance of a base for protein–DNA interaction or whether the bulky group
is just a steric hindrance is difficult to determine.
2. The effect on the DNA structure because of the eliminated base. The missing
nucleoside is a center of enhanced flexibility in the DNA. Therefore, the effect of
246 Schickor, Zaychikov, and Heumann
DNA flexibility on the protein–DNA interaction can be studied. The modifica-
tion of the DNA by hydroxyl radicals is a very fast and highly reproducible
experiment in contrast to other methods. The reagents needed are easily available.
1.1. Generation and Action of Hydroxyl Radicals
Hydroxyl radicals introduce randomly distributed nucleoside eliminations
and associated backbone cleavages in the DNA. The generation and action of
hydroxyl radicals is described in detail in Chapter 5.
1.2. Principle of the Procedure
A DNA fragment labeled either at the 3' or the 5' end is subjected to hydroxyl
radical treatment. The concentration of the hydroxyl radicals is adjusted so that
the number of base eliminations is less than one per DNA, this means that only
approx 10% of the DNA fragments will be modified. This population of ran-
domly modified DNA molecules is incubated with the protein under study (see
Fig. 1). Those DNA molecules that are still able to bind the protein can be
separated from those DNA molecules that are no longer able to bind the pro-
tein by nondenaturing gel electrophoresis or electrophoretic mobility shift. The
bands containing free DNA and the complexed DNA (Fig. 2) are eluted and,
after denaturation, are applied on a sequencing gel in order to determine the
positions of the modifications (Fig. 3). The relative effect on the binding
strength caused by a single-base elimination can be quantified by determining
the intensity change of the different bands by densitometric scanning.
1.3. Interpretation of the Interference Pattern
The interpretation of an electrophoresis pattern obtained by hydroxyl radi-
cal interference studies (Fig. 3) is not as straightforward as hydroxyl radical
footprinting studies. The reason is that a single-base elimination generated by
hydroxyl radicals can cause two effects on protein binding:
1. Lack of specific contacts between the protein and the DNA. This leads to a
decrease in the binding strength.
2. Enhanced flexibility of the DNA at the position of the base elimination and back-
bone cleavage. This can lead to an increase or a decrease of the binding strength.
A quantitative interpretation of the interference pattern is not always pos-
sible, as the intensity of the bands of the interference pattern reflects the sum of
the different effects contributing to the binding strength. Additional informa-
tion concerning the protection of the DNA by the protein (e.g., by using
hydroxyl radical footprinting) is necessary in order to differentiate between the
two effects. The examples in the following subheadings may be used as a guide-
line for interpretation.
Hydroxyl Radical Interference 247
1.4. Examples of the Application of Hydroxyl Radicals
as Interference Probes
1.4.1. Transcription Factors
The interference of binding of some eubacterial and eukaryotic transcrip-
tion factors was investigated using hydroxyl radical pretreated DNA-binding
sequences. In all cases, the single-base elimination led to a decrease of the
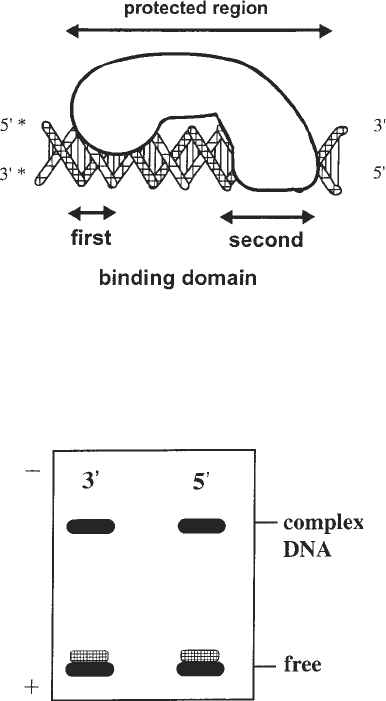
Fig. 1. A schematic representation of a protein–DNA complex with two distinct
interaction sites. At the first binding site, the protein interacts with only one side of the
DNA. At the second binding site, the protein wraps fully around the DNA. The aster-
isk indicates the position of the radioactive label.
Fig. 2. Nondenaturing gel electrophoresis of the complex with hydroxyl radical
pretreated DNA as target for the protein binding. The two lines show the same com-
plex labeled at the 3' and the 5' ends, as indicated in Fig. 1. Note: The single-base
elimination leads to an enhanced flexibility of the DNA indicated by the “smear” of
the band representing the free DNA.

248 Schickor, Zaychikov, and Heumann
binding strength indicating a loss of contacts. Examples are the progesterone
receptor (2), λ-repressor, cro-protein (3), necrosis factor-κB (4), and GCN4
transcription factor (5).
1.4.2. RNA Polymerase-Promoter Interaction
A strong eubacterial promoter was subjected to hydroxyl radical treatment
in order to investigate the influence of single-base eliminations on the binding
of Escherichia coli RNA polymerase (6). This study revealed three patterns of
interaction that could be attributed to different sites of the promoter:
Fig. 3. A putative interference pattern of the protein DNA complex (Fig. 1) obtained
after separation of “free” DNA and “complexed” DNA by a nondenaturing gel elec-
trophoresis (Fig. 2). Lanes I and 4 show the pattern of the “free” DNA labeled at the 3'
and the 5' ends, respectively. Lanes 2 and 3 show the pattern of the “complexed”
DNA. Lanes G contain the length standards obtained by a G-specific Maxam–Gilbert
sequencing reaction.
Hydroxyl Radical Interference 249
1. Direct base contact with the template strand in the “–35 region”: This was con-
cluded from the reduced affinity of the polymerase for a promoter having a base
eliminations in this region together in combination with the results of hydroxyl
radical footprinting studies (7), which revealed close contacts between the bases
of this sequence and the protein. This is an example in which a base elimination
leads to missing contacts between protein and DNA.
2. A DNA-structure-dependent interaction in the “–10 region”: This was inferred
from the increased binding affinity of the polymerase for a promoter having base
eliminations in this region in conjunction with footprinting studies that revealed
that this region is in close contact with the protein (7). This is an example in
which a base elimination leads to enhanced flexibility of the DNA favoring pro-
tein binding and suggests that the DNA adopts a particular conformation when
bound to the protein.
3. An interaction that is based on a defined spatial relationship between the “–35
region” and the “–10 region” domains. This conclusion was drawn from the
following findings:
a. Base elimination within the promoter region between the two domains reduces
the binding affinity of RNA polymerase.
b. The eliminated bases had no contact with the protein (as shown by their
accessibility to hydroxyl radical footprinting studies [7]).
c. The effect was observed for both DNA strands. This is an example in
which base elimination leads to a loss of the defined spatial relationship
between two functionally important sites because of an enhancement in
DNA flexibility.
2. Materials
2.1. The Cutting Reaction
Prepare the following solutions separately (see Note 1):
1. 0.1 M dithiotreitol (DTT).
2. 1% Hydrogen peroxide.
3. Iron(II)–EDTA-mix: Mix equal volumes of 2 mM ammonium iron(II) sulfate
hexahydrate ([NH
4
]
2
Fe[SO
4
]
2
·6H
2
O) and 4 mM EDTA.
4. Stop mix: 4% glycerol, and 0.6 M sodium acetate.
2.2. The Sequencing Gel
1. Urea (ultra pure).
2. 20X TBE: 1 M Tris base, 1 M boric acid, and 20 mM EDTA.
3. Acrylamide solution: 40% acrylamide, and 0.66% bis-acrylamide (see Note 2).
4. 10% Sodium persulfate (see Note 3).
5. 10% TEMED.
6. Sequencing gel (8%): 21 g urea, 2.5 mL of 20X TBE, and 10 mL of 40%
acrylamide solution are made up to 50 mL with bidistilled H
2
O and stirred under
mild heating until urea is dissolved. The solution is filtered (filter pore size: 0.2 µm)
250 Schickor, Zaychikov, and Heumann
and degassed for 5 min. Then, 0.3 mL of 10% sodium persulfate and 0.3 mL of
10% TEMED are added immediately before pouring the solution between the
glass plates.
7. Loading buffer for the sequencing gel (stock solution): 100 mL formamide
(deionized), 30 mg xylenecyanol FF, 30 mg bromophenol blue, and 750 mg
EDTA.
8. Electrophoresis buffer: 1X TBE.
2.3. The Nondenaturing Gel for DNA Isolation
and Electrophoretic Mobility Shift Assay
1. 20X TBE: as described in Subheading 2.2.
2. Acrylamide solution: 30% acrylamide, and 0.8% bis-acrylamide (see Note 2).
3. 10% Sodium persulfate (see Note 3)
4. 10% N,N,N',N'-tetramethylethylene diamine (TEMED) (aqueous solution).
5. 3% Nondenaturing gel: 1.5 mL of 20X TBE, 3 mL of the acrylamide solution,
and 25.5 mL of bidistilled water are mixed and degassed for 5 min. Then 300 µL
of 10% ammonium persulfate and 300 µL of 10% TEMED are added before
pouring the solution between the glass plates.
6. Loading buffer for the nondenaturing gel (stock solution): 50% glycerol, and
0.1% bromophenol blue.
7. Electrophoresis buffer: 1X TBE.
2.4. Other Items
1. Sequencing gel apparatus.
2. Apparatus for nondenaturing gel electrophoresis.
3. Filters for drop dialysis, VS, 0.025 µm (Millipore, Bedford, MA).
4. SpeedVac concentrator.
5. 1X TE: 10 mM Tris-HCl, pH 7.9, and 1 mM EDTA.
3. Methods
3.1. Establishing the Conditions
for Obtaining Specific Protein–DNA Complexes
The method of establishing the conditions for complex formation using the
electrophoretic mobility-shift assay is described in Chapter 4. The enzyme to
DNA ratio should be adjusted so that the ratio of complexed to free DNA is
about 1:1 (see Note 4).
3.2. Hydroxyl Radical Base Elimination
1. End-label an aliquot of the DNA fragment of interest under standard conditions
(8) at the 5'-position, using T
4
polynucleotide kinase and [γ-
32
P] ATP and end
label a second aliquot at the 3'-position, using the Klenow fragment of DNA
polymerase I and the appropriate [α-
32
P] dNTP. In each case remove one label
end by asymmetric cleavage of the DNA fragment with an appropriate restriction
Hydroxyl Radical Interference 251
endonuclease. Purify the uniquely end-labeled DNA fragments by nondenaturing
gel electrophoresis. The total amount of DNA in an assay of 20 µL should not be
below 100 ng (see Note 5).
2. Dissolve each end-labeled DNA preparation in 20 mL of 1X TE buffer. Add to
both samples 2 µL each of the previously prepared solutions of DTT, hydrogen
peroxide, and the iron(II)–EDTA mix by putting single drops of each solution on
the inner wall of the tube and rapidly mixing the three drops before combining
them with the sample using a micropipet.
3. Incubate for 3–4 min at room temperature.
4. Add 25 mL stop mix and 150 µL of ice-cold 100% ethanol to precipitate the
DNA. Keep the solution at –70°C for 30 min.
5. Recover the DNA by microcentrifugation for 30 min. Wash the pellet with ice-
cold 80% ethanol, dry the pellet under vacuum, and redissolve the pellet in 20–50 mL
of 1X TE buffer.
6. Heat the sample for not longer than 2 min at 90°C and place on ice (see Note 5).
Apply the sample onto a 6–10% sequencing gel (for the analysis of fragments in
the range of 50–150 bases a gel consisting of 8% acrylamide is adequate). Use as
length standards a Maxam–Gilbert sequencing reaction of the 5'- or 3'- labeled
DNA fragment.
7. Run the gel at 50 W at a temperature of 60°C for 1.5–2 h. The gel is ready when
the xylenecyanol dye marker is about 3–5 cm above the bottom of the gel.
8. After electrophoresis, expose the gel to an X-ray film using an intensifying screen
at –70°C overnight. For subsequent experiments choose the time of hydroxy radi-
cal cleavage that provides an even distribution of bands and leaves around 90%
of the DNA uncleaved.
3.3. Interference Studies on Protein-DNA Complexes
1. Prepare two 20-µL samples of the complex to be studied using DNA labeled
respectively at the 3' and the 5' ends and hydroxy radical treated as described in
Subheading 3.2. Use conditions established for optimal complex formation (see
Subheading 3.1.). The total amount of radioactivity in one assay should be
around 60,000–80,000 cpm.
2. Pour 30–40 mL of the dialysis buffer containing 8 mM Tris-HCl, pH 7.9, into a
Petri dish (see Note 6). Place a Millipore filter (see Subheading 2.4.) on the
surface of the buffer, shiny side (hydrophobic side) up. Put the samples contain-
ing the complexes onto the filter for 1 h in order to remove salt (see Note 7).
Remove the samples from the filter by a micropipet and transfer them to a fresh
1.5-mL Eppendorf tube.
3. Separate the complex and the free DNA on a nondenaturing acrylamide gel elec-
trophoresis. The method for separation of complexes from free DNA is described
in Subheading 3.3.2. in Chapter 5.
4. The whole procedure for the recovery of DNA from the gel is described in Sub-
heading 3.3.2. in Chapter 5.
252 Schickor, Zaychikov, and Heumann
5. Adjust the amount of radioactivity and volume in each sample to about 5000–
6000 cpm in 4–5 µL. Heat the samples for not longer than 2 min at 90°C and put
them on ice (see Note 8).
6. Analyze the DNA by denaturing gel electrophoresis. Apply the samples to a
6–10% sequencing gel (for the analysis of fragments in the range of 50–150 bases
a gel consisting of 8% acrylamide is adequate). Load the following samples on
the gel: end-labeled DNA, the Maxam–Gilbert reaction as length standard, the
free DNA recovered from the gel, and the DNA recovered from the complex.
7. Run the gel for 1.5–2 h at about 50 W to obtain a temperature of 60°C.
8. Expose the gel after electrophoresis to an X-ray film using an intensifying screen
at –70°C overnight.
4. Notes
1. The iron(II), iron-EDTA mix, and the H
2
O
2
solutions should be freshly made
before use. The solutions of DTT (0.1 M), EDTA (4 mM), H
2
O
2
(as a 30% stock
solution), and the stop mix are stable for months being stored at –20°C.
2. The acrylamide solutions are stable for months if protected from light and
kept at 4°C.
3. Sodium persulfate has an advantage over routinely used ammonium persulfate of
being much more stable in aqueous solution. The 10% sodium persulfate solution
may be kept at least 1 mo at 4°C without loss of activity.
4. It is advisable to keep the enzyme-to-DNA ratio <1 in order to assure stringent
sequence selection conditions.
5. It is strongly recommended to check the quality of the labeled DNA on a
sequencing gel before use. Nicks in the double strand, which could result from
DNase activities during the preparation procedure, will appear as additional bands
in the sequencing gel. This admixture of bands would spoil the whole footprint,
even when present in only small amounts. Furthermore, it is recommended not to
store the freshly labeled DNA longer than 2 wk because of the danger of radia-
tion damage to the DNA.
6. The buffer conditions can be varied (e.g., the pH), but the ionic strength should
not be too high (a maximum of 50 mM NaCl) in order to obtain sharp bands
during the following electrophoresis. Many protein–DNA complexes are very
stable at low ionic strength (e.g., complexes between RNA polymerase and pro-
moters [6]). Therefore, in most cases the stability of the pH in the following
electrophoresis is the only limitation to lowering the ionic strength.
7. The purpose of the dialysis is the removal of salt, the presence of which would
lower the quality of the electrophoresis pattern. As a rough approximation, one can
remove up to 80–90% of the salt present in the sample within 1 h of drop dialysis.
8. Longer heating or boiling creates additional cuts in the DNA.
References
1. Heumann, H., Metzger, W., and Niehörster, M. (1986) Visualization of interme-
diary transcription states in the complex between Escherichia coli DNA-depen-
Hydroxyl Radical Interference 253
dent RNA polymerase and a promoter-carrying DNA fragment using the gel
retardation method. Eur. J. Biochem. 158, 575–579.
2. Chalepakis, G. and Beato, M. (1989) Hydroxyl radical interference: a new method
for the study of protein–DNA interaction. Nucleic Acids Res. 17, 1783.
3. Hayes, J. J. and Tullius, Th. D. (1989) The missing nucleoside experiment: a new
technique to study recognition of DNA by protein. Biochemistry 28, 9521–9527.
4. Schreck, R., Zorbas, H., Winnacker, E. L., and Baeuerle, P. A. (1990) The
nf-kappa-b transcription factor induces DNA bending which is modulated by its
65-kd subunit. Nucleic Acids Res. 18, 6497–6502.
5. Gartenberg, M. R., Ampe, C., Steitz, T. A., and Crothers, D. M. (1990) Molecular
characterisation of the GCN4-DNA complex. Proc. Natl. Acad. Sci. USA 87,
6034–6038.
6. Werel, W., Schickor, P., and Heumann, H. (1991) Flexibility of the DNA enhances
promoter affinity of Escherichia coli RNA polymerase. EMBO J. 10, 2589–2594.
7. Schickor, P., Metzger, W., Werel, W., Lederer, H., and Heumann, H. (1990)
Topography of intermediates in transcription initiation of E. coli. EMBO J. 9,
2215–2220.
8. Maniatis, T., Fritsch, E. F., and Sambrock, J. (1982) Molecular Cloning. A Labo-
ratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
