Moss Tom. DNA-protein interactions: principles and protocols
Подождите немного. Документ загружается.

222 Shaw and Stewart
fractions are recovered from the gel. Methylated residues are converted into
strand scissions and the free and bound DNA fractions are compared on a
sequencing gel. A complete analysis requires the examination of both strands.
This is accomplished by preparing two DNA probes, each uniquely labeled at
one end, and carrying both probes through the protocols. A binding site char-
acterized by methylation protection will therefore appear as a cluster of altered
DMS reactivities.
In methylation interference (8,9), DNA is first reacted with DMS, purified
and then presented to protein. Under the reaction conditions used methylation
is partial, yielding approximately one methylated base per DNA molecule.
Thus, the protein is presented with a mixture of DNA molecules that differ
with respect to the positions of methyl groups. Some methyl groups will
interfere with protein binding because they lie in or near the binding site. Gel
retardation separates the mixture into two fractions: free DNA, which, as long
as DNA is in excess over binding activity, represents the total profile of methy-
lation reactivity, and bound DNA, which will not contain any molecules with
methyl groups incompatible with binding. Both free and bound DNA fractions
are recovered, methylated residues are converted to strand scissions, and the
fractions are compared on a sequencing gel. The binding site is observed as
the absence of bands in the bound sample corresponding to the positions where
methylation interferes with binding.
It is obvious that these two uses of DMS may not deliver identical results.
For example, Fig. 1 presents a comparison obtained from experiments with the
serum response element binding factors p67
SRF
p62
TCF
and their binding site in
the human c-fos promoter (SRE). Because the use of DMS in vivo for genomic
footprinting is limited to the equivalent of methylation protection, a direct com-
parison between in vivo and in vitro data excludes the more widely used
methylation interference assay.
The two techniques are, however, very similar in practical terms and thus
are presented together. Both techniques rely on preestablished conditions that
permit a protein–DNA complex to be resolved in a gel retardation assay
(Chapter 2) and on chemical DNA sequencing methodologies, for which the
reader is advised to consult ref. 2 for a detailed treatment.
2. Materials
1. Dimethyl sulfate (DMS) (Merck), analytical grade.
2. Piperidine (Sigma), analytical grade; use freshly made 1:10 dilution in double-
distilled water.
3. Phenol/chloroform 50% v/v, buffered with 50 mM Tris-HCl, pH 8.0.
4. NA45 paper (Schleicher & Schuell).
5. 3MM paper (Whatman) or GB 002 paper (Schleicher & Schuell).

Methylation Protection and Interference 223
6. Electrophoresis equipment suitable for gel retardation or EMSA.
7. Electroblotting apparatus for Western transfer (e.g., Bio-Rad Trans-Blot).
8. Standard DNA sequencing gel electrophoresis equipment.
9. Vacuum gel drier (optional).
10. TBE buffer: 89 mM Tris base, 89 mM boric acid, and 2 mM EDTA. Make 10X
stock as 108 g Tris base, 55 g boric acid, and 40 mL of 0.5 M EDTA pH 8.3
per liter.
11. NA45 elution buffer: 10 mM Tris-HCl, pH 8.0, 1 mM EDTA, and 1 M NaCl.
12. Carrier DNA: Salmon testis DNA or calf thymus DNA (Sigma), dissolved at
3 mg/mL in 10 mM Tris-HCl, pH 8.0, and 1 mM EDTA, and sheared.
13. Sequencing loading buffer: 90% formamide, 10 mM EDTA, and 0.1% (w/v)
bromophenol blue, 0.1% (w/v) xylene cyanol blue.
14. Gel retardation loading buffer: 20% Ficoll, 20 mM EDTA, 0.1% w/v bromophe-
nol blue.
15. 2X DMS buffer: 120 mM NaCl, 20 mM Tris-HCl, pH 8.0, 20 mM MgCl
2
, and
2 mM EDTA.
16. DMS stop buffer: 1.5 M NaAc, pH 7.0, and 1 M 2-mercapto-ethanol, store frozen.
17. X-ray film (e.g., Kodak X-Omat) or imaging plate for phosphorimager.
3. METHOD
3.1. Methylation Protection
1. Incubate 300,000 cpm of uniquely end-labeled DNA probe (see Note 1) and a
corresponding amount of protein together in a total volume of 100 µL, as previ-
ously optimized for gel retardation analysis.
2. Add 1 µL of DMS and incubate at room temperature (the incubation time depends
on the length of the DNA probe and is empirical; as a guide for a 200-bp frag-
ment, 1.5 min, for a 50-bp oligonucleotide duplex, 3 min).
3. Add 1/10 vol of 250 mM dithiothreitol (DTT), mix gently, add 1/10 vol of gel
retardation loading buffer, mix gently, load onto a 2-mm-thick retardation gel in
Fig. 1. Comparison of methylation interference and protection patterns formed by
factors binding at the c-fos serum response element (SRE) in vitro and in vivo. G
residues identified by methylation interference (9), methylation protection and in vivo
genomic footprinting (7) are indicated. An additional G on both the upper and lower
strands is implicated in the protein–DNA interaction by methylation protection.
224 Shaw and Stewart
1X TBE (or an alternative buffer as determined to be best for the given complex)
and run as optimized for analytical gels. The load may need to be spread over up
to 10 times more well area as compared with an analytical retardation assay (see
Note 2).
4. After electrophoresis, separate the glass plates carefully so that the gel adheres to
one plate and cover the gel with cling film. Expose to X-ray film long enough to
reveal complexes clearly (i.e., 6 h to overnight). The alignment of the film to the
gel must be reliably marked.
5. Put the developed film on a light box. Remove the gel from the cling film and
realign it on the X-ray film. Cut pieces of NA45 paper sufficiently large to cover
individual complexes in the gel yet small enough to fit into 1.5-mL tubes when
rolled up. Wet the paper pieces in retardation gel running buffer and, with the
help of tweezers, position one over each complex of interest in the gel, as
visualized from the underlying film. Also position a similar sized piece of paper
over (some of) the uncomplexed DNA. NA45 paper can be labeled with pencil
before wetting.
6. Carefully cover the gel and paper pieces with two sheets of 3MM paper wetted in
1X TBE (or alternative gel running buffer from step 3). Lay a ScotchBrite pad
from the electroblotting apparatus on top of the paper and turn the gel over. Care-
fully remove the second glass plate, cover the other side of the gel with 3MM
paper and ScotchBrite as before and insert the package into an electrotransfer
apparatus as described in the manufacturer’s instructions with the NA45 paper
toward the anode. Transfer in 1X TBE (or the alternative retardation gel buffer)
at 80 V for 1.5 h (see Note 3).
7. Stop the transfer, unpack the gel carefully with the NA45 paper on top and trans-
fer each piece to a labeled 1.5-mL tube containing 600 µL of elution buffer (check
that the radioactivity has transferred to the paper). Incubate at 70°C for 1 h.
8. Remove NA45 paper from each tube, check that at least half the radioactivity has
eluted into the buffer (do not expect quantitative elution, but at least 50% should
come off), add 20 µg carrier DNA, extract with phenol/chloroform and precipi-
tate with 1 volume of isopropanol. Wash precipitate once in 70% ethanol and dry
under vacuum. (See Notes 4 and 5.)
9a. To reveal modified Gs: Dilute piperidine 1:10 in water and add 50 µL to each
pellet. Vortex briefly and incubate at 90
o
C for 30 min (tubes must be clamped
or weighted down to prevent the lids opening); then dry under a good vacuum.
Take up the samples in 100 µL of water and repeat the drying process. This
strand scission protocol should not convert methylated A residues into strand
breaks. It is often observed, however, that breakages at As do occur with reason-
able efficiency.
9b. To reveal modified As and Gs: The following modification will produce efficient
cleavage at both methyl-G and methyl-A residues. After the preparative retarda-
tion gel, resuspend the dried, purified DNA in 30 µL of 10 mM sodium phosphate
pH 6.8, and 1 mM EDTA. Incubate for 15 min at 92°C. Then add 3 µL of 1 M
NaOH and incubate for 30 min at 92°C, followed by 320 µL of 500 mM NaCl,
Methylation Protection and Interference 225
50 µg/mL carrier DNA, and 900 µL ethanol. Chill and centrifuge to pellet the
radioactivity. Wash the pellet in 70% ethanol and dry.
10. Measure the Cerenkov counts in each tube, then redissolve the samples in water
(e.g., 10 cpm/µL) and transfer equivalent counts (1000 cpm in each case would
be optimal) from each into fresh tubes. Dry down and redissolve in 5 µL
sequencing loading buffer.
11. Prepare and pre-electrophorese a standard sequencing gel (5–12% acrylamide,
depending on probe length). Denature probes at 95
o
C for 5 min, snap cool in ice
and load onto the gel. Run the gel until optimal separation of sequence is
achieved. (See Notes 6–8.)
12. Stop electrophoresis, remove the gel from the tank and lift off one glass plate. Fix
the gel in 20% ethanol and 10% acetic acid for 10 min. Drain briefly and then
overlay the gel with two sheets of 3MM paper and carefully peel it off the glass
plate. Cover the gel surface with cling film and dry on a vacuum gel drier (see
Note 9). Expose the dry gel to X-ray film with intensifying screens as necessary,
or to an imaging plate.
3.2. Methylation Interference
1. Mix 300,000 cpm of end-labeled probe (see Note 1); 100 µL of 2X DMS buffer
and water to 200 µL. Add 2 µL of DMS and incubate at room temperature (the
same guidelines as given in Subheading 3.1.2. apply for the reaction time). Stop
the reaction by the addition of 50 µL cold DMS stop mix and precipitate with 850 µL
cold ethanol. Redissolve in 200 µL cold 0.3 M NaAc pH 7.0, add 700 µL cold
ethanol, and reprecipitate. Wash twice in 80% ethanol and redissolve the probe
in water or binding buffer (about 20,000 cpm/µL).
2. Incubate the probe with protein for gel retardation as previously optimized for
gel retardation analyses of the complexes in question in a total volume of 100 µL.
3. Add 1/10 vol of gel retardation loading buffer, mix gently and load onto a
2-mm-thick retardation gel in 1X TBE (or alternative buffer); then, run as opti-
mized for analytical gels. However, the load should be spread over up to 10 times
more well area (see Note 2).
4. Continue with step 4 and all subsequent steps as described for methylation pro-
tection (Subheading 3.1.).
4. Notes
1. To have sufficient counts to complete the procedure, proceed with at least 10
times the amount of material required for a simple gel retardation analysis (i.e., at
least 300,000 cpm).
2. A common difficulty with these methods is the persistence of contaminants that
accompany DNA after the preparative retardation gel. These contaminants inter-
fere with the migration of DNA on the sequencing gel, producing blurred and
distorted patterns. In order to minimize this problem it is worth ascertaining the
load limit of the retardation gel so that the protein–DNA complex will not smear
but be well resolved and therefore concentrated in the gel before elution.
226 Shaw and Stewart
3. It is also possible to use a semidry electrotransfer apparatus (e.g., Bio-Rad Trans-
blot SD) to transfer the DNA from the gel retardation gel onto NA45 paper. In
this case, both the transfer time and potential are reduced.
4. In some instances, it may prove difficult to elute the DNA from the NA45 paper,
in which case raising the salt concentration or the temperature may improve elu-
tion. (Extending the incubation time does not seem to help.) If not, the batch of
NA45 may be to blame or it is even conceivable that the DNA–protein complex
in question is adsorbed too tightly onto the paper. It is not possible to phenol
extract the NA45 paper in order to remove bound protein–DNA.
5. Retain the isopropanol supernatants until you are sure the samples have precipi-
tated quantitatively. Add more carrier DNA if required.
6. It is similarly advisable to load as little material onto the sequencing gel as prac-
ticable. With the advent of the phosphorimager, the lower limit for the sequenc-
ing gel is well under 1000 cpm/lane.
7. If the end-labeled DNA fragment is relatively long and multiple binding sites are
to be resolved, a gradient or wedge sequencing gel can be used in step 11 of
Subheading 3.1.
8. An appropriate complement for the final result is to perform the Maxam and
Gilbert G+A reactions on the end-labeled probe. On the sequencing gel, these
reactions should provide unambiguous sequence information and, in case diffi-
culties are encountered, clues as to the steps that are problematic.
9. It is not essential to dry down the sequencing gel because after one glass plate has
been removed, it can be covered with cling film and exposed to X-ray film at
–70°C with one screen. This alternative should only be considered if the signal is
sufficiently strong or if a gel drier is not available.
References
1. Church, G. M. and Gilbert, W. (1984). Genomic sequencing. Proc. Natl. Acad.
Sci. USA 81, 1991–1995.
2. Maxam, A. and Gilbert, W. (1980) Sequencing end-labelled DNA with base-spe-
cific chemical cleavages. Methods Enzymol. 65, 499–560.
3. Fried, A. and Crothers, D. M. (1981) Equilibria and kinetics of lac repressor–
operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 9,
6505–6525.
4. Garner, M. M. and Revzin, A. (1981) A gel electrophoresis method for quantify-
ing the binding of protein to specific DNA regions: application to components of
the E. coli lactose operon regulatory system. Nucleic Acids Res. 9, 3047–3059.
5. Gilbert, W., Maxam, A., and Mirzabekov, A. (1976) Contacts between the LAC
repressor and DNA revealed by methylation. in Control of Ribosome Biosynthe-
sis, Alfred Benzon Symposium IX (Kjelgaard, N. O. and Maaloe, O., eds.), Academic,
New York, pp. 139–148.
6. Johnsrud, L. (1978) Contacts between Escherichia coli RNA polymerase and a
lac operon promoter. Proc. Natl. Acad. Sci. USA 75, 5314–5318.
Methylation Protection and Interference 227
7. Herrera, R. E., Shaw, P. E., and Nordheim, A. (1989). Occupation of the c-fos
serum response element in vivo by a multi-protein complex is unaltered by growth
factor induction. Nature 340, 68–70.
8. Siebenlist, U. and Gilbert, W. (1980) Contacts between E. coli RNA polymerase
and an early promoter of phage T7. Proc. Natl. Acad. Sci. USA 77, 122–126.
9. Shaw, P. E., Schröter, H., and Nordheim, A. (1989). The ability of a ternary com-
plex to form over the serum response element correlates with serum inducibility
of the human c-fos promoter. Cell 56, 563–572.

Ethylation Interference 229
15
Ethylation Interference
Iain W. Manfield and Peter G. Stockley
1. Introduction
Structural studies of DNA–protein complexes have now made it clear that
specific sequence recognition in these systems is accomplished in two ways,
either directly by the formation of hydrogen bonds to base-pair edges from
amino acid side chains located on a DNA-binding motif, such as a helix–turn–
helix, or indirectly as a result of sequence-dependent distortions of the DNA
conformation (1). These contacts occur in the context of oriented complexes
between macromolecules that juxtapose the specific recognition elements. As
part of these processes, proteins make a large number of contacts to the
phosphodiester backbone of DNA, as was predicted from biochemical assays
of the ionic strength dependence of DNA binding.
Contacts to phosphate groups can be inferred by the ethylation interference
technique (2). Ethylnitrosourea (EtNU) reacts with DNA to form, principally,
phosphotriester groups at the nonesterified oxygens of the otherwise
phosphodiester backbone. Minor products are the result of the reactions of
EtNU with oxygen atoms in the nucleotide bases themselves (see Note 1).
Under alkaline conditions and at high temperature, the backbone can be cleaved
at the site of the modification to form a population of molecules carrying either
3'-OH or 3'-ethylphosphate groups.
The length of the ethyl group (approx 4.5 Å) means that at a number of
positions along a DNA molecule encompassing the binding site for a protein,
complex formation will be inhibited by the presence of such a modification. At
other sites, outside the binding site, no interference with protein binding at the
specific site will be observed. Addition of the DNA-binding protein to a
randomly ethylated DNA sample, followed by some procedure to separate the
complexes formed from unbound DNA, will fractionate the DNA sample into
229
From:
Methods in Molecular Biology, vol. 148: DNA–Protein Interactions: Principles and Protocols, 2nd ed.
Edited by: T. Moss © Humana Press Inc., Totowa, NJ
230 Manfield and Stockley
those molecules able to bind protein with high affinity and those for which the
ethylation has lowered the affinity (Fig. 1). In practice, modification at differ-
ent sites produces molecules with a spectrum of affinities for the protein. It is,
therefore, not possible to prove conclusively that a particular phosphate is con-
tacted by the protein, but only that ethylation at that site interferes with com-
plex formation.
Only occasionally are large amounts of pure protein readily available for in
vitro biochemical assay of DNA-binding activity and, often, only small
amounts of crude nuclear extracts are available. In many commonly used
assays, complex formation could not easily be detected in such situations. For
example, using DNase I or hydroxyl radicals, a high level of binding-site occu-
pancy is required for a footprint to be detected. Fractional occupancy is readily
detected by gel retardation of complexes (electrophoretic mobility shift
[EMSA]; see Chapter 2) but offers only limited characterization of the details
of the protein–DNA interaction. Interference techniques, such as the ethylation
and hydroxyl radical interference (see Chapter 16) techniques (3), do allow the
molecular details of complex formation to be studied even when only small
amounts of crude protein are available (4). Whatever the level of saturation,
DNA fragments modified at sites reducing the affinity of protein for DNA are
less likely to form complexes. Therefore, the bound fraction on gel retardation
assays will always give an indication of the sites that do not inhibit complex
formation when modified. The groups on the DNA recognized by the protein
can then be inferred. Ethylation can also be used to analyze RNA–protein com-
plexes (5,6).
We have used the ethylation interference technique to probe the interaction
of the Escherichia coli methionine repressor, MetJ, with its binding site in
vitro. Binding sites for MetJ consist of two, or more, immediately adjacent
copies of an 8-bp site with the consensus sequence 5'-AGACGTCT-3', which
has been termed a “met box.” X-ray crystallography has been used to deter-
mine the structure of the MetJ dimer, the complex with corepressor, S-adenosyl
methionine (SAM), and a complex of the holorepressor with a 19-mer oligo-
nucleotide containing two met boxes (7,8). The structure of the protein–DNA
complex in the crystal reveals two MetJ dimers (one per met box) binding to
the DNA by insertion of a β-ribbon into the major groove. The general features
of this model are corroborated by the results of the ethylation interference
experiments and by data from a range of other footprinting techniques.
2. Materials
2.1. Preparation of Radioactively End-Labeled DNA
1. Plasmid DNA carrying the binding site for a DNA-binding protein on a conve-
nient restriction fragment (usually <200 bp).
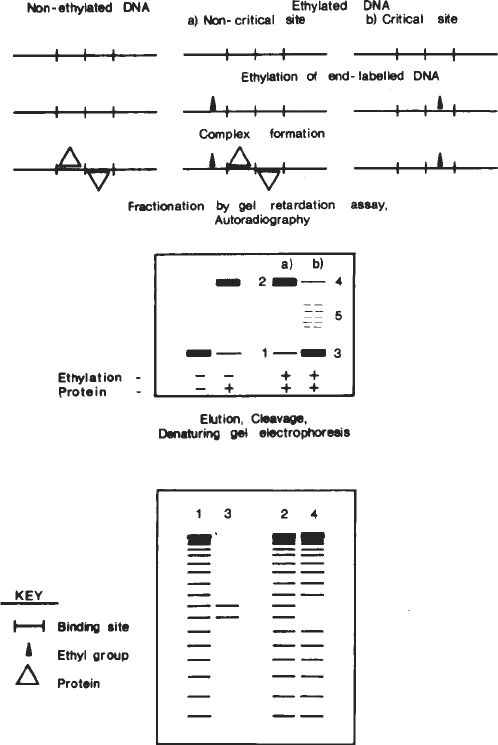
Ethylation Interference 231
Fig. 1. Diagrammatic outline of the ethylation interference experiment. The upper
section shows ethylation of end-labeled DNA (ethyl groups represented by small solid
triangles) and complex formation with protein (represented by large open triangles).
The expected mobility of each species on nondenaturing gels is shown in the middle
section. In idealized form, the pattern of cleavage products that might be expected
from recovery of each species after gel retardation assay is shown in the lower section.
In practice, samples represented by 1 + 3 and 2 + 4 migrate to the same position on the
retardation gel and are, therefore, not separated. The resultant pattern is shown in Fig. 2.
2. Restriction enzymes and the appropriate buffers as recommended by the suppliers.
3. Phenol: redistilled phenol equilibrated with 100 mM Tris-HCl, pH 8.0.
4. Chloroform.
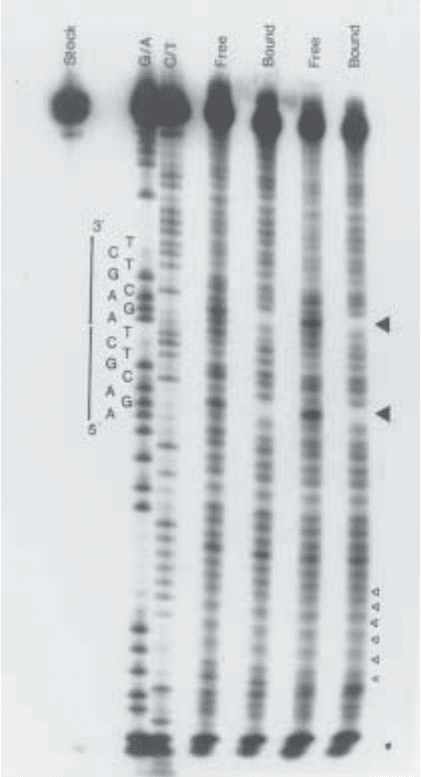
232 Manfield and Stockley
Fig. 2. Ethylation interference of MetJ–DNA interaction. Samples for denaturing
gel electrophoresis were prepared following the methods given here. “Stock” indi-
cates nonethylated DNA that has been through the cleavage reaction. “G/A” and
“C/T” are the products of the purine- and pyrimidine-specific Maxam–Gilbert chemi-
cal cleavage reactions, respectively. “Free” and “Bound” are the DNA fractions that
can and cannot form complexes, respectively. The two sets of data represent results
obtained with DNA ethylated for 30 min (left-hand lanes) or 60 min (right-hand lanes).
The sequence of the MetJ binding site is indicated along the side of the autoradio-
graph. The phosphate ethylation that interferes most strongly with complex formation
is indicated by a large solid triangle. Small, open triangles indicate the minor reaction
products of the cleavage reaction, which, for small fragments, are resolved on these gels.
