Moss Tom. DNA-protein interactions: principles and protocols
Подождите немного. Документ загружается.

160 Buckle and Travers
10. Rougée, M. and Bensasson, R. V. (1986) Détermination des constantes de vitesse
de désactivation de l’oxygène singulet (
1
D
7
) en presénce de biomolécules. Comp.
Rend. Acad. Sci. Paris 302, 1223–1226.
11. Travers, A. A., Lamond, A. I., Mace, H. A. F., and Berman, M. L. (1983) RNA
polymerase interactions with the upstream region of the E. coli tyrT promoters.
Cell 35, 265–273.
12. Maxam, A. M. and Gilbert, W. (1980) Sequencing end-labeled DNA with base-
specific chemical cleavages. Methods Enzymol. 155, 560–568.

UV-Laser Footprinting 161
161
From:
Methods in Molecular Biology, vol. 148: DNA–Protein Interactions: Principles and Protocols, 2nd ed.
Edited by: T. Moss © Humana Press Inc., Totowa, NJ
12
Ultraviolet-Laser Footprinting
Johannes Geiselmann and Frederic Boccard
1. Introduction
1.1. Measurement of DNA–Protein Interactions In Vitro
A large number of processes within the cell, in particular the regulation of
gene expression, rely on the binding of proteins to specific sites on the DNA. A
primary ingredient to understanding these processes is the characterization of
the protein–DNA interaction (1). Such a characterization consists in determin-
ing the position of the binding site on the DNA and measuring the affinity of
the protein for this recognition site. A wide variety of footprinting techniques
can accomplish this task (2,3).
1.2. Footprinting Techniques
These techniques involve the reaction of a footprinting reagent (in the larg-
est sense) with DNA and the subsequent localization and quantification of the
resultant DNA modification. Commonly used footprinting reagents include
DNase I, KMnO
4
, or dimethylsulfate (DMS) (2,3); see Chapters 3–14. Most of
these methods require extended incubation times of the reagent with the DNA–
protein complex, which may lead to artifacts if the footprinting reagent modi-
fies the complex. For example, DMS may react with the protein; DNase I
relaxes a supercoiled plasmid, which may destabilize the DNA–protein com-
plex under investigation. UV-laser footprinting eliminates several of these
disadvantages, but also creates others (see Subheading 1.4.).
1.3. Principle of UV-Laser Footprinting
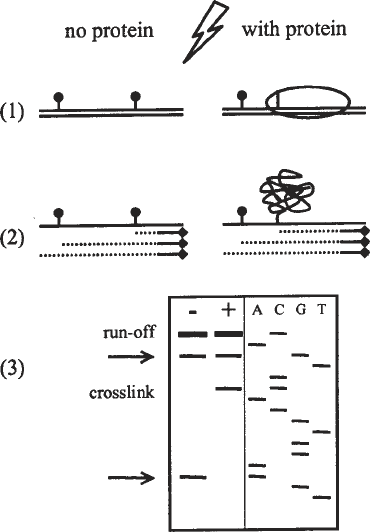
The principle of ultraviolet (UV)-laser footprinting is schematized in Fig. 1.
The sample containing the nucleoprotein complex is irradiated with a short
(less than 10 ns) pulse of UV-laser light. An identical sample, but lacking the
162 Geiselmann and Boccard
protein, is treated in parallel in the same way. The conditions are adjusted such
that the number of photons delivered onto the sample exceeds the number of
absorbing molecules. The nucleic acid bases are excited and undergo photo-
reactions, the nature of which depend exquisitely on the local environment of
the bases. The possible reactions include intrastrand reactions of consecutive
bases (the formation of thymine dimers is the most prominent such reaction),
interstrand reactions (although their quantum efficiency is too low to contrib-
Fig. 1. The principle of UV-laser footprinting. The double-stranded DNA is repre-
sented by the double line, and the protein by the ellipse. (1) A sample of DNA alone,
or the DNA–protein complex is irradiated with one pulse of UV-laser light. The bases
can undergo intramolecular photoreactions, react with the solvent (symbolized by the
circle), or form a crosslink with the bound protein (symbolized by the line connecting
DNA and protein). Only the reactions of the top strand are shown in the figure. (2) The
samples are denatured, a radioactively labeled primer (line with diamond) is annealed
to one stand of the DNA (the top strand) and extended using a DNA polymerase (dot-
ted line). The primer extension stops at damaged bases or at the end of the fragment.
(3) The primer extension products are analyzed on a sequencing gel, along with a
dideoxy-sequencing reaction using the same primer. The location of the photoreac-
tions are marked with an arrow, the crosslink and the runoff are indicated.
UV-Laser Footprinting 163
ute to footprinting signals), reactions with solvent molecules (e.g., with H
2
O),
and crosslinks with the protein (4–8).
In general, it is not possible, but neither is it necessary, to determine the
nature of the photoreaction at a particular base. Because the photoreactions are
highly sensitive to the local environment of the DNA, binding of a protein
changes this environment and thus produces a footprint (i.e., a difference
between the DNA photoreactivity in the absence and presence of the protein).
In the example of Fig. 1, the photoreaction at the left extremity of the DNA
fragment remains constant because the protein does not change the local envi-
ronment of this base. However, the presence of the protein prevents a photo-
reaction toward the right end of the fragment (perhaps by excluding water from
the vicinity of the base) and favors a new photoreaction with an amino acid
side chain, resulting in a covalent bond, a crosslink, between the protein and
the DNA. All such reactions modify the nucleotide base and, hence, impede
the progression of DNA polymerase during subsequent replication of the dam-
aged DNA strand. Arrest of the polymerase is detected in a primer extension
reaction by the appearance of shortened replication products, which then serve
to localize the modified base.
1.4. Advantages and Disadvantages of UV-Laser Footprinting
Ultraviolet-laser footprinting circumvents many potential artifacts of more
classical techniques by trapping the complex under investigation at the time of
irradiation. The signal is acquired very rapidly (on the order of microseconds)
(i.e., faster than typical rearrangements of a DNA–protein complex [on the
order of milliseconds]), and the footprint thereby freezes the initial state of the
complex (9). Laser light, as opposed to ordinary UV light, is needed in order to
limit the “incubation time” to several ns while providing a sufficient number of
photons to excite all nucleotide bases of the sample. The rapidity of signal
acquisition allows one to obtain kinetic structural signals by irradiating a com-
plex at different times after mixing the components. The technique has the
further advantage that it can be transposed to in vivo experiments in a straight-
forward manner because UV light readily penetrates bacterial cells. However,
because of the limited sample size of in vivo experiments, the present version
of the technique is only suitable for studying DNA–protein interactions in bac-
teria when using a binding site carried on a multicopy plasmid.
Using the UV-laser technology, it is possible to follow the kinetics of the
assembly of DNA–protein complexes. This technique can be used to determine
the order in which protein–DNA contacts are established in a multiprotein–DNA
complex. Proteins and DNA are mixed at time zero and the sample is irradiated
at a specific time interval after mixing (10). Modern mixing techniques allow a
time resolution on the order of a several milliseconds (11).
164 Geiselmann and Boccard
The main disadvantage of UV-laser footprinting is the unpredictability of
the footprinting signal. It should be noted that the footprint is not the result of
the protein shielding the DNA from the UV irradiation. The presence of the
protein merely changes the probability of certain photoreactions by excluding
solvent, changing the conformation of the DNA, or juxtaposing a reactive
amino acid and a particular base. A structural interpretation of the footprinting
signal is therefore, in general, not possible. A more practical limitation of the
technique is the need for a relatively expensive laser.
We describe the experiment for the particular case of the binding of an
Escherichia coli protein, the integration host factor (IHF), to one of its specific
binding sites, the yjbE site (12), in vitro and in vivo. IHF is a small,
heterodimeric protein (molecular weight [MW] of the dimer is approx 19 kDa)
that binds to specific sites on the DNA (for a review, see ref. 13). Upon bind-
ing to DNA IHF bends the DNA by about 180º (14). This DNA bending gives
rise to a very strong UV-laser footprinting signal, probably because two con-
secutive pyrimidines are brought into optimal alignment for the formation of a
pyrimidine dimer (15). Variations of this basic protocol, applicable to the study
of any DNA–protein interaction, are described in Subheading 4.
2. Materials
1. Ultraviolet-laser: The most commonly used lasers are YAG lasers, e.g., the Spec-
tra-Physics Quanta-Ray GCR series lasers, which emit infrared light of 1064 nm.
Two consecutive passes through a frequency-doubling crystal yields high-inten-
sity light of 266 nm, which is sufficiently close to the absorption maximum of
nucleic acid bases. Frequency doublers are a standard add-on for virtually all
commercially available YAG lasers. The laser power should be between 30 and
50 mJ per pulse at 266 nm and a pulse duration of 5 ns is standard. The energy of
one 30-mJ pulse represents about 4 × 10
16
photons (i.e., 67 nmol photons).
2. Power meter to measure the energy of the laser beam.
3. Thermostated water bath.
4. Spectrophotometer.
5. Water pump and 0.45-µM filter device for washing E. coli cells.
6. Phosphoimager: A phospho-storage device is ideal for quantifying sequencing
gels. The most commonly used instruments are sold by Molecular Dynamics,
Fuji, or by Bio-Rad.
7. IHF binding buffer: 50 mM Tris-HCl, pH 7.5, 70 mM KCl, 7 mM MgCl
2
, 3 mM
CaCl
2
, 1 mM EDTA, 10% glycerol, 200 mg/mL bovine serum albumin (BSA),
and 1 mM β-mercaptoethanol.
8. IHF: working stock solution at 3–5 µM in 50 mM Tris-HCl, pH 7.4, 800 mM KCl,
40 mM K-phosphate, 2 mg/mL BSA, and 10% glycerol.
9. Plasmid pBluescriptII (Strategene) containing the IHF binding site cloned into
the multicloning site (MCS). Stock solution in water at 100 nM.
UV-Laser Footprinting 165
10. Primer extension reaction. Annealing buffer: 1 M Tris-HCl, pH 7.6, 100 mM
MgCl
2
, and 160 mM dithiothreitol (DTT). Elongation mix for 2 µL: 2 U of T7
DNA polymerase in 2.4 mM of each deoxyribonucleotide, 3 mM Tris-HCl,
pH 7.5, 0.75 mM DTT, 15 mg/mL BSA, and 0.75% glycerol (see Subheading 3.
for the annealing and elongation steps). Stop solution: 95% formamide, 20 mM
EDTA, 0.05% bromophenol blue, and 0.05% xylene cyanol.
11. Sequencing reaction. Enzyme dilution buffer: 20 mM Tris-HCl, pH 7.5, 5 mM DTT,
0.1 mg/mL BSA, and 5% glycerol. Stop solution: 95% formamide, 1 mM EDTA,
0.05% bromophenol blue, 0.05% xylene cyanol.
12. Sequencing gel: Sequencing reactions are analyzed on 40-cm-long denaturing
(7 M urea) 8% polyacrylamide (ratio acrylamide:bis acrylamide 19:1) gels in 90 mM
Tris–borate, and 2 mM EDTA (TBE), and TBE used as running buffer. Gels were
transferred onto Whatman (3MM Chr) paper and dried at 80°C for 40 min in a gel
dryer (Bio-Rad model 583) linked to a vacuum pump.
13. Extraction of plasmid DNA. Solution I: 100 mM Tris-HCl, pH 7.5, 10 mM EDTA,
400 µg/mL RNase I. Solution II: 0.2 N NaOH, and 1% sodium dodecyl sulfate
(SDS) made freshly. Solution III: 3 M potassium, and 5 M acetate solution, made
by adding 11.5 mL of glacial acetic acid and 28.5 mL of H
2
O to 60 mL of 5 M
potassium acetate.
14. LB and minimal M9 media are used to grow and wash E. coli cells, respectively (16).
3. Methods
3.1. In Vitro UV-Laser Footprinting
1. Arrange the laser beam, using appropriate mirrors, such that it is directed verti-
cally into a water bath.
2. Align, and fix firmly, an Eppendorf holder in the water bath such that the laser
beam enters precisely in the center of an open Eppendorf tube. A piece of black
paper stuck into the bottom of the Eppendorf tube can help align the tube with the
laser beam; the impact of the laser light is very audible and “burns” the site of
impact, whitening the otherwise black paper.
3. Operate the laser in repetition mode (see Note 1) for at least 10 min and adjust the
doubling crystals to obtain a laser power at 266 nm of at least 30 mJ per pulse.
This laser power is measured during the warm-up period with an appropriate
power meter, before the actual footprinting reaction.
4. Incubate IHF at the desired concentration with 5 nM plasmid DNA in 40 µL
binding buffer for 20 min at 25°C (see Note 2). It is best to use a flat-bottomed
Eppendorf tube, but regular 1.5-mL Eppendorf tubes are adequate. The laser beam
generally has a diameter of 5 mm, and for maximal use of the light energy, the
sample should have roughly the same dimensions. Care should be taken to ensure
that all of the sample is irradiated by the laser beam.
5. Place the sample under the laser beam and irradiate with one pulse of UV-laser
light. It is best to operate the laser in repetition mode (i.e., continuously emitting
around 10 pulses per second). Most lasers also have the possibility to emit a
single pulse of light. However, the power of such a pulse is not very well con-
166 Geiselmann and Boccard
trolled, because the yield of the doubling crystals is extremely sensitive to
temperature. Continuous emission of laser pulses at a frequency of about 10 Hz
ensures a constant temperature of the crystals and therefore a stable pulse energy.
An electronically controlled shutter is used to obstruct the beam. The shutter
opening is coordinated with the emission of the laser pulses to ensure that only
one pulse of laser light passes for each opening of the shutter.
6. After irradiation, remove the protein from the irradiated DNA by incubating with
50 µg/mL proteinase K for 15 min at 50°C. Extract the samples with half a vol-
ume of a phenol/chloroform solution (made by adding equal volumes of phenol
pH 8 and chloroform), precipitate with 2 vol of ethanol, and resuspend the DNA
in 18.5 µL of H
2
O.
7. Primer extension (see Note 3): Add 2 µL of a solution of 0.2 µM radiolabeled
primer (5'-[
32
P] labeled using T4-kinase) to 18.5 mL of DNA. (Increasing the
primer concentration beyond this twofold excess over template will increase the
strength of the primer extension signals only marginally.) Denature the samples
by heating to 100°C for 3 min and chill on ice for 5 min. After the addition of 2.5 µL
of annealing buffer, incubate the samples for 3 min at 50°C (to anneal primer)
and chill again for 5 min on ice. Add 2 µL of the elongation mix (see Note 4) and
incubate the reaction for 10 min at 37°C. The mix is prepared freshly but can be
kept on ice for several hours.
8. Precipitate the DNA by adding 150 µL of ethanol and incubating for 10 min at
–20°C. Centrifuge for 10 min at full speed (about 12,000g) in a microcentrifuge.
Resuspend the DNA in 10 µL of loading dye.
9. Analyze 3 µL by gel electrophoresis on a denaturing 8% polyacrylamide sequenc-
ing gel. After electrophoresis, transfer the gel onto Whatman paper and vacuum
dry at 80°C for 40 min.
10. To determine precisely the location of the footprint, a reference ladder is gener-
ated by sequencing the same plasmid DNA using the same radiolabeled primer
(see Note 5). Denature 15 nM of plasmid DNA in a volume of 8 µL by heating to
100°C for 2 min. Chill on ice for 5 min. Add 1 µL of radiolabeled primer (0.25 µM)
and 1 µL of annealing buffer. Incubate for 3 min at 50°C. Chill annealing reac-
tion on ice for 5 min. Add 2.8 µL of each Deaza G/A T7 Sequencing™ Mixes
(Pharmacia) to four termination reaction tubes and prewarm at 37°C. Dilute 1 µL
of T7 DNA polymerase with 4 µL of enzyme dilution buffer. Add 2 µL of diluted
T7 DNA polymerase to the annealing reaction. Dispense 2.8 µL of annealing
reaction in each of the termination tubes. Incubate for 5 min at 37°C. Add 4 µL of
the stop solution.
11. Expose the dried gel to a phospoimager screen overnight and scan the screen
using the phospho-imager and its associated software (e.g., ImageQuant from
Molecular Dynamics).
12. Deduce a line profile of the different lanes using the Phospho-Imager software.
13. Transfer the data to Microsoft Excel and superimpose the scans on the same graph
in order to visualize and quantify the footprint (see Notes 6–11).
UV-Laser Footprinting 167
3.2. In Vivo UV-Laser Footprinting
1. Grow cultures of IHF
+
(W3110) and IHF
–
(W3110 hip) strains (12) transformed
with the plasmid carrying the ihf site in LB medium to the desired OD
600
(0.6 or
to saturation).
2. Wash the cells in minimal M9 medium (optically transparent buffer), resuspend
in minimal M9 medium to a final OD
600
of 1, and incubate the cells at 37°C (see
Note 12).
3. Irradiate as described above a large number (40–60), of 50-µL cell aliquots (see
Note 13). Freeze the cells immediately after irradiation in a dry-ice bath.
4. Pool the cells and extract plasmid DNA from 2–3 mL of cells using the following
alkaline lysis procedure. Centrifuge the bacteria for 5 min in a tabletop Eppendorf
centrifuge. Resuspend the pellet in 100 µL of solution I and add 100 µL of solu-
tion II. Mix by inversion several times. Add 100 µL of solution III and mix by
inverting the tube several times. Centrifuge the tubes at full speed in a tabletop
Eppendorf centrifuge (>10,000g) for 5 min. Transfer the supernatant to a clean
tube and add 210 µL of isopropanol. Incubate for 5 min at room temperature and
centrifuge the tubes at full speed for 10 min. Wash the pellet with 70% ethanol
and dissolved the DNA in 37 µL of H
2
O.
5. For primer extension, add 2 µL of a solution of 0.2 µM primer (5'-[
32
P] labeled)
to 18.5 mL of DNA and process and analyze the samples in the same way as for
the in vitro reactions, steps 7–13 of Subheading 3.1. (see Notes 10 and 11).
4. Notes
1. Laser setup. As mentioned in the Subheading 3. in order to obtain a stable laser
power it is best to operate the laser in repetition mode. If a single-pulse mode is
used the energy of a particular pulse is ill-defined and the absence of a
footprinting signal may simply be the consequence of diminishing laser power.
All lasers provide an electrical signal that allows external equipment to be
coordinated with the laser pulse. To our knowledge, shutters are not commer-
cially available. However, it is an easy task for a good mechanics shop to con-
struct such a shutter. We used a shutter made of a small sheet of blackened Teflon
obstructing a hole of approx 8-mm in diameter through which the laser beam had
to pass in order to reach the sample. Any material can be used, but it should be
kept in mind that the laser will eventually burn a hole into the material and that it
is best to use a black material in order to minimize hazardous reflections of the
laser light. A simple electronic circuit controlled the opening of the shutter by
activating an electomagnet that pulled the Teflon sheet (via an attached piece of
metal) away from the hole. After the light had passed, the current to the magnet
was cut and a spring pulled the sheet back over the hole.
2. The binding buffer described is the standard buffer used for measuring DNA
binding of IHF. Other proteins may require different buffer conditions. The buffer
may be adjusted with certain limitations. The salt concentration should not be too
low in order to avoid nonspecific binding of the protein to DNA. The buffer
should not include a high concentration of reagents that absorb at 266 nm. For
168 Geiselmann and Boccard
example, the interaction of the cyclic AMP receptor protein (CRP) with DNA
requires the presence of cAMP in the buffer (17). Keep the concentration of such
nucleotides below 100 µM. As mentioned in Subheading 3., one laser pulse con-
tains the equivalent of about 100 nmol in photons. A typical reaction volume is
50 µL; therefore 100 µM ATP absorbs about 5 nmol of photons. A more physi-
ological concentration of ATP in the millimolar range would dramatically
decrease the yield of the photoreaction.
3. The procedure describes the footprinting reaction for only one strand of DNA.
Evidently, the other strand can be analyzed in the same way using the appropriate
primer. The primers should be chosen such that the region of interest is within
200 nucleotides from the primer. Most photoreactions have a quantum efficiency
below 1%, the formation of thymine dimers reaching several percent. Therefore,
on average, there will be roughly 1 photoreaction per 100 base pairs (bp).
Considering only the 100-bp region downstream of a primer assures single-hit
conditions. If the DNA carries too many photoreactions, the primer extension
will stop at the first defect and the signal of a photoreaction further downstream
will pass undetected.
4. The detection of photoreactions. All DNA polymerases are very sensitive to dam-
aged bases. It is not important to use T7 DNA polymerase in the primer extension
reaction, any other DNA polymerase (e.g., Klenow, Taq) gives equivalent sig-
nals. However, signals obtained with different polymerases may not necessarily
be identical because some may be more sensitive to particular photodamaged
bases than others. Even RNA polymerases can be used if the template harbors an
appropriate promoter. We have successfully used T7 RNA polymerase to tran-
scribe the region of interest from a T7 promoter located on the vector DNA. The
major signal of the IHF footprint remained unchanged, but, instead of a single
band, T7 RNA polymerase generates a doublet of bands (15).
5. To determine the location of termination sites precisely, we generated a refer-
ence DNA ladder consisting of a sequencing reaction of the same DNA region
(see Subheading 3.). In general, it is assumed that the primer extension reaction
of the irradiated DNA stops just before the modified base. For example, if the
sequence of the DNA read on the gel is 5'-GGAC-3' and the primer extension
reaction of the irradiated DNA shows a band at the position of the A in the
sequencing reaction (run in parallel on the gel), then the photoreaction most likely
took place on the following base pair (the C in the above sequence). Because the
primer extension reaction reads the opposite strand, the photoreaction actually
damaged the G marked with an asterisk:
5'-GGAC-3'
3'-CCTG
*
-5'
6. There are several possible reasons for not detecting a UV-laser footprinting sig-
nal. The most obvious problem is that the protein does not bind to the DNA.
Generally, we verify binding by a gel retardation assay. The second reason may
be that protein binding does not change the photoreactivity of the bases in the
recognition site. This is the case, for example, for CRP, which yields very weak
UV-Laser Footprinting 169
signals in UV-laser footprinting despite a strong interaction measured by other
techniques (J. Geiselmann, unpublished results). Because photoreactivity
depends on the sequence, a particular site might not be photosensitive; for
example, the main IHF signal had not been observed for the ssb site, probably
because the sequence of this site does not contain the highly reactive TC pyrimi-
dine doublet (15).
7. A trivial reason for not detecting a photoreaction is that the laser did not hit the
sample. The best control, and a control to include in all laser footprinting reac-
tions, is to analyze the primer extension reaction of DNA alone. Two DNA-alone
samples should always be included: one irradiated sample and an identical sample
that has not been exposed to UV-laser light. The irradiated sample should yield
readily visible bands all along the lane (Fig. 2, lane f), whereas the nonirradiated
sample should show no elongation arrests (Fig. 2, lane e).
8. The irradiated DNA does not give any elongation arrests with T7 DNA poly-
merase. Verify the primer extension mix. Perform a control primer extension
reaction using the nonirradiated plasmid template, but cut about 100 bp down-
stream of the primer with a convenient restriction enzyme. Primer extension using
this template should give a very strong band corresponding to the elongation
reaction reaching the end of the fragment.
9. Artifactual footprinting signals. A contaminated template preparation or a dam-
aged DNA template could be misinterpreted as giving a footprinting signal. It is
very important to verify that the nonirradiated template does not produce any
bands during the primer extension reaction. This is particularly important for in
vivo reactions because the plasmid preparation could partially damage the plas-
mid and lead to artifactual bands (Fig. 2, lanes c and d).
10. Comparing signals obtained under different conditions in vitro, or comparing in
vitro to in vivo signals. The efficiency of sample preparation or of the primer exten-
sion reaction can vary from sample to sample. In order to compare different lanes
we run a small portion (10%) of the primer extension reactions on a sequencing gel
and quantify the lanes using a phosphoimager. We then load the same samples on a
second sequencing gel, but equilibrating the amounts loaded according to the sig-
nal intensities on the first gel. For example, if one of the reactions was only half as
efficient as the other ones, we load two times more of this sample on the second
gel. This readjustment should not be allowed to exceed a factor of 2.
11. Quantifying lanes. Once an intensity equilibrated gel exposure is obtained, we
obtain line profiles of all lanes using the Phospo-Imager software and compare
lanes by superimposing the line profiles in Microsoft Excel. Remaining small
(several percent) differences in the intensities of the lanes should be normalized
by multiplying the scans with a scaling factor between 0.9 and 1.1 that is deter-
mined subjectively by the user in such a way that the global patterns superim-
pose. Lanes containing a high background of nonspecific radioactivity cannot be
quantified with confidence.
12. It is important to keep in vivo samples at the physiological temperature (37°C for
E. coli) to ensure optimal binding.
