Lewin Benjamin (ed.) Genes IX
Подождите немного. Документ загружается.

lzL
sVNU eprng fq
pepar16
aE uel 6u$!pl
VNU
II'lZ
ilxo
erql
yo
aruanbas
aql
01 elrlplJJ
UrqsJrueJJ
e seq uraloJd
1
lrunqns
espprxo aurorqro]zb
er{1
Jo
aruanbas
Jqt
'pJJJAoJsrp
eq ot
JSef,
tsJrJ
Jql uI
'eupuoq)olrrrr
JrrrosouedL4;o
saua8
IeraAJS
yo
spnpord aqt ur a)uJnbas ur sa8ueqr
JrlerupJp Lq
papanar
sr Surlrpa
yo
ad,{1 JJqlouV
'ase6q
yp1
pue
'r{1mr1re
asere;suer11r{pr.ln
leululal
'aspallnuopua
1o
xalduor
e
r\q pazr{1e1er st 6ur1Lp3
o
'saulpun
1o
(uoqelep
'ua4o
ssal
to)
uorlr.ppe
ro;
a1e1due1 aq1 sapLnord
ypg
aprnb
eq1
o
'pa1pa
aq o1 uorbar oql
Jo
sapts
qloq
uo
ypg
eprn6 e
qluu
srred aspq
VNU
alerlsQns
e{f
r
'outpun
Jo
suoqalap.lo suoqJasur [q srnrro eupuoq]oltul
auosouedfi.r1 ur 6urlrpa
![!
aAtsua]xl r
svNU aplng
fiq
paparr6
aB uPl
6u!}tpl
vtrtu
'seleJlsqns
Jleql uI
.dtr;gtrads
aruanbas
JoJ
tuaruJrrnbar;o
sadfu
luaJaJJIp
J^eq
,{eru
sruals,{.s Surtrpa
lueJeJJrp
snql'uorttuSolar
rqoads ro1 zi:essarau sr uo6ar
xaldnp arp uIqlIM
Surrredsrru
Jo
uratted
v'uoJlur
rupansumop
aql
ur aluanbas ,{reluauralduroJ e
pup
uoxe
Jql uI
uor8ar
petpa
aql uJJMlJq
peuroJ
s1 atys
ta8rer
Jql
Jo
uopgSorar ro;
Lressarau sI
teqt
uor8ar
parred-aseq
e
'vNg
S-UnlC
er{t
Io
aseJ
aql
q
lpql
sMoqs
#ts"J.d *'it{'l*Ji
'le8Jel
IUJIJIJJnS
e sapnord
alts
Surlrpa aql Surpunorrns
(saseq
9g-)
aruanbas
gerus
.{lanrlplal
e
teqt
slsaSSns
luana
Sutltpa
g-ode
aqt
ro; rua1s,{.s oJitr ul ue
;o
tuarudolenep
aq1
'aruanbes
JprloalJnu e azruSorar
,{lnarlp
plnor
Jo
'sarudzua
SurLyrporu-yNgt
01 snoSo
-leue
rJuueur e ur eJnlf,nJls
Lrepuoras
yo
uor3ar
relnrrued e azruSolar Leru xaldruoJ
eqJ
'y1qg
roldarJr
uruol
-oJ3S
P Ur JnJJO S1UJAJ
relrurs
pue
'YNU
JOl
-dara:
aleruetnlS Jql
q
salrs
la8ret
Jql sJZIU
'uorbar
xaldnp
yp6
parred
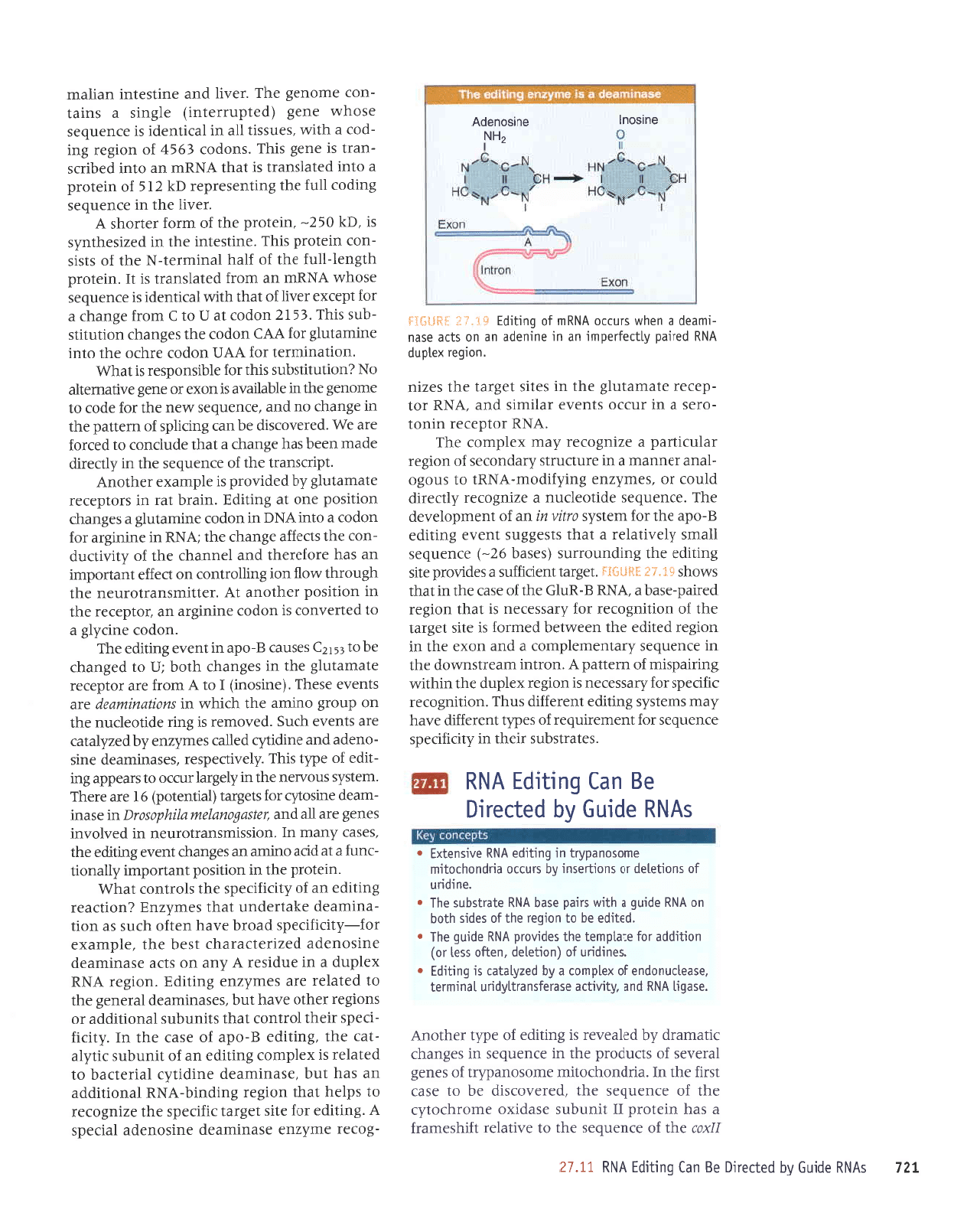
l{lpayadur up ur auruapp
up uo sltp
aseu
-rurPop
P uoqM slnrlo
vNUru
to
6uL1rp3
6'r-.14 *Hfi*Ec
-3orar
arur{zua
Jseulrupap
JuISouepP
lenads
y
'3ur1rpa
roJ J1IS
la8:el
rtyrads
aql aztuSolar
o1 sdlaq
lBql
uot8ar
Sutputq-ypg
leuolltppe
ue seq
tnq
'aseulueap
aurptlr{J
IPIJJDeq
ol
peleleJ
sr
xaldtuor SuIlIpa
ue
Jo
llunqns
lrtz(1e
-teJ
Jql
'3ur1rpa
g-ode
Jo
asp)
eqt uI
'^tpIJ
-nads
rraql
IoJtuoJ
lpqt
slpnqns
IPuoIlIppP
Jo
suorEar
raqto
elpq
]nq'ses€ulueap
prauaE
aq1
01
patplar are saru.{.zua
Sulrlpg
'uor8ar
ygg
xaldnp
e uI enplsJr
y
due uo sl)e
Jseulureap
Jursouapp
pezrJatf,erpqJ
rsaq
aql
'aldruexa
;oy-d1nr;trads
peorq JApq ualJo
qJns
se uoll
-Purlueap
J>lPuapun
leqt
seru,{zug
zuol}f,eJJ
Surlrpa
ue;o
z{ltrqtrads
aqt
sloJluo)
teqM
'uralord
Jq1 ut
uolllsod
lueuodrur
z(1euop
-)unJ
e
le
pDp
ourure
up saEueqt
luaura
Sql1pa aql
'saser
Lueru
uI
'uolsslursueJlornau
uI
pJAIoAUI
saua8
a.re
[e
pue
'ta1su6oua7aw
ap4dosotq
ur aseur
-ueep
arnsoft
roy sta3rur
(ppualod)
91
ale Jreql
'rualsk
snoAJJu
eql
rn,{.1a8re1rrnro
ol
sreadde 3rn
-llpa
Jo
ad,{1 srql
,{.1a.nuradsar
'sJseulureep
auIS
-ouape
pue
aurpp,{r
palpr
saru,{zua,{q
paz,{prer
aJe slualJ
q)ns
'pe^ouar
sr Surr
Jppoapnu
eql
uo dnor8
oulure
Jql
qllqM
ur
sultloutuffiap
Jte
sluJAJ
asaql
'(aursour)
1
or
v
uorJ
a:B roldarar
aleurelnlS
eqt
ul saSueqt
qloq 1n
ot
pa8ueqr
aq
01
€EIzJ
sJsnpf,
g-ode
ur
lua,ra
Suntpa
"qJ-
--
'uopo)
JurJAIb e
01
pauJluoJ
sI
uopoJ
aurur8re
uP toldelJJ
eql
ur
uoplsod
Jer{toue
lV
'rJlllursueJloJneu
aql
qSnorqt,r,tog uot 8ut11o:luol
uo
peJJJ
lueuodrut
ue sPq eJoJeJaql
puP
IJuueqJ
:ql
;o
,(lr.tr1;np
-uo)
aq1 sDaJJe
a8ueqr
aql
IYNU ut
aurut3re
ro;
uopor
e otul
VNo
ul uopoJ
aunue1n13
e
sa8ueql
uoltrsod
auo
le
Sutrpg
'uIPJq
lPJ
ut sroldarar
ateurelnp
u(q
papr,lord
st aldruexa
JeqlouY
'ldtnsuerl
aql
Jo
JJuJnbas
aqt
ur ,{prarp
appur
uaeq
seq a8ueq;
p
lPqt
apnlJuo)
ol
peJJoJ
JJe e1,1
'pareloJslp
eq
uPJ Suortds;o
uralled aq]
ur a8ueqr
ou
pup
'atuanbas
,lrau
eql roJ apoJ
01
aruouaS
aql
rn alqelle^e
sI
uoxa ro
auaS anuerrrJ{e
oN
Zuountpsqns
slqr
roJ alqtsuodsar
q
lPqM
'uorleurrural
JoJ
wo
uopoJ
aJqJo
aq1 otul
aururelnlS
ro;
yy)
uopo)
aqt sa8ueqr
uonnllls
-qns
srqJ'€EIz
uopol
le
|l
01
)
IxorJ
aSueql
e
ro;
]datxa
JJAIT
Jo
teql
qll
^
IeJIluJpI
sI J)uJnbes
JsoqM
VNUrx
ue uroJJ
pelelsueJl
sl
U
'ulalord
qr3ua1-11ny aql
Jo
JIeq
l€ulurel-N
eqr
Jo
slsls
-uor
uralord
sIqJ
'eullselut
eql
ut
paztsaqlur(s
sl
'(I
gEZ-
'utatord
JI{t
Jo
IrrJoJ
Jauoqs
Y
_
'Je^rl
eql
uI eJuanDJS
Surpor
ilnJ
aql
Suuuasardar
q1{
Z
I
g
1o
utalord
e o1ul
pelPlsuPrl sI
leql
VNUIU
uP
olul
paqlrJs
-upr1
sI aua8
srql
'suopoJ
f.9SV
lo
uor8ar 3ut
-poJ
e
qlllvt
'sanssll
IIP
uI
IeJlluJpl
sl JJUJnbJS
JSoqm
aua8
(pardn;ra1ut) a13uts
e
sulel
-uol
JrrrouJ8
aql
'rarrg
pue eullsatul
ueIIPur
tur'j
eursoul
oulsouapv
gene.
The
sequences of the
gene
andprotein
given
in
iii,i:*l
:.;.**
are
conserved in
several try-
panosome
species. How
does this
gene
function?
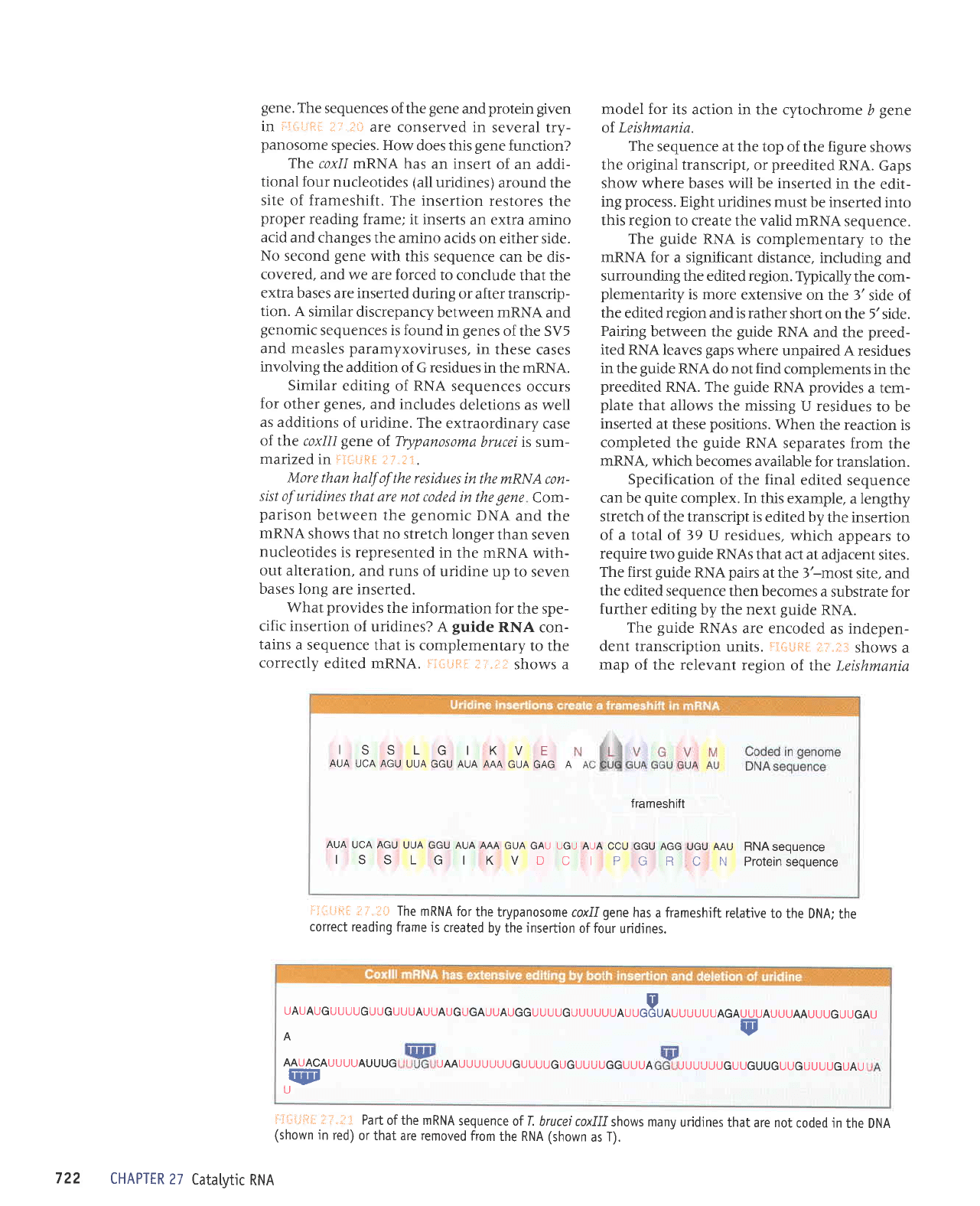
The
coxll mRNA
has an insert
of an addi-
tional four
nucleotides
(all
uridines)
around the
site of frameshift.
The insertion
restores
the
proper
reading
frame;
it inserts
an extra amino
acid
and changes
the amino
acids on either
side.
No second
gene
with this
sequence can
be dis-
covered, and
we are forced
to conclude
that the
extra
bases are inserted
during
or after transcrip-
tion.
A similar
discrepancy
between nRNA
and
genomic
sequences is found
in
genes
of the SV5
and measles paramyxoviruses,
in these
cases
involving
the
addition of G residues
in
the
mRNA.
Similar
editing of RNA
sequences
occurs
for
other
genes,
and includes
deletions as
well
as additions
of uridine. The
extraordinary
case
of the coxIII
gene
of Trypanosoma
brucei is sum-
marized
in
ai*:iRl
iI.I:.
More than
half of
the residues
in the wRNA
con-
sist of uridines
that
are not coded
in the
gene
Com-
parison
between
the
genomic
DNA
and the
mRNA
shows
that no stretch
longer
than seven
nucleotides
is represented
in
the mRNA
with-
out alteration,
and runs
of
uridine up
to seven
bases long
are inserted.
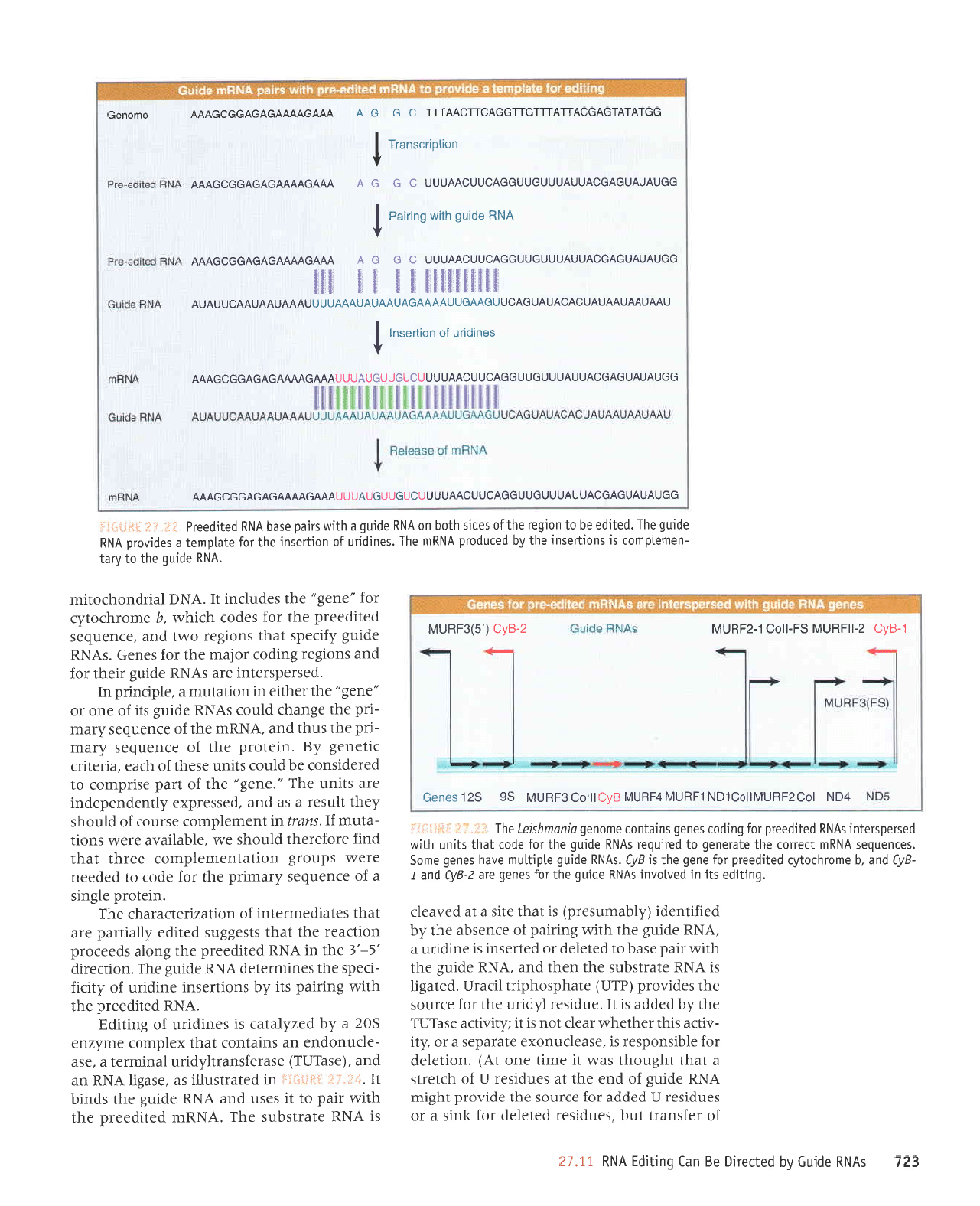
What provides
the information
for
the spe-
cific insertion
of
uridines? A
guide
RNA
con-
tains
a sequence
that is complementary
to the
correctly
edited
mRNA.
l'i*ijFlt
i,r.ii-i
shows a
model for its
action in the cytochrome
b
gene
of
Leishmania.
The sequence at
the top of the figure
shows
the original transcript,
or
preedited
RNA.
Gaps
show
where
bases will be inserted
in the
edit-
ing
process.
Eight uridines
must be inserted
into
this region to
create the valid mRNA
sequence.
The
guide
RNA is complementary
to the
mRNA for
a significant distance,
including
and
surrounding the edited region.
T\zpically
the
com-
plementarity
is more
extensive
on the 3'side
of
the
edited region and is rather
shon
on the 5'side.
Pairing
between
the
guide
RNA
and the
preed-
ited RNA leaves
gaps
where
unpaired
A residues
in the
guide
RNA do not find
complements
in
the
preedited
RNA. The
guide
RNA
provides
a tem-
plate
that
allows the missing
U residues
to
be
inserted
at these
positions.
When
the reaction
is
completed
the
guide
RNA
separates
from
the
mRNA,
which becomes available
for
translation.
Specification of
the
final
edited
sequence
can be
quite
complex.
In this example,
a lengthy
stretch of
the transcript is
edited by
the insertion
of a total
of
l9
U residues,
which
appears
to
require
two
guide
RNAs
that act
at adjacent
sites.
The first
guide
RNA
pairs
at the
3'-most
site, and
the edited sequence
then becomes
a
substrate for
further
editing by the next
guide
RNA.
The
guide
RNAs
are encoded
as indepen-
dent transcription
units.
fg*LlFtfl
;,-:.i::
shows
a
map of the relevant
region
of
the Leishmania
r:'i:r-iii{
il,!li: The
mRNA for
the
trypanosome
coxll
gene
has
a
frameshift
retative
to
the DNA;
the
correct reading
frame is
created
by the insertion
of four
uridines.
i;if;l.ifii;-:
rr
1'"1:1 Partof
themRNAsequence
of T.brucei
coxllf
showsmanyuridinesthatarenotcodedintheDNA
(shown
in
red)
or that are removed
from
the RNA
(shown
as
T).
ISSLGIKVE
AUA
UCA AGU
UUA GGU AUA
AAA GUA
GAG A
frameshift
AUA
UCA AGU
UUA GGU AUA
AAA GUA
GAU UGU AUA
CCU GGU AGG
UGU AAU
RNASEOUENCE
I
S S L
G I K V D
C I P
G R
C N Proternseouence
UAUAUGUUUUGUUGUUUAUUAUGUGAUUAUGGUUUUGUUUUUUAu,J,o,,,,,UAGAUUUAUUUAAUUUGUUGAU
A
AAUACAUUUUAUUUG
UAAUUUUUUUGUU
qEE
U
ru
UUGUGUUUUGGUUUA
UUUUUUGUUGUUGUUGUUUUGUAU
722
CHAPTER
27
Catatytic
RNA
EZL
sVNU aplng riq
paparr6
ag uel
6ulflpt
VNU
tI'lZ
Jo
JJJSuerl
lnq
'sanprsJJ
pJlalep
roJ >lurs
e ro
sanprsJJ
n
peppe
JoI J)Jnos aql apr.r.ord
rq8tu
y1qg
aprn8
Jo
pua
aqt
tp
sJnprsar
n
Jo
q)tals
e
tpql tq8noqr
seM
tr
Jurrt Juo
ty)
'uor1a1ap
roy
alqrsuodsJr sr
'Jseel)nuoxe
aleredas
e ro
Llt
-Arpe
srql JJqleqM JeJIJ
lou
sr
tr
:Ilr^rlJp
JSeJnI
aqt.{q
pappe
sl
tI
'enplser
1[prrn
Jqr JoJ
eJrnos
aqt sapno;d
(afn)
areqdsoqdrrl
prern
'pate311
sI
vNu
rtertsqns
Jqt uJql
pue
'vNu
aprn8
aql
qttl:red
eseq ol
pJlelep
Jo
pauJsur
sr
JurprJn
e
'y1..1g
aprn8 eqt
qtrm
SurrrBd
Jo
JJuJsqe
aqt r{q
pJrJrtuepr
(,{lqerunsard)
sl
teqt
Jtrs e
tp
pJ^palr
'6ur1Lpa
slr ur
po^lo^ur
sypX apLnb eq1 ro1 seuab
arc
7-9fi
pue
y
-g[j
pue'q
aurorqrolAr
palrpeard
rol
aueb aq1 s1
gfj
'sypX
aprnb a1dL11nu aneq
sauab
auo5
'seluanbas
VNUU
lrelor
aq1 elereueb
o1
perrnbar
sygX aprn6 aql lol apol
leql
slrun
qltM
pasradsrelursypgpalLpeerdrol
6urporsaua6sureluoreuouabDru?wqsLa].ogl_
l,l'r,:
1:tji!1,:ili
sl
vNu
alerlsqns aqJ
'YNutu
pallpeeJ0
Jql
qtp,r
rred 01
1r
sJsn
pue
YNU
aptnS
aqt sputq
1I
'r:,i]";..:l
:iiJi]l-11:i
uI
pJleJlsnl[
Se
'aseStl
vNu
uP
pue
'(ase1n1)
aseraysuenl.dpr.rn
IeuIIuJJl
e
'ase
-alJnuopua
ue suleluoJ
leql
xaldruor aruz{.zua
S0Z
e
Iq
paztrlewt sI seulplrn
;o
Sutltpg
'YNU
palparro
rql
qlpr,l Sur;red slr
Lq suoluasul
euIpIJn
;o
,{.1nt;
-nads
aql
sJuturJtrp
y1trg
aptn8 eqJ
'uopterlp
,s-,€
etfl ul
YNu
patrpaard
aql Suop
spaalord
uortleJr
Jqt
teqt
s1sa33ns
pJllpe .dlerued are
lPql
selelpJruJJluI
Jo
uollezlJelf,PJeqJ
eqI
'ura1o.rd
a13urs
e;o
aruanbas
.drerutrd
Jqt JoJ epoJ
ol
pJpaeu
a,raa,r sdnor8
uorleluarualdruot
eerqt
leq1
purJ
JJoleJJql
plnoqs J/!r
'elqelle^P
eJel!{ suoll
-plnru
lI'suau
ut
tuarualdruoJ
JSJnoJ
Jo
plnoqs
.daqt
11nsar
e
se
pue
'passardxa.dltuapuadaput
aJe slrun
aq1
,,'aua8,,
aql
Jo
lred
astrdruor
o1
psJeprsuoJ Jq
plnoJ
sllun JSJqI
JO
q)eJ
'PIJJIIJJ
rrtauaS
,(g
'ularord
eql
Jo
aruanbas
.,{.reru
-rrd
aqt snql
pup
'VNutu
Jql
Jo
JJuJnbas
ui.reut
-r,rd
aqt
a8ueqr
plnol
sVNU
aptn8
slt
Jo
euo Jo
,,aua?,,
JI{t
JJqlIa ul uollPlnlu
P
'aldnurrd
u1
'pasradsralul
Jre
syNU aprn8
rraql ro1
pue
suor8ar
3urpor
roleru
Jql JoJ
sJue5
'sVNU
aprn8
d;oads
leqt
suorSal
o,rtl
pue
'aruanbas
parpaard
aqt
roJ sJpoJ
qJIqM
'4
aruo.lqrolLr
ro;
,,aua8,,
aqt
sJpnlJul
tI
'VN11
Ielrpuoqlollu
'yp5
eprn6 aq1 o1
&e1
-uauralduor
sr suorlosur eq1 Aq
parnpotd
VNUur
aq1
'saurpun
Jo
uotilasut aq1
tol a1e1dLue1
e saptnord
yflg
aprnbeql'polrpaaqoluoLbaraqlJosaptsqloquoypXapLnEeqlrmsrLedaseqVNUpaltpoeld:li'l'it, Iil{:Lri:i
toN toczlunv{ilocf
oN HUnn
rlunu\
EASllloc elunt/\
s6
szt
sauae
t-s^c z-illHnu{
sJ-ltoc
t-zlunl/\
z-8^c
(,g)eJUnnl
eenvnvnevecvnnvnnn0n
noevcnncvvnnn
ncnen n0nvnn
nwvewwev9v99cevw
eenvnvnevecvnnvnnnenn
eevcnncwn
nn
ncnennenvnnnwvevwvevev00cewv
+
sourpun
1o
uotuesul
I
NWNWNWNVNCVCVNVNEVCNNEWENNVVVVEVNVVNVNVWNNNNVVVNVVNVVCNNVNV
flilf;fiilfriilfifltililf;frHfiil
9envnvnevocvnnvnnnenn9evcnncwnnn
3 3 e
v
vwewwevev99cewv
+
VNU
eplnb
qltlt
6uP;e3
|
egnvnvnevecvnnvnnnenne9vcnncwnnn
c e
e v
wveww9v9vecS9vvv
+
uotlducsuell
I
I
getvlvtevecvt-Lvlltel-]eevSl-tcvv-Ll-l
ce
e v
vwevwveveveeccvw
auouaS
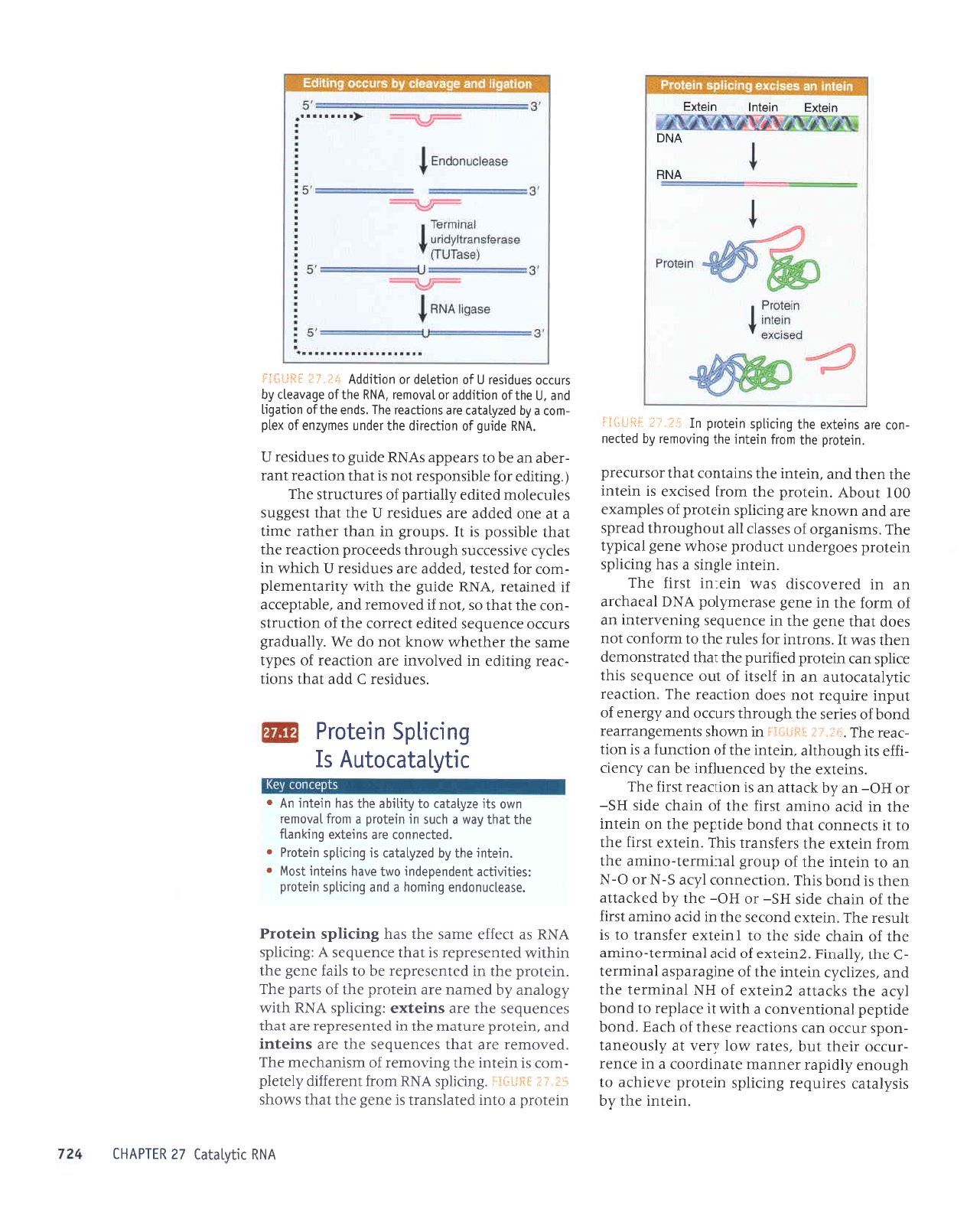
'urJlur
JLII
^q
srs^lplpJ sJrrnbaJ
Suorlds
uratord
JAerqJe ol
q8noua
dlprder
-rJuueur
aleurprooJ
p
ur J)ue-r
-rnf,Jo
rrJqt
lnq
'salpt
.t,rol
Lra.L
le
.{1snoaue1
-uods
rnllo
uef,
suorlJpal
esJql
Jo
qteA
.puoq
JplldJd
IPuoIluaAuoJ
e
qll,11
1t
aleldar
ot
puoq
1'{re
aqf
s>lJelte
ZuretxJ
Jo
HN
IpuruJet
Jql
pue
'sazrpi{r
urelur
aql
1o
aurSeredse
leurrural
-)
aql Llleurg'Zurelxe
Jo
prJe
Ieururrel-ourrue
eql
Jo
ureqJ
aprs
Jql 01
IurelxJ
lJJSueJl
o1 sr
llnsJr
JqI
'urJlxJ
puof,Js
eql u
plf,p
ourue
lsJrJ
rql
Jo
urerlr
Jpls
HS-
ro
Ho-
aqr ,{q
pJ>lJelle
uar{t sr
puoq
srqJ
'uort)Juuo)
1,{re
5-p
ro
O-N
ue
ol uratur aql
1o
dnorS
lpu[uJel-oullue
eql
ruoJJ
urJlxa
aql sJJJSupJl
srql
'urelxJ
lsJrJ
aql
ot
tr
slJeuuoJ
leql
puoq
aprldJd
eqt
uo urJtur
eql ur
pr)e
ourue
lsrrJ
eq1
Io
ureqJ epls
HS-
ro
HO-
ue Lq
1re11e
ue sr uortJeal
tsrrJ
JqJ
'surJlxa
Jql
^q
pJJuJnlJur
eq upJ Lruao
-IJJa
sll
q8noqtle
'urJlur
Jql
Jo
uortJunJ
e sr uorl
-)eJJ
eqJ
'+l',:
i:
r,i*lil:;.
j
ur rruoqs
sluarua8uer.rear
puoq
Jo
sJrrJS
aqt
q8norqr
sJnJJo
pue
,{3raua yo
lndur
arrnbal
lou
seop
uorlf,pJJ
Jr{J
'uorlf,eJr
rrldlelerolnp
ue ur
JIJSIr
Jo
1no
aruanbas
srql
arr1ds uet
uralord
pagrrnd
aql
reql
peleJtsuorxap
uJql
sPM
1I
'SUOJlur
IOJ
SelnJ
aql o1 ruJoluoJ
lou
saop
leql
aua8 aqt
ur aruanbas
Suruanralur
ue
Io
ruJoJ
aql ur aua8
aseraru,i(1od
VNO
IeJeqJJp
ue ur
peJa^oJsrp
Se^l. urJlur
lsJrJ
JqJ
'urJtur
a13urs
e seq Sunrlds
uralord
sao8rapun
pnpord
JSoq,tr
aua8
lerrdz(1
aql'susrue8ro;o
sassep
1e
tnoq8norqt
peards
JJe
pue
u.lrou>l
a.re
Suoqds
uratord;o
saldruexa
00I
tnoqy
'uratord
aqt uoJJ pJSr)xJ
sr uratul
aql ueql
pue
'ulJlur
aql
sureluoJ
leql
rosrntard
'uLelord
aql
uo.lJ uralur
aql
burnouar r\q
papau
-uol
ale sutalxe
oql 6urcLlds
uralotd u1
!i.i
i _;iirit*i,j
vNu
lrl^lPlel
/z
ultdvHl
uleloJd
p
olur
pJlelsueJl
sr auaS aql
1eq]
sMoqs
*I'ir
$li**td
'SuDrIds
\/Nu
ruorJ
lurreJlrp
,{1a1a1d
-uroJ
sr urJlur
eqt Surnoruar
Jo
rrrsrueqJau
JqJ
'pa^oruer
ere
leql
saruanbas
Jqt Jrp sulelul
pue
'ura1o.rd
JJnleur
Jr{l ul
peluasa;dar
JJp
leql
saruanbas
Jql
aJp sulalxe
:Sunqds
yNU,ItW
.{3opue
Lq
paueu
rre ualoJd
aqt;o sued
aql
'utalord
Jqt
ur
pJtuasardar
eq ot
slleJ aua8 aql
ulqll1!\
paluasardar
sr
tPqt
aruanbas
y
:SuDqds
vNu
sP
DaJJe
arues eql spq
Sultqds
ulelord
'aspallnuopua
6uruoq
e
pue
6uoqds uralord
:saqtAtllP
luepua0aput
oMl aAPq sutelUt
]S0[4]
r
'uralur
aql [q
paz[1e1er
sr
6upLlds ura]ot/
o
'pallouuot
elp
sutolxa 6uo1ue1;
aql
leql
felln
e
qons
ur urelord
p
uo4
le^oue]
u/v\o
slr azr\ie1er o1 r\1qrqe
aql seq uroJur u!
o
lrl^lPtPlolnv
sI
6uortdS
uralold
'senprseJ
)
ppe
leq1
suorl
-rear
Surlrpa ur
pellolur
aJe uorlJeJr;o
sad,{1
rlues
eql rJqlJqM
Mou>l
tou
op e14'.{11enper8
srnJJo aruenbas pellpJ
lJaJroJ
Jql
Jo
uorDnJls
-uoJ
JI{t
teql
os
'tou
Jr
paloruJr
pue
'alqeldale
Jr
pJurelJr
'y1qg
aprn8 eql
qlrM
dlrreluaruald
-ruoJ
roJ
p31sJ1
'pJppp
JJe senprsal
n
q)rrlM
ur
sap.{r
anrssa>ns
q8norql
spaarord
uorlJeJr aql
reqt
alqlssod
sl
U
'sdnorS
ur ueql rJqtel
Jturl
e
le
auo
pJppe
JJe sJnprseJ
n
aqt
teql 1se33ns
selnleloru
pallpJ
.dgerued;o
saJntJnrts
aqJ
('3uppa
roy alqrsuodsar
1ou
sr
teql
uolpeJJ
tueJ
-Jaqp
ue aq
ot s:eadde sypg
aprn8 ot sJnprseJ
n
'ypX
aprnb
Jo
uorllalrp
oql.lapun sauAzue
1o
xald
-uor
e fq
pazfleler
ele suoqlpa.l
aql'spue aq11o
uoLlebrl
pue
'n
aqlJo uorlrppp
lo
lp^oruol
'VNU
oqlJo abenealr Aq
slnllo senptsal
n
J0
uotlolap
l0 uotltppv
?*'liJ
S#**:.:
VNH
VNO
ureuS
utolul
uteuS
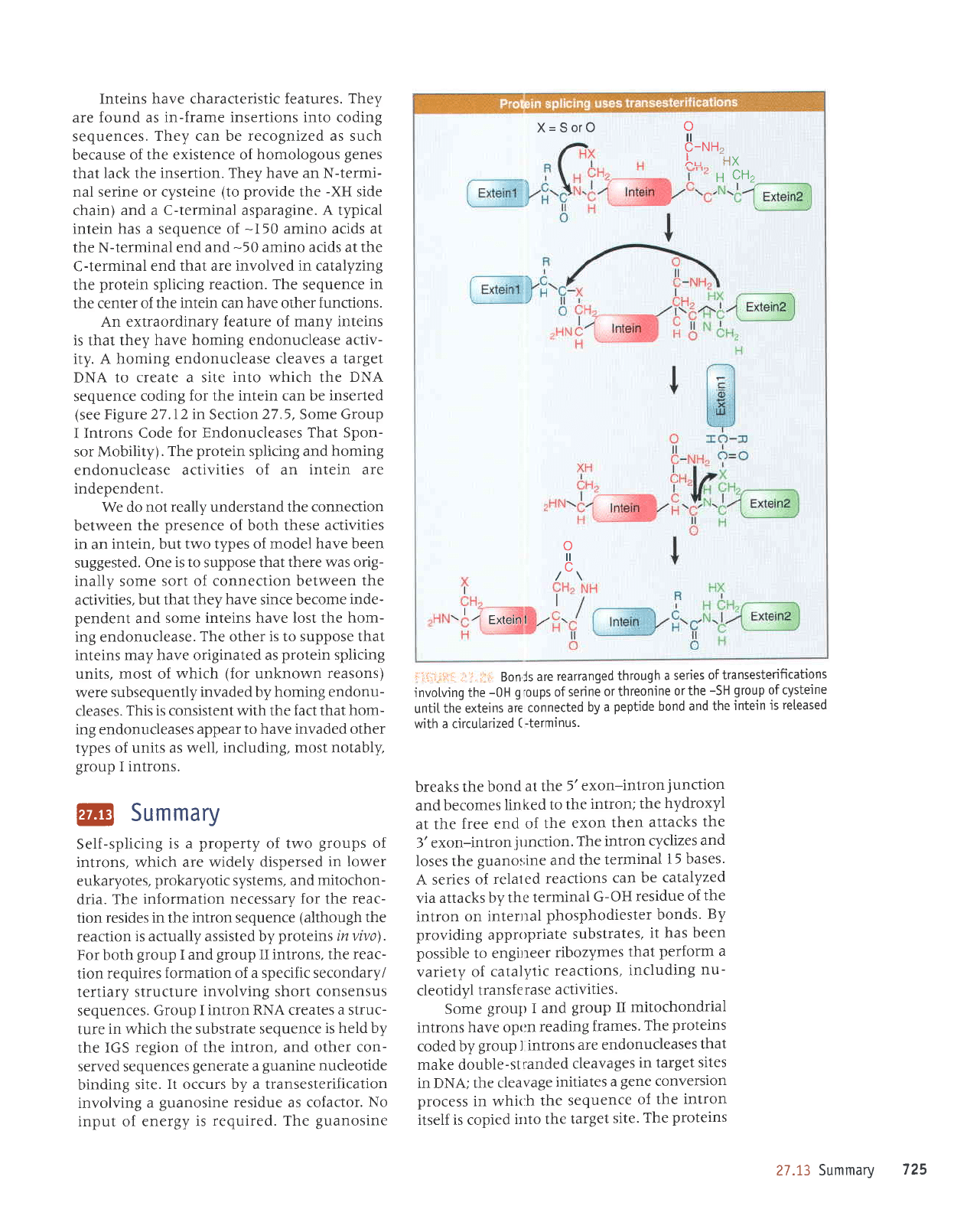
Inteins have characteristic leatures. They
are found as
in-frame insertions into coding
sequences.
They can be recognized as such
because
of the existence of
homologous
genes
that lack the
insertion. They have an N-termi-
nal serine or cysteine
(to provide
the
-XH
side
chain)
and a C-terminal asparagine.
A
typical
intein has a sequence
of
-I50
amino acids at
the N-terminal end and
-50
amino acids at the
C-terminal
end that are involved in catalyzing
the
protein
splicing
reaction. The
sequence
in
the center
of the intein can have other
functions.
An extraordinary
feature
of
many inteins
is that they
have homing endonuclease
activ-
ity. A homing
endonuclease cleaves a target
DNA to
create a site
into
which the
DNA
sequence
coding
for the intein can be inserted
(see
Figure
27.l2in
Section
27.5,
Some
Group
I Introns
Code for Endonucleases
That Spon-
sor Mobility).
The
protein
splicing and
homing
endonuclease
activities of an
intein are
independent.
We do not
really
understand the
connection
between
the
presence
of both these
activities
in an
intein, but two types of model
have been
suggested.
One
is to
suppose
that there was orig-
inally some
sort oI connection between
the
activities,
but that they
have
since become
inde-
pendent
and
some inteins have lost the
hom-
ing
endonuclease.
The
other
is to suppose that
inteins
may have originated as
protein
splicing
units, most of
which
(for
unknown
reasons)
were
subsequently
invaded by homing endonu-
cleases.
This is consistent with the
fact that hom-
ing endonucleases
appear to have invaded other
types of
units as well,
including, most notably,
group I introns.
l@
Summary
Self-splicing
is a
property
of two
groups
of
introns,
which are widely
dispersed in
lower
eukaryotes,
prokaryotic
systems, and
mitochon-
dria.
The
information necessary
for
the
reac-
tion
resides in the intron sequence
(although
the
reaction
is actually assisted by
proteins
in vivo)
.
For both
group
I and
group
II introns,
the reac-
tion
requires
formation of a specific secondary/
tertiary
structure
involving shott consensus
sequences.
Group
I intron RNA creates
a struc-
ture
in
which
the substrate
sequence
is held by
the
IGS region
of the intron, and
other con-
served sequences
generate
a
guanine
nucleotide
binding
site.
It occurs by a transesterification
involving
a
guanosine
residue as cofactor.
No
input of energy
is
required. The
guanosine
i:i{rilli:i
i:lr.r
l
Bonls
are
rearranged
through
a series
oftransesterifications
invotving the
-0H
g'oups
of
serine
or threonine
or
the
-SH
group
of cysteine
untjL
the exteins
ar€ connected
by
a
peptide
bond
and the
intein
is
reteased
with a circularized
(-terminus.
breaks the
bond
at the
5'exon-intron
junction
and
becomes
linked
to
the
intron;
the
hydroxyl
at the
free end
of the
exon
then
attacks
the
3'exon-intron
jtrnction.
The
intron
cyclizes
and
loses the
guanot;ine and
the
terminal
l5
bases'
A series
of relaled
reactions
can be
catalyzed
via
attacks
by the
terminal
G-OH
residue
of
the
intron
on intental
phosphodiester
bonds.
By
providing
appropriate
substrates,
it has
been
possible
to
engitreer
ribozymes
that
perform a
variety
of
catalytic
reactions,
including
nu-
cleotidyl
transferase
activities.
Some
group I and
group II
mitochondrial
introns
have opt:n
reading
frames.
The
proteins
coded by
group
J introns
ate endonucleases
that
make double-stranded
cleavages
in
target
sites
in DNA;
the cleavage
initiates
a
gene
converslon
process in whi<:h
the
sequence
of
the
intron
itself is copied
ilrto the
target
site.
The
proteins
X=SorO
o
tl
c-NH2
j.-
trx
H
CHz
I
\
/'
Exteinl
/
tl
27.13 Summarv
725

coded by
group
II introns
include
an endonu-
clease
activity
that initiates
the
transposition
process,
and
a
reverse
transcriptase
that
enables
an RNA
copy
of the intron
to
be copied into
the
target
site.
These
types of introns probably
orig-
inated
by insertion
events. The
proteins
coded
by both
groups
of
introns
may
include
maturase
activities
that assist
splicing
of the intron
by
sta-
bilizing
the formation
of the
secondary/tertiary
structure
of the active
site.
Catalytic
reactions
are
undertaken
by
fhe
RNA
component
of the RNAase
P ribonu-
cleoprotein.
Virusoid
RNAs
can undertake
self-cleavage
at a
"hammerhead"
structure.
Hammerhead
structures
can form
between
a
substrate
RNA
and a ribozyme
RNA, which
allows
cleavage
to be directed
at
highly
specific
sequences.
These
reactions
support
the view
that
RNA
can form
specific
active
sites that have
catalytic
activity.
RNA
editing
changes
the
sequence
of an
RNA
after
or during
its
transcription.
The
changes
are required
to create
a meaningful
coding
sequence.
Substitutions
of individual
bases
occur in
mammalian
systems;
they take
the form
of
deaminations
in
which
C is con-
verted
to
U or A is
converted
to L
A catalytic
subunit
related
to
cytidine
or adenosine
deam-
inase
functions
as
part
of
a larger
complex
that
has
specificity
for
a
particular
target
sequence.
Additions
and
deletions
(most
often
of uri-
dine)
occur
in trypanosome
mitochondria
and
in
paramyxoviruses.
Extensive
editing reactions
occur in
trypanosome
s in which
as many
as half
of the
bases in
an mRNA
are
derived
from
edit-
ing. The
editing
reaction
uses a
template
con-
sisting
of
a
guide
RNA
that
is complementary
to
the nRNA
sequence.
The reaction
is
catalyzed
by an
enzyme
complex
that includes
an endonu-
clease,
terminal
uridyltransferase,
and RNA li-
gase,
using free
nucleotides
as
the source
for
additions,
or releasing
cleaved
nucleotides
fol-
Iowing
deletion.
Protein
splicing
is an
autocatalytic
reaction
that
occurs
by bond
transfer
reactions
and input
of energy
is
not required.
The
intein
catalyzes
its
own
splicing
out
of the flanking
exteins.
Many
inteins
have
a homing
endonuclease
activity
that is
independent
of the
protein
splic-
ing
activity.
References
Group I Introns
Undertake
Setf-Spticing
by Transesterification
Reviews
Cech, T. R.
(1985).
Self-splicing
RNA: implicarions
for
evolution.
Int. Rev.
Cytol 91,
3-22.
Cech, T. R.
(
1987)
. The chemistry
of
self
-splicing
RNA
and RNA
enzymes.
Science
236,
t532-t5j9.
Resea rch
Been, M. D.
and Cech, T.
R.
(1986).
One
binding
site
determines
sequence
specificity
of
Te
tr ahymena
pre
-rRNA
self
-
splicing,
/rans-
splicing, and RNA
enzyme
activity.
Cell 47,
207-216.
Belfort,
M., Pedersen-Lane,
J., West,
D., Ehren-
man, K.,
Maley,
G., Chu, F.,
and Maley,
F.
(1985).
Processing
of the intron-containing
thymidylate
synthase (td) gene
of
phage
T4 is
at
the RNA level.
Cell 41,
)75-J82.
Cech, T.
R. et al.
(
I 981
).
In vitro
splicing
of
the
rRNA precursor
of Tetrahymena'.
involvement
of
a
guanosine
nucleotide
in
the
excision
of
the intervening
sequence.
Cell 27,
487-496.
I(ruger,
I(.,
Grabowski, P.
J.,
Zatg,
A.
J., Sands,
J.,
Gottschling,
D. E., and
Cech, T.
R.
(1982).
Self-splicing
RNA: autoexcision
and
autocy-
clization
of the ribosomal
RNA
intervening
sequence
of Tetrahymena
Cell 11,
147-1i7.
Myers,
C. A., I(uhla,
B., Cusack,
S., and
Lam-
bowitz,
A. M.
(2002).
tRNA-like
recognirion
of
group
I introns
by a tyrosyl-tRNA
syn-
thetase.
Proc
Natl. Acad.
Sci.
\JSA
99.
2630-26)5.
Group I Introns
Form
a Characteristic
Secondary
Structure
Resea rc
h
Burke,
J. M. et
al.
(1986).
Role
of
conserved
sequence
elements 9L
and 2 in
self-splicing
of
the Tetrahymena
ribosomal
RNA
precursor.
Cell 45,167-176.
Michel,
F. and
Wetshof, E.
(
I 990).
Modeting
of
the
three-dimensional
architecture
of
group
I
catalytic
introns
based on
comparative
sequence
analysis.
J. Mol
Biol. 216,
iB5-610.
Ribozymes
Have
Various
Catatytic
Activities
Cech, T.
R.
(1990).
Self-splicing
of
group
I introns.
Annu
Rev. Biochem
59,54)-568.
Review
726
CHAPTER
27
Catal.ytic
RNA

Re sea
rc h
Winkler, W. C., Nahvi,
A., Roth, A.,
Collins, J.
A.
and Breaker, R. R.
(2004).
Control of
gene
expre ssion by a natural metabolite-responsive
ribozyme. Nature 428, 281-286.
@
Some Group I Introns Code for
Endonucleases That Sponsor Mobil.ity
Review
Belfort, M. and Roberts, R. J.
(1997).
Homing
endonucleases: keeping the house in order.
Nucleic Acids Res. 25. J)79-)388.
Group
II Introns May Code
for Multifunction
Proteins
Reviews
Lambowitz, A. M. and Belfort, M.(1993).
Introns
as
mobile
genetic
elements. Annu- Rev.
Biochem. 62, 587-622.
Lambowitz, A.
M.
and
Zimmerly,
S.
(2004).
Mobile
group
II introns. Annu. Rev.
Genet
38,
l-35.
Resea
rc h
Dickson,
L., Huang, H. R., Liu, L., Matsuura,
M.,
Lambowitz,
A. M.,
and
Perlman, P. S.
(2001).
Retrotransposition of a
yeast group
II intron
occurs
by reverse splicing directly into ectopic
DNA
sites.
Proc Natl. Acad. Sci USA 98,
r)207-rj2r2.
Zimmerly, S. et
al.
(1995).
Group
II intron mobil-
ity occurs by target DNA-primed
reverse tran-
scription. Cell
82, 545-554.
Zimmerly, S. et
al.
(1995).
A
group
II intron
is
a
catalytic
component of a DNA endonuclease
involved
in intron mobility. Cell 8j,
529-5)8.
Some
Autosplicing Introns Require
Maturases
Resea rch
Bolduc et al.
(2003).
Structural
and biochemical
analyses of
DNA
and
RNA binding by a
bifunctional
homing endonuclease and
group
I
splicing
factor. Genes. Dev. 17, 2875-2888.
Carignani, G. et
al.
(1983).
An RNA maturase
is
encoded by the
first intron of the mitochon-
drial
gene
for
the subunit
I of cytochrome oxi-
dase in S. cerevisiae. Cell
]5, 7j3-7 42.
Henke, R. M., Butow,
R. A.,
and
Perlman, P. S.
(
1995
).
Maturase and endonuclease
func-
tions
depend on separate conserved
domains
of the bifunctional
protein
encoded by the
group
I intron aI4 alpha of
yeast
mitochon-
drial
DNA. EMBO J. 14, 5094-5099.
Matsuura, M., No;rh,
J. W.,
and Lambowitz,
A. M.
(200 l). Meclranism
o[
malurase-promoted
group
II intron splicing.
EMBO J
20,
7259-7270.
Viroids
Have Catatytic
ActivitY
Reviews
Doherty, E.
A.
ani
Doudna,
J.
A.
(2000).
Ribozyme strlrctures
and
mechanisms.
Annu Rev.
Biochem.
69,597-615.
Symons,
R. H.
(
I 992
).
Small
catalytic
RNAs.
Annu Rev.
Biochem. 61,641-671.
Resea rc h
Forster, A. C. and
Symons,
R.
H.
(1987). Self-
cleavage
of
virusoid
RNA is
performed
by
the
proposed
55-nucleotide
active site.
Cell 50,
9-t6.
Guerrier-Takada,
tl.,
Gardiner,
K., Marsh,
T., Pace,
N., andAltman,
S.
(1983).
The RNAmoiety
of
ribonuclear;e
P is the
catalytic
subunit
of the
enzyme.
Cell )5,
849-857.
Scott, W. G.,
FinclL,
J. T.,
and
ICug, A.
(I995).
The
crystal
structrtre
of an
all-RNA
hammerhead
ribozyme:
a
proposed mechanism
for
RNA
catalytic
cleavage.
Cell
81, 991-1002.
RNA
Editing
Occurs
at
IndividuaI
Bases
Resea
rc h
Higuchi, M. et al.
(19931.
RNA
editing
of AMPA
receptor subunit
GluR-B:
a
base-paired
intron-exon
structure
determines
position
and
efficiency. CeU
75, l)61-l)70.
Navaratnam,
N e:.
al.
(
I 995
).
Evolutionary
origins
of apoB
mRNA editing:
catalysis
by a cytidine
deaminase
that
has acquired
a
novel
RNA-
binding
motit at
its active
site. Cell
81,
r87-195.
Powell. L.
M.. Wattis,
S. C.,
Pease,
R. J.,
Edwards,
Y. H., Knott,
'f.
J., and
Scott,
J.
(1987). A
novel form ol
tissue-specific
RNA
processing
produces
apolipoprotein-B48
in
intestine.
Cell
50,83I-840.
Sommer,
B. etal.
(1991).
RNAeditinginbrain
controls
a delerminant
of ion
flow
in
gluta-
mate-gated
c.eannels.
Cell
67
,
ll-19.
RNA Editing
Can
Be
Directed
bv
Guide
RNAs
Aphasizhev
R.,
Slticego,
S.,
Peris,
M., Jang,
S.
H.,
Aphasizheva,
I., Simpson,
A.
M., Rivlin,
A',
and Simpson ,
L.
l2OO2).
Ttlpanosome
mito-
chondrial
3'
lerminal
uridylyl
transferase
(TUTase):the
key enzyme
in U-insertion/
deletion
RN,a.
editine.
Cell
108,
637-648.
Researc
h
References
727

Benne,
R., Van
den Burg J., Brakenhoff,
J. P.,
Sloof, P., Van Boom,
J. H.,
and Tfomp, M.
C.
(I986).
Major
transcript
of the frameshifted
coxll
gene
from
trypanosome
mitochondria
contains four
nucleotides
that are not
encoded
in
the DNA.
CeU 46,819-826.
Blum,
B., Bakalara,
N., and
Simpson, L.
(1990).
A
model for RNA
editing in kinetoplastid
mito-
chondria:
"guide"
RNA
molecules
transcribed
from
maxicircle
DNA
provide
the edited infor-
mation.
Cell 60, 189-198.
Feagin,
J. E., Abraham,
J. M.,
and Stuart, K.
(1988).
Extensive
editing
of the cytochrome
c
oxidase III
transcript in Trypanosoma
brucei.
Cell
51,4I)422.
Seiwert,
S. D., Heidmann,
S.
and
Sruarr, K.
(1996).
Direct
visualization
of
uridylate deletion
in
vilro
suggests
a mechanism for
kinetoplastid
editing.
Cell
84,831-841.
Protein
Spticing Is Autocatalytic
Review
Paulus,
H.
(2000).
Protein
splicing
and related
forms of
protein
autoprocessing.
Annu Rev.
Biochem. 69,447496.
Research
Derbyshire, V.,
Wood, D.
W., Wu, W.,
Dansereau,
J. T., Dalgaard,
J.2., andBelfort,
M.
(1997).
Genetic definition
of a
protein-splicing
domain: functional
mini-inteins
support
structure
predictions
and
a model for
intein
evolution.
Proc. Natl. Acad.
Sci.
USA 94.
tt466-tt47t.
Perler,
F. B.
et al.
(L992).
Intervening
sequences in
an Archaea
DNA
polymerase
gene.
Proc. Natl.
Acad
Sct. USA 89, 5577-5581.
Xu, M.
Q.,
Southworth, M.
W., Mersha,
F. B.,
Hornstra,
L. J., and Perler,
F. B.
(19931
. in
vitro
prorcin
splicing
of
purified
precursor
and
the identification
of a branched
intermediate.
Cell
75,
liTl-1377.
728
CHAPTER
27
Catatytic
RNA

6ZL
aODd
lrau
uo
panuquoJ
'solnqnlorrrru
0l uorllauuol aql
apr^old
Aeu saxaqduor
oMl asaql
lrouuol 1eq1
suralold eq1
r
'uorllunJ
luoruorluor r0J
lprtuassa
s!
III-lCl
ol spurq
1eq1
xaldLuor
urelord
gg93
aq1
r
'II-l0l
]P
pau.loJ
sr alnllnrls urlPurolql
lPnsn
aql
ol anrlpurallp up sr
1eq1
xaldLuor uralord
pazrlerrads
y
o
xalduol uralord
p
spur.€ alauorluel eqf
'11-363
uorbar Llrp-I-V aql
)uplJ
tpq]
III-I0l
pue
I-l0l
soluonbos
pa^rasuol
iloqs
oql
lo
lslsuol
sluauala
Nll
o
'srsolrru
1e
[1e1elnrre ale6erEas o1
pruseld
e rvrolle
o1
fi1L1Lqe eq1
f,q
aostaatals u!
pagquepr
a.lp sluouala
[!]l
r
anLst^dil
's
ur saluanbas
vNo
iloqs
a^eH salauollual
'uMoul
lou
sr
yp6
enrlLladar oql
Jo
uor]lunJ aLlf
o
'yp6
enqLladar
lo
slunoue abrel
uleluol
seuosouro.lql rrlofile4na raq6rq uL se]euor]uol
o
vN0
a^qqaoau ureluol APhl salau.lorlual
'salu0n00s
vN0
alrtleles
ur
qlu
sr.
leql
urleur0rql0ralaq a^Pq uolJo s0.r0ruo.llu0l
o
'uorDal
lueuorlual slr ur suuoJ
lPql
eroqrolauq aql ol salnqnlo.llrur
Jo
luauqrPlle
oql
r\q
elpuLds
lrlolru aql uo
plaq
sr auosorxo.lql rrlofire1na
y
o
alnao uoqe6ar6as e sI aurosoruolql lqo^rPln3 aql
,,'sgnd,.
e,rLb o1
puedxe
sauos
-ourolql
euefiod uo
uoLssardxa auab
1o
salrs ale
]p{}
spuefl
r
uorssaldxl
auag
Jo
salrs
le
puedxl
sauosourolLll
eual^lod
'deu
1err6o1o1Ar
e se
pasn
oq upl
1eq1
spupq
lo
soues e eneq suereldLp
Jo
sauosoruo.lql ouolnlod
.
spuPg rulol saurosoruorql aual,{lod
'srxp
leu0s0ul0rql
aql ulorJ
popualxa
ale
leql
s00ol
MOqS seuosouroiqr
qsnlqduel
uo uoLsserdxa auab
Jo
solr$
r
papuaul
arv sauosouorql
LlsnlqouPl
'spueqlolut
qlP-l-9
aql ut
polellualuol
alP sOUO!
r
'spuPqlalut
aql ueql
}uolu0l
l-!
ut iaMol elP spuPq oql
r
'spueq-9
pallpr
ale
qlrqM
'suorlpuls
Jo
sauas
p
Jo
oluereadde aql
a^pq ol sauosoruo.lql
aq] asnPl sanbruqlal 6ururels ur.P|.lol
I
sulailed 6uLpueg a^eH sauosorxorql
'asPq0lelut
ln0
-qbnorql
palred Alasuap utPual
ugeuolqlolaloq;o
suor6eX
.
's0ruosou0rql
llloltul
upql
pelred A11q6q ssel
st
qrtqM
'utlsuolqlna
Jo
ulloJ
aql
ur sr. urleuorlll
Jo
sseu
lelauab
aql
'asPqdlalut
6uun6
r
'srsolrur
6uunp
r{1uo uaes
oq uPl
seuosouto.l!r
lPnpLnLpul
r
u tlPtuoJ
LllolalaH
pue
utlPtx0lqlnl
olut
papl^]o sI ullPtu0lql
'0luenDos
sns
-uesuol
llJDads
fiue aneq
lou
op
]nq
ql!-.l-V
ore
sl!fi otlf
r
'suvs
ro suvt/\l
pollPl
sesuanbes
llJt]eds
lP
xllleu .lPallnu
eql 01
poqrPne
s!
vNo
r
xulew aseqdralul
ue
o1
vNo
qlPnv
saluanbas
llJllads
'peqrellP
aie
vN0
paltoriadns
1o
sdool eq1
qrtq/v\
01
ploljels utalold P
aAeq sauosotllo.ltll
ose{oP}e}rl
o
'qI
98-
Jo
sutPulop
luepuadoput
olur
palrolradns [1art1e6au st
utlPtuo.lq]
asPqdra]ut
J0
vN0
.
plo#Prs
e
o]
paqlP]lv suteuo0
puP
sdool sPH
vN0
lqo,{rP)nl
'dq
gg1/rorredns
I-
sr. butttor.redns
;o
flrsuap
a6erane
aq1
r
'sureu0p
polrolladns [1anL1e6au
]uapuedaput
ool-
sPq
ptoellnu
0q1
.
paltolredns
{
auiouo!
leuape8
aqt
'polJquapt
uaoq
lou
e^eq
VN6
eql 6uLsuepuor
loJ alqtsuodsal
ale
lPql
suralotd
aql
r
'uralold
lo
VNU
uo
llP leql
sluabe
^q
paploJun
oq uel
pue
ssPu
nq
vNo
%08-
st
ptoallnu
lPua]leQ
aLll
o
ptoallnN
e sI auoua9
lPuellP8
otll
'llaqs
ut010lo
polquasseald P olut
vNo
oql
uesul
sasnlt^
vNo
lsluoqds
.
'1r
punole
llaqspeeq
aql alquassP
Aeql se
euouab
VNU
aqt
asuapuol
sasur^
VNU
sno]ueulelll
r
'pasuapuol
filaualya
st
lleqspPaq
aql uttlltM
plle
ltelrnI
o
'lloqspPaq
eq]
Jo
alnllnlls
oql
fq
paltultl
sr snlh
P olut
polPlod.lolu!
eq uPl
lPql
vNo;o
q16ua1 eql
r
sleol
ltaql olut
pabellPd arv
sauoua!
lPlL^
u0qlnpollul
3Nrtrn0
u3l-dvH3
rflfir
wm
Telomeres
Have
Simpte Repeating
Sequences
o
The
te[omere is required
for
the stabitity
of the
chromosome end.
r
A
tetomere consists
of a simpte repeat
where
a C + A-rich
strand
has
the seouence
cr(A/r)F4.
Te[omeres
Sea[ the
Chromosome Ends
.
The
protein
TRF2
catalyzes
a
reactjon
in
which
the 3'
repeating
unit of the G
+
T-
rich
strand
forms
a
loop by disptacing its
homotog
in an
upstream region
of the
telomere.
Telomeres Are
Synthesized
by a
Ribonucteoprotein Enzyme
o
Tetomerase
uses the 3'-0H
of the G + T
telomeric strand to
prime
synthesis of tan-
dem
TTGGGG reoeats.
.
The RNA component
of telomerase has
a
sequence that
pairs
with the
C
+
A-rich
repeats.
r
One of the
protein
subunits
is
a reverse
transcriptase
that uses the RNA
as temptate
to synthesize
the G
+ T-rich
sequence.
Te[omeres Are
EssentiaI for
SurvivaI
Summarv
@
Introduction
A
general
principle
is
evident in
the organiza-
tion of all
cellular
genetic
material. It
exists as
a compact
mass that is
confined
to a limited vol-
ume,
and its various
activities,
such as replica-
tion and
transcription,
must
be accomplished
within
this space. The
organization
of this mate-
rial
must accommodate
transitions
between
inactive
and
active states.
The
condensed
state of nucleic
acid results
from its
binding
to basic
proteins.
The
positive
charges
of these
proteins
neutralize
the nega-
tive
charges
of the nucleic
acid. The
structure
of
the nucleoprotein
complex
is determined
by
the interactions
of the
proteins
with the DNA
(or
RNA).
A common problem
is
presented
by the
packaging
of DNA
into
phages,
viruses,
bacre-
rial
cells, and
eukaryotic
nuclei. The
length
of
the DNA
as an extended
molecule
would vastly
exceed the
dimensions
of the
compartment
that
contains
it. the DNA
(or
in
the case
of some
viruses,
the RNA) must
be compressed
exceed-
ingly
tightly to fit into
the
space avallable.
Thus
in contrastwith
the customary
picture
of DNA
as an
extended doubk helix,
structural deformation
of DNA
to bend or
fold
it into
a mlre clmpact
form
is the rule
rather than
exceotion.
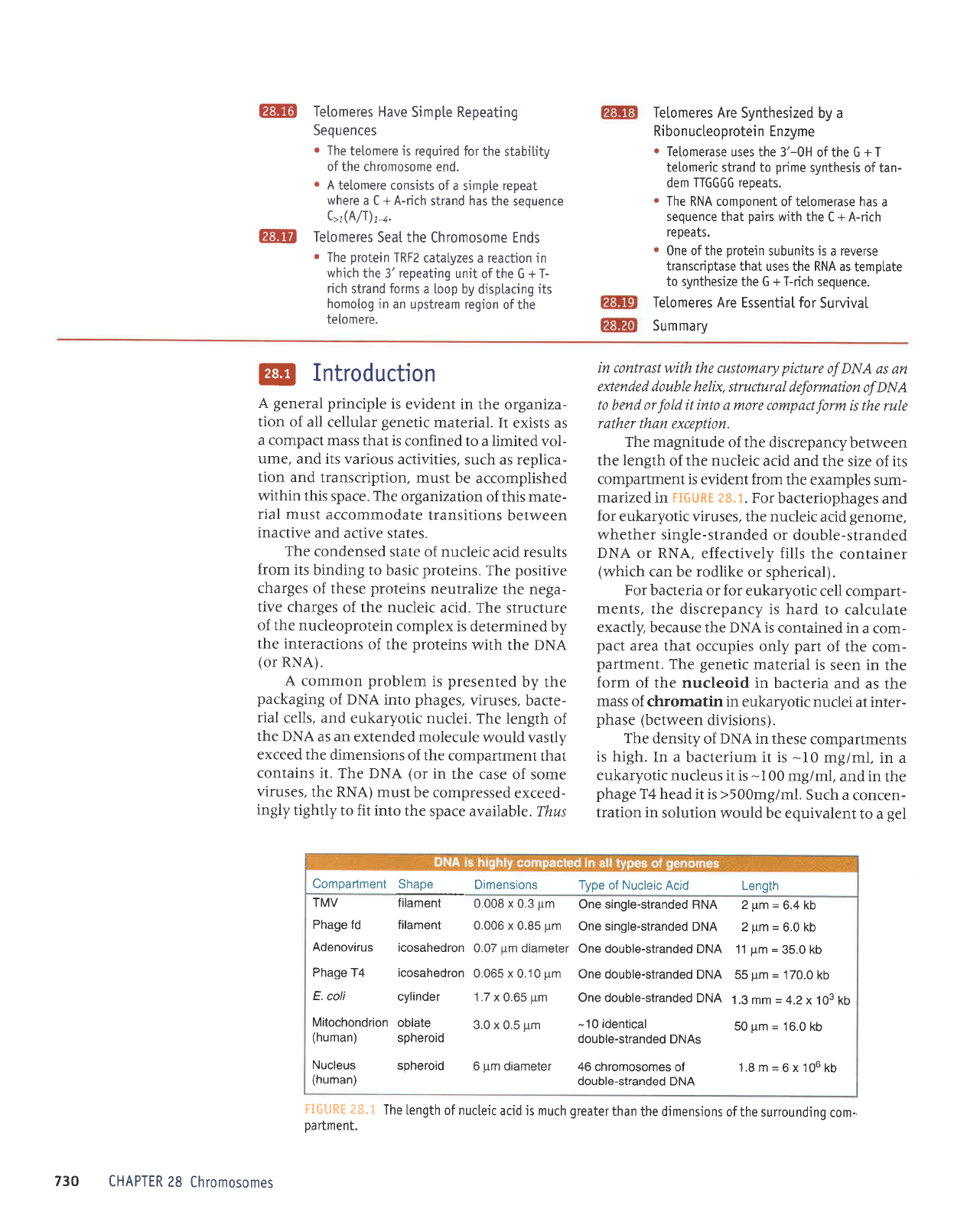
The magnitude
of the
discrepancy
between
the
length
of the nucleic
acid and
the size
of
its
compartment
is evident ftom
the examples
sum-
marized
in
FIGUHt
eS.1.
For bacteriophages
and
for eukaryotic viruses,
the nucleic
acid
genome,
whether
single-stranded or
double-stranded
DNA
or
RNA,
effectively fills
the
container
(which
can be rodlike
or spherical).
For
bacteria or for
eukaryotic
cell compart-
ments, the
discrepancy is hard
to calculate
exactly,
because the DNA is
contained in
a com-
pact
area
that occupies
only
part
of the com-
partment.
The
genetic
material
is
seen in
the
form
of the nucleoid
in bacteria
and
as the
mass
of chromatin in eukaryotic
nuclei
at inter-
phase (between
divisions).
The density
of DNA in these
compartments
is
high. In a
bacterium it is
-10
mg/ml,
in
a
eukaryotic
nucleus it is
-100
mg/ml,
and in
the
phage
T4 head
it is
>5O0mg/ml.
Such
a concen-
tration
in solution
would be equivalent
to a
gel
ftfi'-lHHf;S.t
TheLengthofnucleicacidismuchgreaterthanthedjmensionsofthesurroundingcom-
Dartment.
Compartment
TMV
Phage fd
Adenovirus
Phage T4
E.
coli
Mitochondrion
(human)
Nucleus
(human)
Shape
filament
filament
icosahedron
icosahedron
cylinder
oblate
spheroid
spheroid
0.008 x
0.3
pm
0.006 x 0.85
pLm
0.07
pm
diameter
0.065 x
0.10
pm
1.7 x
0.65
pm
3.0 x
0.5
pm
6
pm
diameter
Dimensions Type
of Nucleic
Acid
Length
One single-stranded RNA
One single-stranded
DNA
One
double-stranded DNA
One
double-stranded DNA
One double-stranded
DNA
-10
identical
double-stranded
DNAs
46 chromosomes
of
double-stranded
DNA
2pm=6.4kb
2pm=6.0kb
11
pm
=
35.0 kb
55
pm
=
170.0
kb
1.3mm=4.2x103kb
50
pm
=
16.9 16
1.8 m
=
6 x 1Oo kb
730
CHAPTER
28
Chromosomes
