Lewin Benjamin (ed.) Genes IX
Подождите немного. Документ загружается.

Incubate
with
+
Gel electrophoresis
I
The
Splicing
Endonuclease
Recognizes
tRNA
o
An endonuctease
cteaves
the IRNA
precursors
at
both ends
of the
intron-
o
The
yeast
endonuclease is
a
heterotetramer
with
two
(reLated)
catatytic subunits.
r
It
uses a
measuring
mechanism
to determine
the
sites of cteavage by their
positions
retative
to a
ooint
in
the IRNA
structure.
r
The
archaeal
nuctease
has
a simpter structure
and
recognizes
a butge-hetix-butge
structuraI motif in
the substrate.
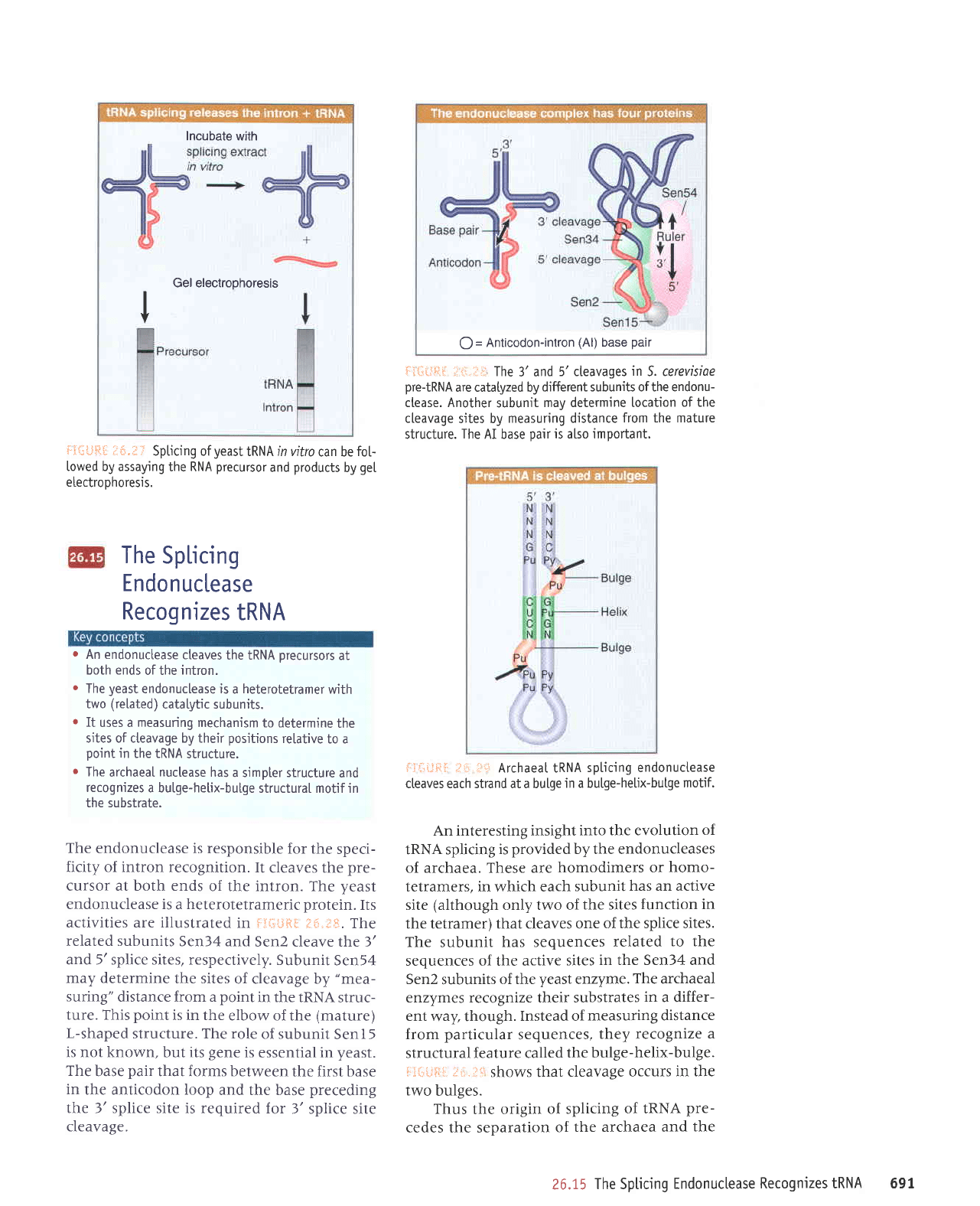
The
endonuclease is responsible
for the
speci-
ficity of intron recognition.
It
cleaves the
pre-
cursor at both ends
of the intron. The
yeast
endonuclease is
a
heterotetrameric protein.
Its
activities are illustrated
in fl{iLiH[ t*, ;|*.
The
related subunits
Sen34 and
Sen2 cleave the 3'
and 5'splice sites, respectively.
Subunit
Sen54
may determine
the sites
of cleavage by
"mea-
suring"
distance from a
point
in the IRNA
struc-
ture. This
point
is in the
elbow of the
(mature)
L-shaped
structure. The role
of subunit
Sen
I
5
is not known,
but
its
gene
is
essential in
yeast.
The
base
pair
that forms between
the first base
in
the anticodon loop
and the base
preceding
the 3' splice site is required
for
3' splice site
cleavage.
O=
Anticodon-intron
(Al)
base
pair
r*tfl*tit
llrl.iii;': The
3' and
5' cleavages
in
S. cerevisiae
pre-tRNA
are catatyzed by
different subunits ofthe endonu-
ctease. Another
subunit
may determine location
of the
cleavage
sites by
measuring distance
from the mature
structure.
The
AI base oair
is also important.
fl I
ii
iJ
F
lr
i:l
:::
.
i.'
i,i
A rc h a ea t t
R N A s
p
[i ci
n
g
e
n
do
n u clease
cleaves each strand at a bulge
in a bulge-helix-butge
motif.
An interesting insight
into the evolution
of
IRNA splicing
is
provided
by the endonucleases
of archaea. These are homodimers
or
homo-
tetramers, in which each
subunit
has an active
site
(although
only two of
the sites
function in
the tetramer) that cleaves one
of the splice sites.
The
subunit
has sequences
related
to the
sequences of the active
sites
in the Sen34 and
Sen2
subunits
of the
yeast
enzyme.
The
archaeal
enzymes recognize their substrates
in
a differ-
ent
way. though.
Instead of
measuring distance
from
particular
sequences,
they recognize
a
structural feature called
the bulge-helix-bulge.
l,I:;li"illtr
:,:,.i.iii shows that
cleavage occurs
in the
two
bulges.
Thus
the origin
of splicing
of IRNA
pre-
cedes the separation
of
the archaea and
the
Fl*LJfrf
i.:{i.t.r
Spticing of
yeast
tRNA in
yifro
can be
fol
lowed by assaying
the RNA
precursor
and
products
by
gel
etectrophoresis.
26.15
The Spticing
Endonuclease
Recognizes IRNA
69t
eukaryotes. If it originated by insertion of the
intron into
tRNAs, this
must have
been
a very
ancient event.
Each 5'terminus ends
in
a
hydroxyl
group;
each
3' terminus ends
in a 2',3'-cyclic
phosphate
group.
(All
other known RNA splicing enzymes
cleave on the other side
of the
phosphate
bond.)
The two half-tRNAs base
pair
to form a
tRNA-like structure. When
AIP is
added.
the
second reaction occurs.
Both
of the unusual
ends
generated
by the endonuclease must be
altered.
The cyclic
phosphate
group
is opened to
generate
a 2'-phosphate terminus. This reac-
tion requires cyclic
phosphodiesterase
activity.
The
product
has a 2'-phosphate
group
and a
3'-OH
group.
The 5'-OH
group generated
by the
nuclease
must be
phosphorylated
to
give
a 5'-phosphate.
This
generates
a site
in
which the 3'-OH is next
to the 5'-phosphate. Covalent integrity of the
polynucleotide
chain is then restored
by
ligase
activity.
AII three activities-phosphodiesterase,
pollnucleotide
kinase, and adenylate synthetase
(which provides
the ligase function)-are
arranged in different functional domains
on
a
single
protein.
They
act sequentially to
join
the
two IRNA
halves.
The spliced molecule is now uninterrupted,
with a 5'-3'phosphate linkage at the
site of
splicing, but
it
also
has a 2'-phosphate
group
marking
the event.
The
surplus
group
must be
removed by a
phosphatase.
Generation of
a2',3'-cyclic
phosphate
also
occurs during the IRNA-splicing reaction in
plants
and mammals. The reaction in
plants
seems to be the same as in
yeast,
but the detailed
chemical
reactions are different in
mammals.
The
yeast
IRNA
precursors
also can be
spliced
in
an extract obtained from the
germi-
nal
vesicle
(nucleus)
of. Xenopus
oocytes. This
shows that the reaction is not
species-specific.
Xenopus must have enzymes able
to recognize
the introns in the
yeast
tRNAs.
The
ability to splice the
products
of IRNA
genes
is therefore
well conserved, but is likely
to
have
a different origin
from
the other
splic-
ing reactions
(such
as that of nuclear
pre-
mRNA). The
IRNA-splicing reaction
uses
cleavage and synthesis of bonds
and is deter-
mined
by sequences that are external
to the
intron.
Other splicing reactions use
transester-
ification,
in which bonds are transferred
directly,
and the sequences required for
the reaction lie
within the intron.
l@
IRNA Cleavage and
Ligation Are
Separate
Reactions
.
Release of the intron
generates
two half-tRNAs
that
pair
to form the
mature
structure.
.
The
halves have the unusual ends 5' hydroxyl and
2'-3'
cyctic
phosphate.
.
The
5'-0H end
js
phosphorytated
by a
potynucteotide
kinase, the cyclic
phosphate group
is
opened by
phosphodiesterase
to
generate
a
2'-phosphate
terminus and 3'-0H
group,
exon
ends are
joined
by an RNA ligase,
and the
2'-phosphate is removed by a
phosphatase.
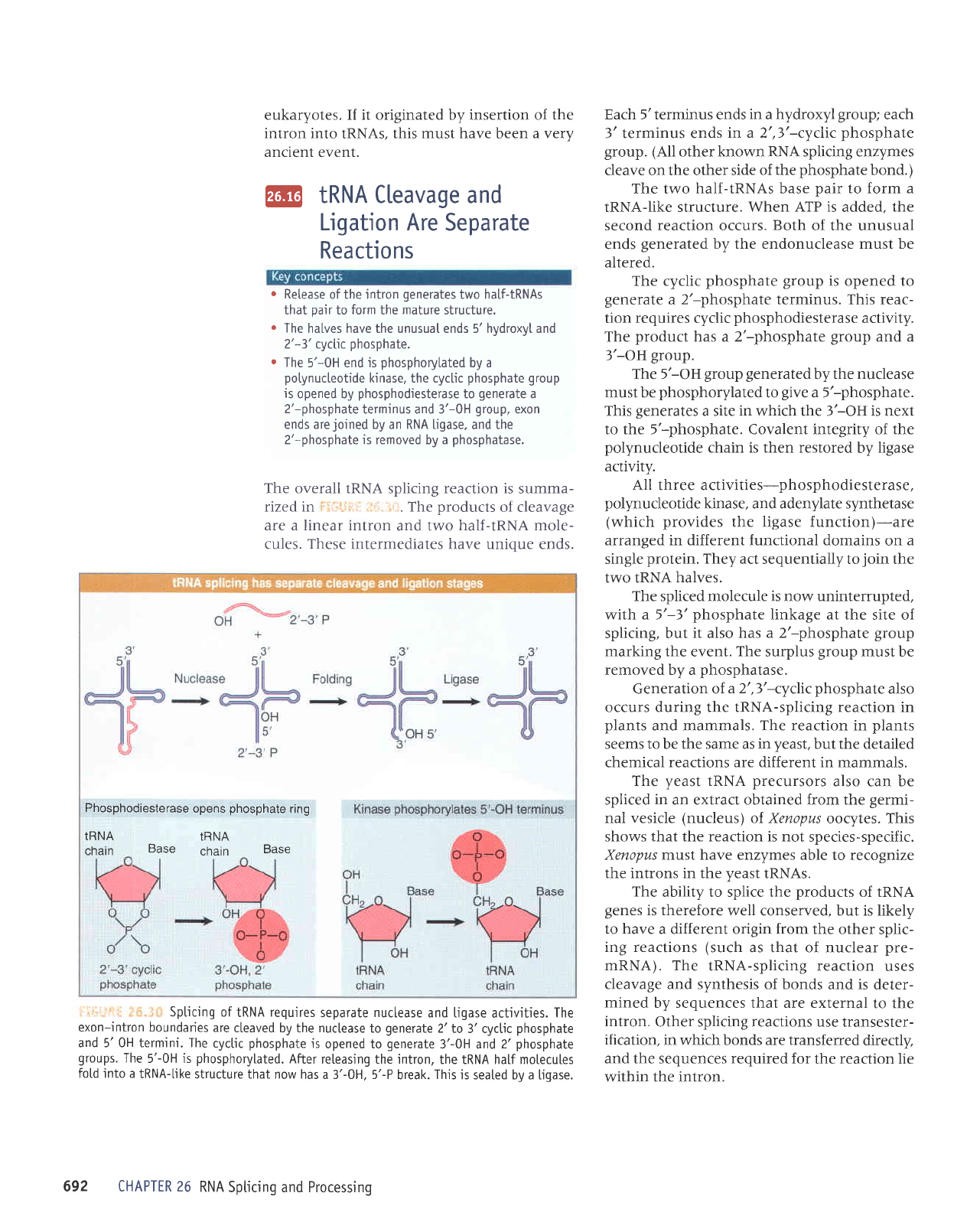
The overall IRNA
splicing
reaction is
summa-
rized in
'
,r,,:ii,
.r.-:.-:ir. The
products
of cleavage
are a linear intron
and two half-tRNA
mole-
cules.
These
intermediates have unicue ends.
itl,,i::,;
Spticing of
tRNA
requires
separate
nuclease
and ligase
actjvities.
The
exon-intron boundarjes
are
cteaved by the nuclease
to
generate
2'to
3'cyctic
phosphate
and 5'0H termini. The
cyclic
phosphate
is
opened to
generate
3'-0H
and
2'phosphate
groups.
The
5'-0H
js
phosphorylated.
After
releasing the intron,
the IRNA half motecules
fotd into a
tRNA-Like
structure that now
has a 3'-0H. 5'-P
break.
This
is seated by a ligase.
Phosphodiesterase
opens
phosphate
ring
tRNA
IRNA
chain
Base
chain
Base
692 CHAPTER 26
RNA
Spticing and Processing
The
Unfolded
Protein
Response
Is
Related
to IRNA
Splicing
o
Irelp
is an inner nuctear
membrane
protein
with
its N-terminaI
domain in
the ER [umen,
and
its
C-
terminal
domain in the nucteus.
.
Binding
of an unfotded
protein
to
the N-terminaI
domain
activates the
C-termjnal nuclease
by
a uto
p
hosp
horytation.
o
The
activated nuclease
cleaves
Hacl mRNA to
retease
an intron
and
generate
exons that are
tigated by a IRNA ligase.
.
The
spticed Hacl mRNA
codes for
a transcription
factor
that activates
genes
coding for
chaperones
that
hetp
to fotd unfolded
proteins.
An
unusual splicing
system that
is related to
IRNA splicing mediates
the response
to unfolded
proteins
in
yeast.
The accumulation
of unfolded
proteins
in
the lumen
of the endoplasmic
retic-
ulum
(ER)
triggers
a response
pathway
that
Ieads
to increased transcription
of
genes
coding
for
chaperones that
assist
protein
folding in the
ER. A
signal must therefore
be transmitted
from
the lumen of the ER
to the nucleus.
The sensor
that activates
the
pathway
is
the
protein
Irelp.
It is an integral
membrane
pro-
tein
(Ser/Thr)
kinase
that has
domains on
each
side of the ER membrane.
The
N-terminal
domain in the lumen
of the ER
detects the
pres-
ence
of unfolded
proteins,
presumably
by bind-
ing
to exposed motifs.
This causes
aggregation
of monomers
and activates
the C-terminal
domain on the other
side of the membrane
by
autophosphorylation.
Genes that are activated
by this
pathway
have
a common
promoter
element
called the
UPRE
(unfolded
protein
response
element). The
transcription factor Haclp
binds to
the UPRE,
and is
produced
in
response
to accumulation
ol
unfolded
proteins.
The
trigger for
production
of Haclp is the
action of Irelp
on Hacl nRNA.
The
operation of the
pathway
is summa-
rized in
; li
l;'::i
: .
.: t.
Under normal
conditions,
when the
pathway
is not
activated, Hacl mRNA
is translated into
a
protein
rhat is rapidly
degraded. The activation
of Irelp results in
the
splicing of the Hacl
mRNA to
change the
sequence
of the
protein
to a more
stable form.
This form
provides
the functional
transcription
factor that activates
genes
with the UPRE.
Unusual splicing components
are involved
in
this
reaction. IrelP has an endonuclease
activ-
ity
that acts directly on
Hacl mRNA to cleave
the two
splicing
junctions.
The
two
junctions
are ligated by the IRNA
ligase that acts
in
the
IRNA splicing
pathway.
The endonuclease
reac-
tion resembles
the
cleavage of IRNA during
splicing.
Where
does
the modification of
Hacl nRNA
occur? Irelp is
probably
located in the
inner
nuclear membrane, with the N-terminal
sensor
domain in
the
ER lumen, and the
C-terminal
kinase/nuclease domain
in the nucleus.
This
would enable
it
to
act directly on
Hacl RNA
before it is exported to
the cytoplasm.
It
also
would allow easy access
by the IRNA
ligase.
There
is no apparent
relationship between
the
Irelp nuclease activity
and the IRNA splicing
endonuclease, so
it is not obvious
how this spe-
cialized system would
have evolved.
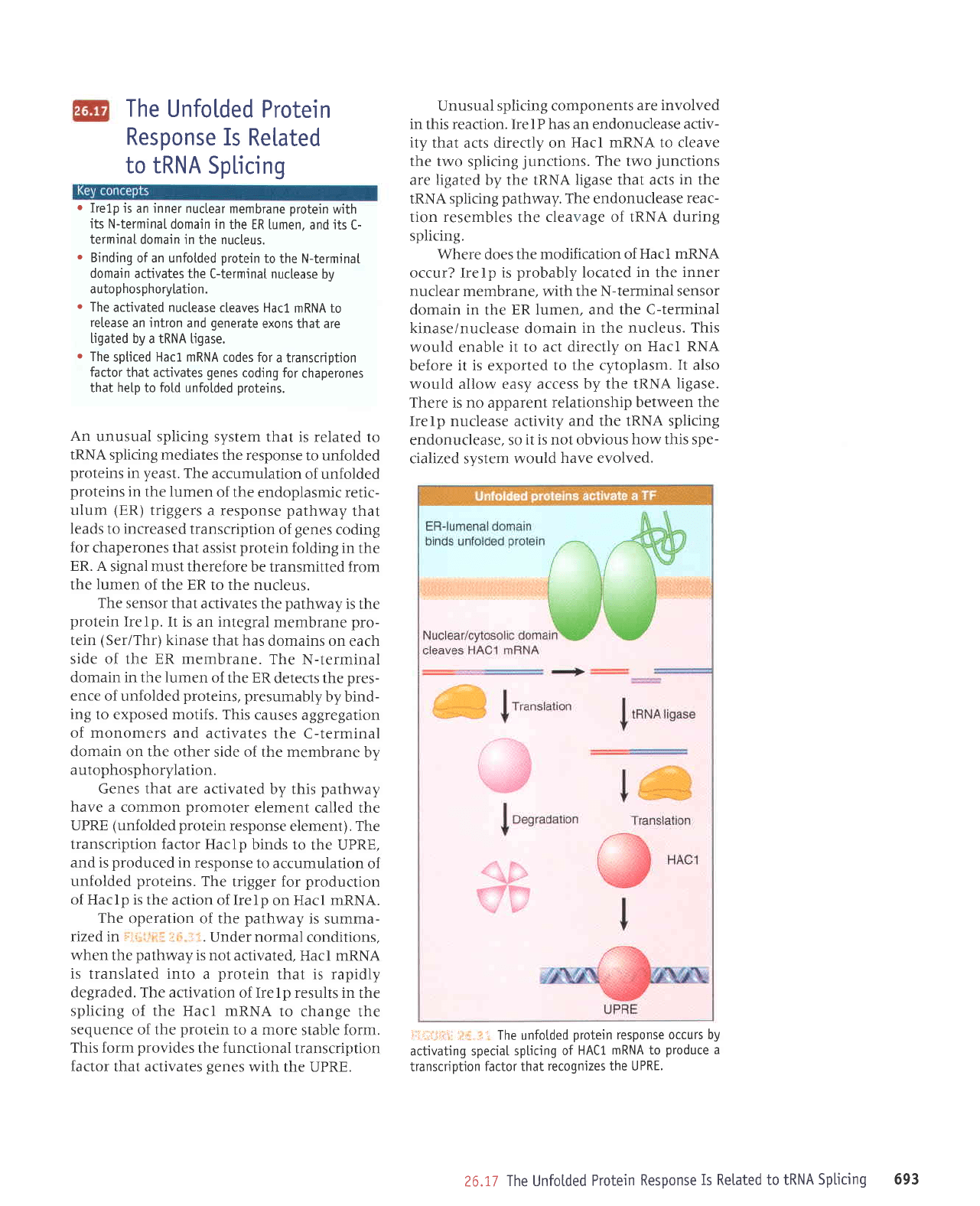
i
ir,ritir
,,
,:,
-,'r
The unfotded
protein
response
occurs by
activating speciaI
spticing of
HACl
mRNA to
produce
a
transcription
factor that
recognizes the UPRE.
26.1.7
lhe Unfotded
Protein
Response
Is Related to IRNA
Spticing 693
i:*tJ*f
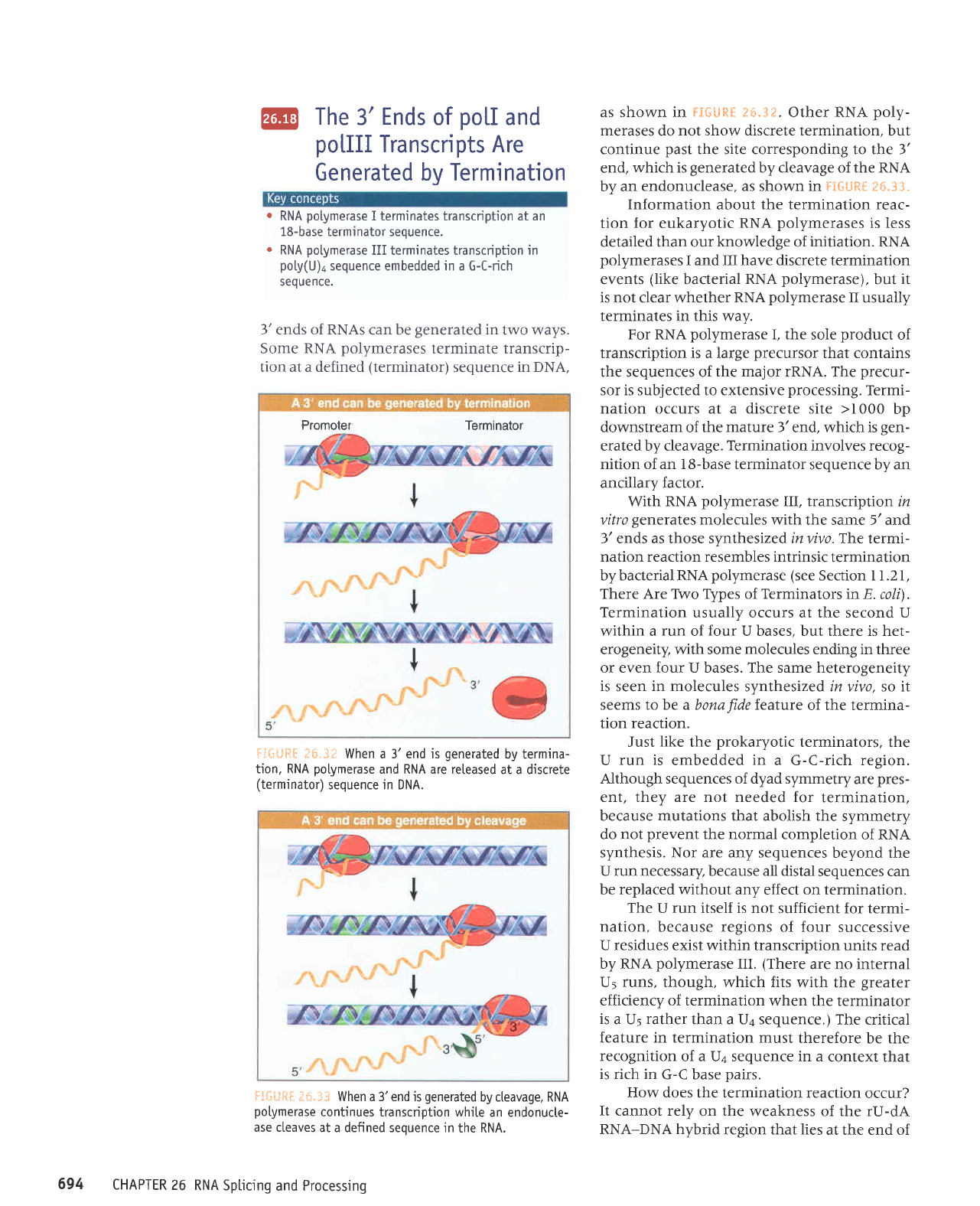
J*-.iF
When a 3' end is
generated
by termina-
tion, RNA
potymerase
and RNA are released
at a discrete
(terminator)
sequence in DNA.
F;*liftf.
iri"3t When a 3'end is
generated
by cteavage, RNA
polymerase
continues transcription while
an endonucte-
ase cteaves
at a defined sequence in
the RNA.
CHAPTER
26
RNA SpLicing
and Processing
as shown in FI*[Jftil
e*.3*.
Other RNA
poly-
merases
do
not show discrete termination,
but
continue
past
the site corresponding to the 3'
end,
which
is
generated
by cleavage of the RNA
by an
endonuclease, as shown in FfSLiRE 3S.33.
Information
about
the termination reac-
tion for eukaryotic RNA
polymerases
is less
detailed than our
knowledge
of
initiation.
RNA
polymerases
I and III have discrete termination
events
(like
bacterial
RNA
polymerase),
but it
is not clear whether RNA
polymerase
II usually
terminates in this way.
For RNA
polymerase
I, the
sole
product
of
transcription is a large
precursor
that contains
the sequences of
the major rRNA.
The
precur-
sor is subjected to extensive
processing.
Termi-
nation occurs at a discrete site
>1000
bp
downstream of the
mature f'end,
which is
gen-
erated by cleavage. Termination involves recog-
nition of an 18-base terminator
seouence bv an
ancillary factor.
With RNA
polymerase
III, transcription
in
vitro
generates
molecules with
the same 5'and
3' ends as those synthesized in vivo. The termi-
nation reaction resembles intrinsic
termination
by bacterial RNA
polymerase (see
Section I I.2I,
There Are Two
\pes
of Terminators
in E. coli).
Termination
usually occurs at the second U
within a
run
of
four
U bases, but there is het-
erogeneity, with some
molecules
ending in three
or even four U bases. The same heterogeneity
is
seen
in molecules
synthesized in vivo, so it
seems to be a bona
fide
feature
of the termina-
tion
reaction.
Just
like
the
prokaryotic
terminators,
the
U
run is
embedded
in
a G-C-rich region.
Although sequences of dyad symmetry
are
pres-
ent, they are not needed for
termination,
because mutations that abolish the
symmetry
do not
prevent
the normal completion
of RNA
synthesis. Nor are any sequences
beyond the
U run necessary because all distal
sequences can
be replaced without
any effect on termination.
The U run itself is not
sufficient for termi-
nation,
because regions of four
successive
U residues exist within transcription
units read
by RNA
polymerase
III.
(There
are no internal
U5 runs, though, which fits
with the
greater
efficiency of termination
when the terminator
is
a U5
rather
than a Ua seeuence.)
The critical
feature in termination
must therefore
be the
recognition
of a Ua sequence in a
context that
is rich in
G-C base
pairs.
How does
the termination reaction
occur?
It cannot rely
on
the
weakness of the rU-dA
RNA-DNA hybrid
region that lies
at the end of
@
The
3'
Ends
of
po[I
and
pol.III
Transcri
pts
Are
Generated by Termination
.
RNA
polymerase
I terminates transcription at an
18-base terminator sequence.
r
RNA
potymerase
III terminates transcription in
poty(U)a
sequence embedded in a G-C-rich
Seouence.
J'ends of RNAs
can be
generated
in
two ways.
Some RNA
polymerases
terminate transcrip-
tion at a defined
(terminator)
sequence
in DNA,
Promoter Terminator
694

'uorlel^uopeAlod
ro;
pua
,6
e alerauaE o1 a6enealr
rol fressarau
s!
VVV1VV
atuanbas
oql
ti
.ti.
:iHli*T:J
uorlelr{uaper{1od
pue
a6p^eall
[q
paletauag
alv sVNUrx
Jo
spu]
,Eaql
6I.gZ
eJnlJuls
leurrulJl
,f
radord
Jql
Jo
uorleJJ
-uJt
'i-;i::'it::
:i-$lifji,,{
uI
palPJlsnlll
are Sulsse)ord
,€
sJ>lPuJpun
]eq]
xalduror
Jql
Jo
suorl)un}
pup
uorleuro}
JqJ
'suort)eeJ
eql Surz^leue ol
elnoJ
eql
peuJdo
ltltrur
sJnJJo
uortelLuape,{lod
qrlqM
ur ruJts^s
e
Jo
tueudole^ap
eq;.
'uoqe1.,(uape,{1od
pue
a8eneap
qloq
loJ
prpJeu
sr
1eu8rs
aq1
'pue
,€
patelLuapeLlod
aql
Jo
uorte
-Jeue8
slua.Lard.rarupxaq
Wynyv
eql
Jo
uoll
-ptnur
Jo uortJlr(I
'uorlrppe
(y),{1od
Jo
rtrs eql
yo
rueartsdn sJprloJlrnu
0€
ot
I I
tuoJJ uor8a-r
Jqt uI
11vvnyy
aruanbas
pelresuoJ,{1q3rq
aql
;o
aruasard
aql sr
(tsear{
ur
tou 1nq)
sarol.re>1na
raq8rq
ur sVNUtu
Jo.
JJntpeJ
uorutuo)
V
vNu
rql
Jo
pur,€
rql ol
SurpuodsarroJ
elrs aql
Jo
ueJrlsuMop uor8ar
3uo1
e urqlrm
suorleJol SuilrBrr
le
rnrro .{eru
tnq'peurJep
IIJM
lou
erp
II
JSeJJu.{1od
y1.1g
roy
sJlrs uorteururral
aql Lq,r,r
sureldxa srql
'(;ryo14
rolf,e{ oqu
seoo /1'oH
'ee'
II
uorlJas Jes)
aseraru.dlod
VNU
IprJaDeq
Lq
uortdursue,rt
3ur
-IEUIIUIA]
UI OqJ
JO
EIOI
Jql JOJ
]Pql
O1 JEIIIUIS
SI
Iepou
IIeJaAo
JqJ
'eteururat
ot uouduf,suerl
Sursnel
'VN(
ruoJJ
aseraruLlod
VNU
Jo
Jseelar
aqt
s:a33rr1 uolt)eralur
slqt
pup
'aserauLlod
Jql
Jo
ureuop
IPurruJJl-zkoqrer
Jqr ol
punoq
Jre
leql
sutalord,{.:elIrue qllm
sDeJelut ueql
11
'aseraruLlod
vNU
qrpu
dn seqJte)
tl tpqt
os
'paztsaqlu.ds
sr
tr
upql JatspJ
VNU
aql
saper8ap
11
'a8e,tealr
rJUp
pJqrrJsueJl
Jq ol SurnurluoJ sr
]eq]
vNu
aql
Jo
puJ
,E
Jql o1 spurq eseelJnuoxe
uv
'JI
eseJau,{1od
y55
Lq
uorleururral ro; ra8
-3In
rrarpul
ue saprnord
luane
e8pneelJ JqJ
_
'pJlf,alOJOun
sr aBpAeJIJ aqt ,{q
paleraua8
sr
leql
puJ
,S
Jql
lnq
'a3errea1)
aqt JeUe uolldlJlsueJl senurluoJ
aseraru,{1od
y1qg
'der
aqr Lq
pazlgqels
dpea:p
sr
pue
,s
sll
'pue
,€
eql IUOJJ uoBepBrSap
lsureSe
vN6ru
Jql sJzllrqels uo4elluape,{1od aq1
'uor1
-e1Luape.{1od
pue
Surpnr rrp
qloq
sa>lpuapun
xaldruor
Surssarord a18urs
y'uor1e1,,{uape,{1od
,{q
pa,uo11oy
1nr
rrl.dloalJnuopua ue ro;
sta8
-rpl
se
pazruSof,JJ
aJe
VNU
eql ur
sJruenbes
pue
'pua
,€
eql ot SulpuodsarJoJ elrs Jql
lsed
saquJsuerl JspJaru^lod
VNU't,l{'sH
3Hft. Ls
ur
pJleJlsnllr
sr
pua
,€
aql
Jo
uorleJauJ)
'vNu
realJnu ruoJJ
VNUUr
Jo
uorternlpu Jql rc1 ftassanu sr
uorlpl
-.{uape.{1od
tpqt
sMoqs srql
'useldoldr
aq1 ur
VNUIU
;o
aruereadde aql stuanard uortrppp str
'VNU
Jeepnu
yo
uorldrnsueJt Jqt dots
tou
saop
urdar.dpror
q8noqrlV
'uydart(plo)
se
umou>l
osle sr
qJIr{M
'Jursouepe^xoep-,€
3o1eue aql
.dq
paluanard
aq uer
vNU
reelrnu or
(y)L1od
Jo
uolt1ppv
'uone1.duape,{.1od
.{q
pa^l,ro11oy
a8e
-,tea1r
.dq
pateraua8
eJp svNgru
Jo
spuJ
,€
aqJ
'lueudolarap
rruoA:qua
sn douay
6u1tnp
uoqelrtuapeep ro uoqel^uape^lod rruuseldo1^r
lo.lluor lrPl ,€
aql u! saluanDas
ull-r-fl-v
r
'pua,E
aql o1
filantsserord sanptsal
V
002-
ppe
aseraufi1od
(y)A1od pup
ro]rpJ flrqoads aq1
r
'vvvnvv
Jo
r!PorlsuMop
VNU
onealr aspollnuopua
pue
rolrpj AlLrrloeds
eq1
r
'aseraur{1od
(y)ri1od
pup
'eseepnuopua
up lotlpJ
r{lLrgLrads
e sulpluol
1eq1
xalduor urelord e sarrnbel uogrea.l a{f
r
'pe1e1r\uapeAlod
sr
1eq1
VNUrU
Jo
puo
,t
p
alereuab
o1 e6e,reep ro;
leu6rs
e sl
VVV1VV
aruenbes aq1
o
uorlPl/tuepP^lod
pue
abeneall
Aq
palerauag
alv
svNUr.u
Jo
spul
,€
aql
';1as1r
arurlzue eql
Jo
uorpunJ e eq o1 surJJs uorleuruJJl
'auo1e
qsrtd
-uroJJe
touuef,
11
aserarudlod
vNu
q)rqM
'uot1
-f,eeJ
uorlerlrur eql
qtl.tr
tspJluor
u1
'pu8rs
goqs
e
qlns
ot Llerryrads os
puodsar
uBr
aruu(zua
eql
Moq
lnoqp
pJIZZnd
ureruar eM
'uollpurtu
-rat
rnol;e>lord ur
paalonur
urd-neq aql ot
uedral
-uno)
e
Jq
ol ruJas
1ou
sJop JJaq]
lnq
'aru,{zua
eqt
uMop Suunols uI elor e s,{e1d
uor8a-r
qlrr
-)-D
aq1 sdeqra4
'pJqlJlsueJl
eJe sanplsJJ
1l
o^1.l
lsJr}
aqt
r(1uo
ualJo
JsneJJq
'ldrDsuerl
aq1
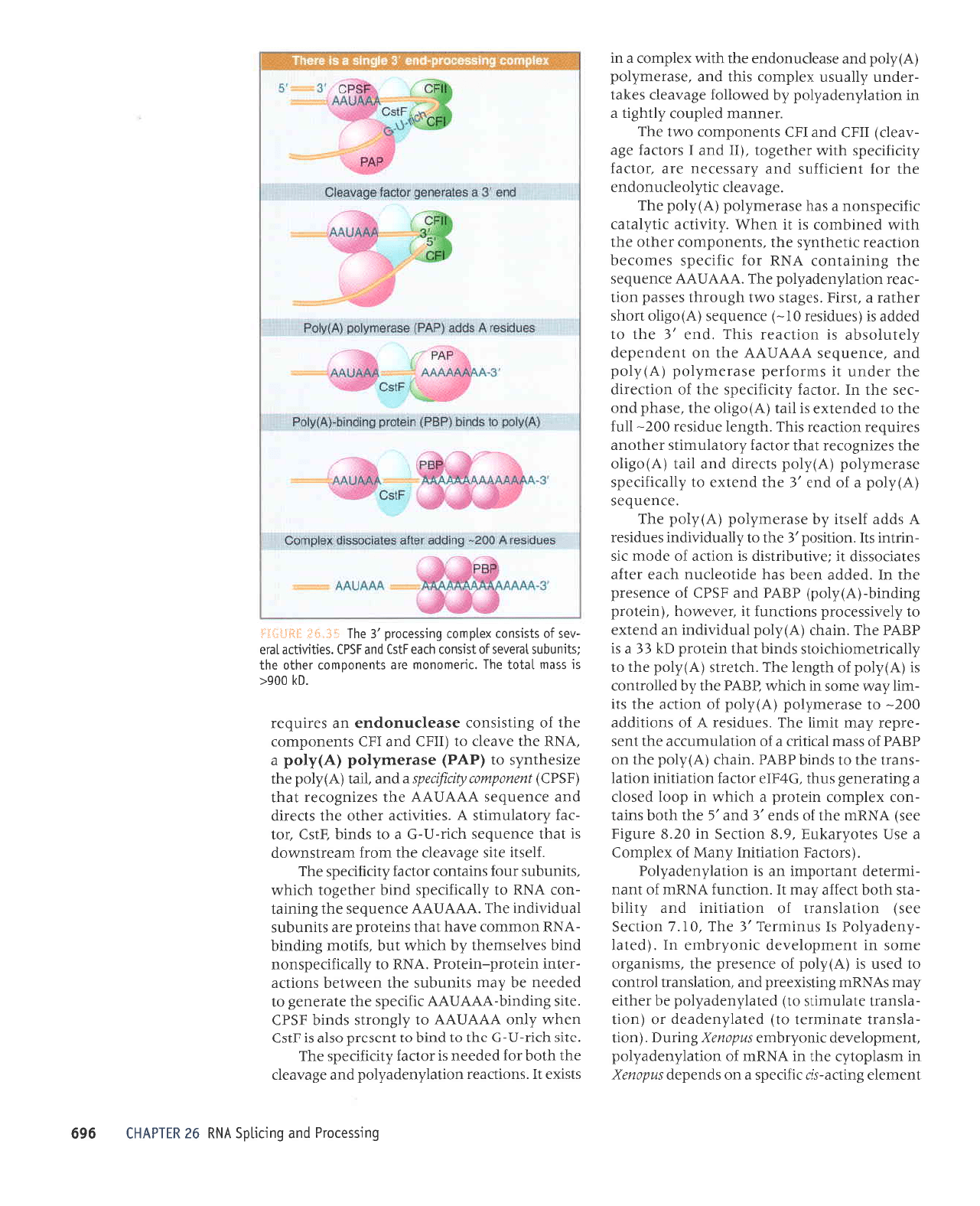
luJruJle
Suttre-sr l;nads e uo spuadap sndouay
ur useldo1{f, eql ur
VNUUr
Jo
uortelduapedlod
'luaurdolanap
rruozkqura
sndouay Surtnq'
(uor1
-elsuert
ateururlJt ot)
pareltruapeap
ro
(uor1
-elsueJl
Jlelnurts ot)
pa1e1.{uapez(1od
eq rrqtrr
Leru syggru Surlsxaard
pue
'uorlplsuerl
IoJluoJ
01
pJSn
sr
(y).{1od;o
aruasard Jq1
'srusrup8ro
Jruos ur
luarudola,Lap
rruo,{rqrua u1
'(pare1
-,{uaped1o4
sI
snuruJel
,€
JqI
'0
I'4
uortlJS
aas) uollplsuen
Jo
uortertrul
pup
,{tItQ
-p1s
qtoq
pa;;e.{eru
tI
'uortlunJ
VNUUr
Jo
tueu
-rrxJJlJp
tuetrodurr
ue sr uorlelr(uape.{1o4
'
(s.ropeg
uortertrul,{ue14
;o
xalduo3
e asn
sato^,i:e>lng
'6'8
uorlJes ul
0Z'g
a;n8r4
aas)
yggru
Jql
Jo
spue
,€
pue
,S
eqt
qloq
surel
-uor
xaldruor uratord
p
qJrqM
ur dool
pesol)
e Surteraua8
snqt
'g7CIJ
JolJeJ uorteplq uortel
-suen
eqt ot spurq
dgvd
'urpqr
(y)d1od
rqt uo
dgYd
Jo
sseru
IPf,rtuJ
P
Io
uorlPlnunJJe eql
lues
-ardar
^eru
tlrull
aqJ
'sJnprsJr
V
Jo
suortrppe
002-
ot aseraru.dlod
(y)d1od
Jo
uortrp aql sll
-ru11
zle.r,t eruos ur
qrlq1!1.
agvd
Jql
Lq
palonuor
sr
(y).{1od
yo qr3ua1
eql
'qJlarls
(y)z(1od
aqr ot
,{llerrnaruorq)rols spurq
teqt
urato:d
O>l €€
e sl
dgvd
rqJ'ureq)
(y),{1od
Ienpurpu
ue
purtxa
o1 z{.la,rrssarord suorlrun;
ll
'Jelellroq '(uratord
Supurq-(y)z(1od)
4gy4
pue
dsdl
;o
aruasard
Jql
uI
'prppe
urJq
seq epllorlrnu
qrea
rrlJP
selelJosslp
ll
iJ^rlnqulsrp
sr uorlJp
Jo
Jporu Jrs
-urrlur
s11
'uorlrsod,€
Jql ol
,{genprnrpur
sJnprsJJ
v
sppe
;1asrr
Lq
aseraruLlod
(y),{1od
aq1_
.JJUENDES
(y),{1od
p
Jo
pue
,€
rqt
purtxa
o1
,{11e1;oads
aseraru,{1od
(y).{1od
stf,arrp
pue
Irel
(y)o311o
aql sazruSorJJ
leql
roDe; r{rolelnrurls raqloue
sartnbar uollJpeJ srql
'qr8ual
JnprsJr
002-
IInJ
eql ot
pepuetxe
sr
Iret
(y)o8r1o
aqt
'aseqd
puo
-Jes
Jql uI
'JollPI
,{tlrl}pads
Jql
Jo
uortf,JJrp
aI{]
repun
11
sruro;rad aseraruLlod
(y),i1od
pue
'rJuJnbas
yyynyv
eqt uo
tuapuadap
,{latnlosqe
sr uorlJpal srqJ
'pua
,€
eqt ol
peppp
sr
(sanprsar g1-)
atuanbas
(y)o31o
rroqs
raqleJ e
'tsrld 'sa8els
out
qSnoJql
sassed uorl
-rear
uouel.duapeLlod
aqI'Wvnyy aruanbas
aql SututeluoJ
17NU
ro;
rryrlads seuroJeq
uorl)eeJ rllaqtur(s
aql
'sluauoduoJ
Jaqlo Jql
qllz\{ paulquoJ
sI
1l
uarl6
',
ltntlre
rtt.{1eter
rr;oadsuou e
seq aserJru.{1od
(y)d1od
aq1
'
aBeneap rqLloapnuopua
Jql JoJ
luer)rJJns
pue
,{ressarJu
Jrp
'Jot)eJ
.d1nr;nads qlp,l
raqtaSol
'(g
pue
1
sroDe; a8e
-zreap)
IICf,
pup
193
sluauodurol
oMl eqJ
'reuueur
paldnor
,{lrq8lt e
ur uorlelLuapez(1od
.{q
pa,uo11oy
a8eneap sa>1e1
-JJpun
dllensn
xaldruor
slqt
pue
'aseraruLlod
(y).{1od
pue
eseJlJnuopur
aqt
qUlvr
xalduol
e ur
6uLssaror6
pue
6uLrqd5
VNU
9Z
UlIdVHl
stsrxJ
tI
'suortJeJJ
uouel,{uape,{1od
pue
a8eneap
rr-lt
qtoq
roJ
pJpJJu
sI rolrpJ
Atl.rllt:ads aqJ
'Jtrs
qlrr-ll-D
eql 01
pulq
ol
lueserd
oslp sI
{lsJ
uaq.,rzr.
^dpo
VVvn\/V
ot
.{.l3uorls spulq
dsd)
'a1rs
Surpurq-Wv1vy
rr;nads aql
alerauaS o1
pJpJJu
aq ,{.eru strunqns
Jql ueeMlJq suollJe
-JJtur
uretoJd-uralo.rd'VNU
ot
Llprt;nadsuou
purq
srnlrsruaqt.dq
qllqM
tnq
'sJlloru
Surpurq
-VNU
uoruuroJ
JAeq
leql
sulatord
JJp sllunqns
Ienprlrpur
aql'Vvynvy
aruenbas aqt
Sututel
-uor
yNU
ot
.{11err;nads
pqq
raqlaSot
qrlqm
'slrunqns
rnoJ sureluor ropeJ,{loryoads
aq1
'Jleslr
Jlrs aSenealr Jqt
ruoJJ rueeJlsumop
sr
leql
aruanbas
qrlr-n-c
e 01 spurq
dls)
Jol
-re1
fuolelnrurts
V
'sJrlrlrt)p
rJqlo eql sDerlp
pue
aruanbas
VVV1VV
aqt saztuSoJJr
leql
(SSaf
)
yauodwoc
[1tcpfnadsepue
1u
(y)d1od
aql
azrsaqtuLs ot
(dvd)
aseraru,{.1od
(y)I1od
e
'VNU
Jqt J^eJIf,
ot
(IId)
pue
Idf,
stuauodruor
Jql
Jo
Surlsrsuor esEalJnuopue
ue sa:rnbar
'01
006<
sr sseu
,lP101
aql
'lr.ieu0uour
0lP sluau00ul0r
ieqlo 0ql
lslrunqns
lPla^os J0
lslsuor
qrPa
llsl
puP
lsdl
'saqnqlP
lela
-^os
Jo
slsrsuor
xaldLuor
6ulssarord
,€
oql
i;{'i.iil
31;{1:}-1.d
969
(the
CPE) in the
3'tail. This
is
another
AU-rich
sequence,
UUUUUAU.
In Xenopus
embryos
at least
two
type of cls-
acting
sequences
found in
the 3'tail
can trigger
deadenylation. EDEN (embryonic
deadenyla-
tion
element) is a I7-nucleotide
sequence.
ARE
elements are AU-rich
and
usually
contain
tan-
dem repeats
of AUUUA.
There
is a
poly(A)-spe-
cific RNAase
(PARN)
that
could
be involved in
the degradation.
Of
course,
deadenylation
is
not always
triggered
by specific
elements;
in
some situations
(including
the normal
degra-
dation
of
mRNA
as it
ages),
poly(A)
is degraded
unless it is
specifically
stabilized.
\cc
u
U
A
A
u
U
Cleavage
of the
3' End
of Histone
mRNA
May
Require
a
Sma[[ RNA
o
Histone
mRNAs
are
not
potyadenytated;
their 3'
ends are
generated
by a cleavage
reaction
that
depends on the structure
ofthe mRNA.
.
The cleavage reaction
requires
the
SLBP to bjnd to
a stem-toop
structure
and the U7
snRNA to
pair
with
an adjacent
single-stranded region.
Some mRNAs
are not
polyadenylated.
The for-
mation
of their
3' ends is
therefore
different
f rom the
coordinated
cleavage
/polyadenylation
reaction.
The
most
prominent
members
of this
mRNA
class are the mRNAs
coding
for histones
that are
synthesized
during DNA
replication.
Formation
of their
3' ends
depends upon
sec-
ondary structure.
The structure
at the
3'termi-
nus is
a highly conserved
stem-loop
structure,
with
a stem of 6
bp and
a loop of four
nucleotides.
Cleavage occurs
four
to
five
bases
downstream
of the
stem-loop. TWo
factors are
required
for the cleavage
reaction:
The stem-
loop
binding
protein
(SLBP)
recognizes
rhe
structure,
and the
U7 snRNA
pairs
with a
purine-rich
sequence
(the
histone
downstream
element,
or
HDE)
located
-10
nucleotides
down-
stream
of
the
cleavage
site.
Mutations
that
prevent
formation
of the
duplex stem of the
stem-loop
prevent
formation
of the
end of the RNA.
Secondary
mutations
that restore duplex
structure
(though
not nec-
essarily
the original sequence)
behave
as
rever-
tants. This
suggests thal
formation
of the
secondary
structure is more important
than the
exact sequence.
The
SLBP binds to
the stem-loop
and then inter-
acts
with U7 snRNP to
enhance its interaction
with the downstream
bindine
site
for
U7 snRNA.
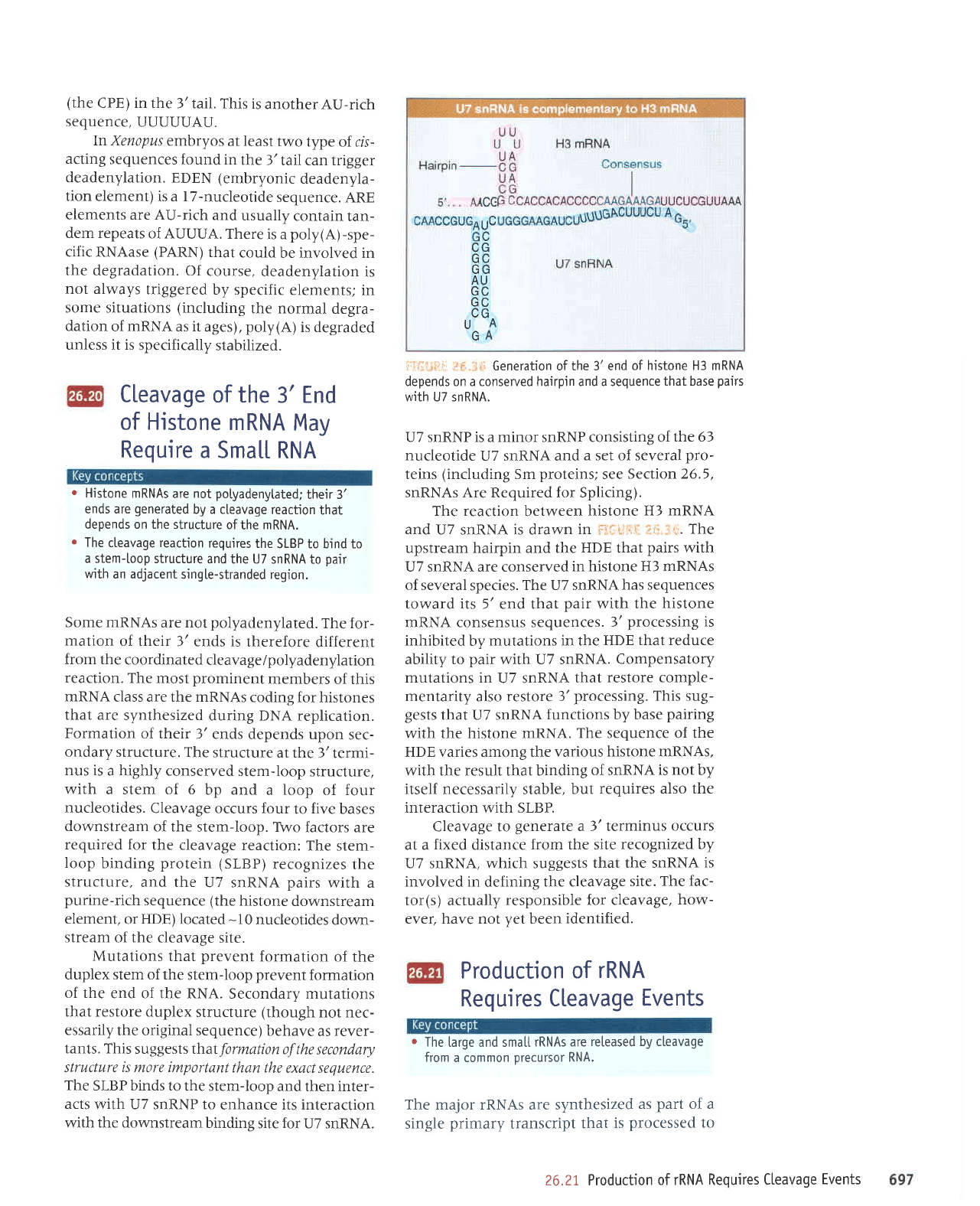
i
ir'l':rrir
,,
;
I
":
Generation of the 3' end
of histone H3 mRNA
depends on a conserved hairpin and a sequence
that base
pairs
with U7 snRNA.
U7 snRNP is a minor snRNP consisting of
the 63
nucleotide
U7 snRNA and
a set of several
pro-
teins
(including
Sm
proteins;
see
Section 26.5,
snRNAs Are Required for Splicing).
The reaction
between
histone
H3 mRNA
and
U7 SnRNA iS drawn
in
:,,,ttiir:
,,i..
r,,,,.
The
upstream hairpin and the HDE that
pairs
with
U7 snRNA are conserved
in histone H3 mRNAs
of several
species.
The U7 snRNA
has sequences
toward its 5' end that
pair
with the
histone
mRNA consensus sequences.
3'processing
is
inhibited
by
mutations in the
HDE that reduce
ability to
pair
with U7 snRNA.
Compensatory
mutations in
U7 snRNA
that
restore comple-
mentarity
also restore
3'processing.
This
sug-
gests
that U7 snRNA
functions by base
pairing
with the histone mRNA.
The sequence of the
HDE varies among the various
histone mRNAs,
with the result that binding
of snRNA
is not
by
itself
necessarily stable,
but requires
also the
interaction with SLBP.
Cleavage to
generate
a J'terminus
occurs
at a fixed distance
from the site
recognized by
U7 snRNA, which suggests
that the snRNA
is
involved in
defining
the cleavage
site. The
fac-
tor(s) actually responsible
for cleavage,
how-
ever, have not
yet
been
identified.
Production
of
rRNA
Requires
Cleavage
Events
o
The
large and smatl
rRNAs are
reteased by cleavage
from
a common
precursor
RNA,
The major rRNAs are synthesized
as
part
of a
single
primary
transcript
that
is
processed
to
26.2I Production
of rRNA Requires
Cteavage
Events
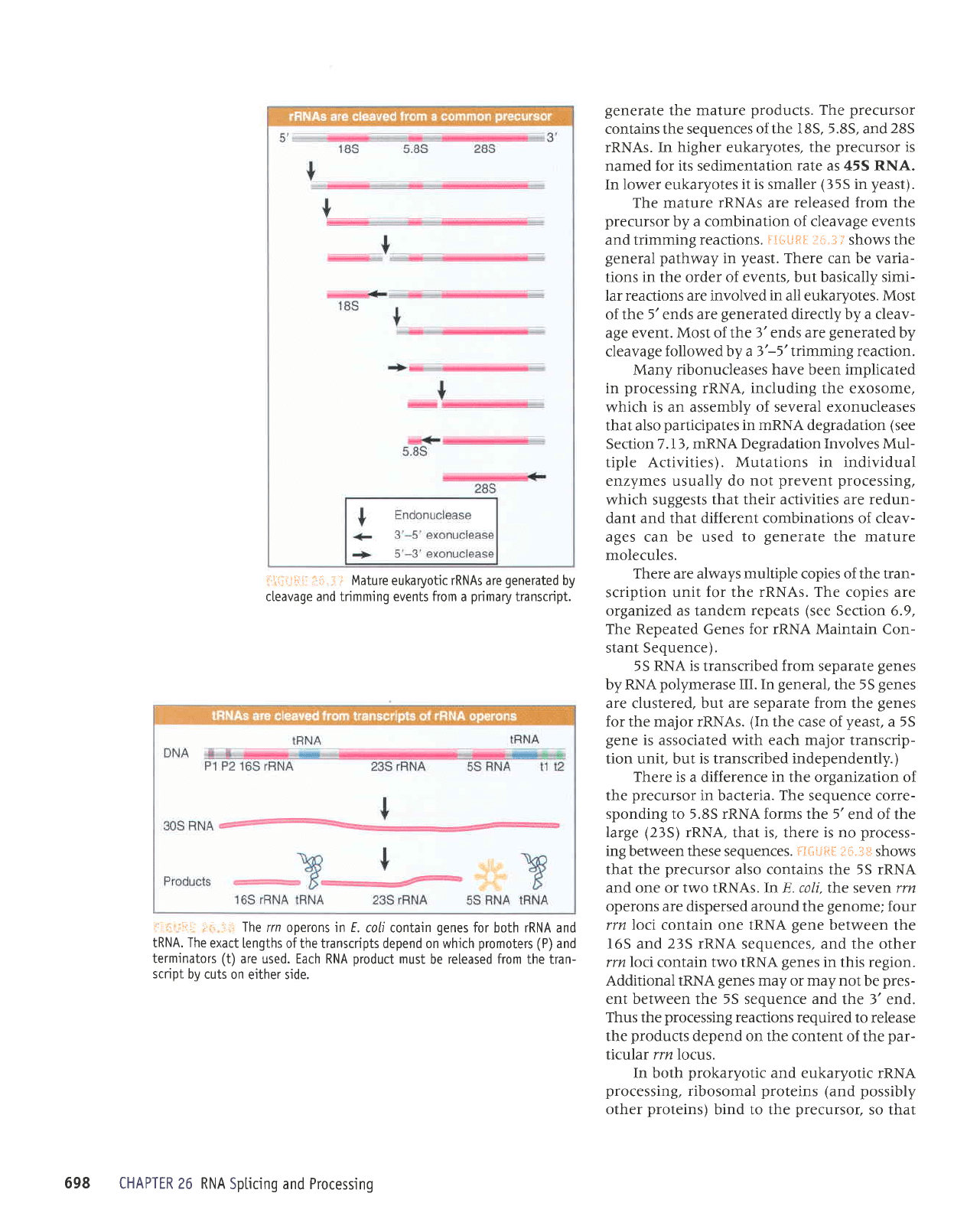
i
:
i:
i
i
i.:
i: .t'
i-::.
"i
.;' Mature eu karyotic rRNAs a re
generated
by
cleavage and trimming events from
a
primary
transcript.
:
i,.i;:;ri
.::,
:::.
The
rn operons in E.
coli contain
genes
for both rRNA and
tRNA. The
exact [engths
ofthe transcripts depend on which
promoters
(P)
and
terminators
(t)
are used. Each RNA
product
must
be released from the tran-
script
by cuts
on either side.
generate
the mature
products.
The
precursor
contains the sequences
of the 185, 5.8S, and
28S
rRNAs. In higher eukaryotes,
the
precursor
is
named for its sedimentation
rate
as 45S RNA.
In lower eukaryotes
it is smaller
(35S
in
yeast).
The mature
rRNAs
are
released from the
precursor
by a combination of cleavage events
and trimming
reactions.
$ifi#ftEj ili!.:T
shows the
general
pathway
in
yeast.
There
can be varia-
tions
in
the order
of events, but basically simi-
lar reactions are involved
in
all eukaryotes.
Most
of the 5'ends are
generated
directly by a cleav-
age event. Most of the
3'ends are
generated
by
cleavage
followed by a 3'-5' trimming reaction.
Many
ribonucleases have been implicated
in
processing
rRNA, including the exosome,
which
is
an assembly
of several exonucleases
that also
participates
in nRNA degradation
(see
Section 7.13, mRNA
Degradation Involves Mul-
tiple Activities). Mutations in individual
enzymes usually do
not
prevent
processing,
which suggests
that their activities
are
redun-
dant
and that different combinations of cleav-
ages can be used
to
generate
the mature
molecules.
There are always multiple copies of the tran-
scription unit
for the rRNAs. The copies are
organized as tandem
repeats
(see
Section 6.9,
The Repeated
Genes
for rRNA Maintain
Con-
stant Sequence).
5S RNA
is
transcribed
from
separate
genes
by RNA
polymerase
III.
In
general,
the 5S
genes
are clustered, but
are separate from
the
genes
for the major rRNAs.
(In
the case of
yeast,
a 5S
gene
is associated with each major transcrip-
tion unit, but
is
transcribed
independently.)
There is a difference
in
the organization of
the
precursor
in bacteria.
The
sequence corre-
sponding to 5.8S rRNA forms the 5'end
of the
Iarge
(23S)
rRNA,
that
is,
there
is
no
process-
ing between these sequences.
Fli;ilrt*
tti-i3*
shows
that the
precursor
also contains the 5S rRNA
and one or two tRNAs. In E. coli, the seven rrn
operons
are dispersed around
the
genome;
four
rrn Loci contain one tRNA
gene
between
the
165 and 23S rRNA sequences, and the
other
rrnloci contain two IRNA
genes
in this region.
Additional IRNA
genes
may or may not
be
pres-
ent between the 55 sequence and
the 3' end.
Thus the
processing
reactions required
to release
the
products
depend on the content
of the
par-
ticular rrnlocus.
In both
prokaryotic
and eukaryotic
rRNA
processing,
ribosomal
proteins (and
possibly
other
proteins)
bind to the
precursor,
so that
698
CHAPTER 26 RNA
SpLicing and Processing
669
6uLssa:or6
VNUI
lo;
parrnbag
arv SVNU
\ews
ZZ'gZ
are z(aq;
'syNuous
0Z-
p
dnor8
VIV/H
rr{1
sarrnbar
yNUJ
ul
uorleruJoJ
eurprJnopnesd
'reBns
aql ol
paurolar
sr
E)
pue
'pJleloJ
sr Jspq Jq1
'ue>l
-oJq
sr Jsoqu
ol
prJe
11.{.prrn uroJJ
puoq
IN
aql
I{)lqM
uI
",.;1."!;,r1
:jS*:i}I.:J
uI
uMoqs uoIlJeJJ aql
sallolur
JurprrnopnJsd;o
srsaqluzis aq1's1trNUJ
Jlerqapel
ul
00I-
pue
sVNUI
lsea,{
ur sJnprsJr
lt,
gy
arc
JJJqJ
'Jurprrnopnasd;o
srsaqluLs aqt
ur
pJAIoAur
sr syNgous
;o
dno:3
Jeqtouv
'dlrnrtre
aselLqtaru
aqt
Jo
upd
saprnord;1aslr
VNuous
eqt
teqt
sl [r{lqlssod
Juo
lprzuatf,preqJ
uaeq
tou
seq
(s)ase1.{qtrur
JqJ
'rpJ
os
pezrJJl
-JpreqJ
uJJq
JAer{ sVNUous
0t-
lVNUous
tua
'rebns
aq1
01 anrlelal aseq aq1 6ur
-lelol
pup
puoq
le6ns-g3
e
qlm
puoq
te6ns-11r1
aq1 6uLreldet
[q auLpunopnesd
o1
pelenuo]
sr eutpufl
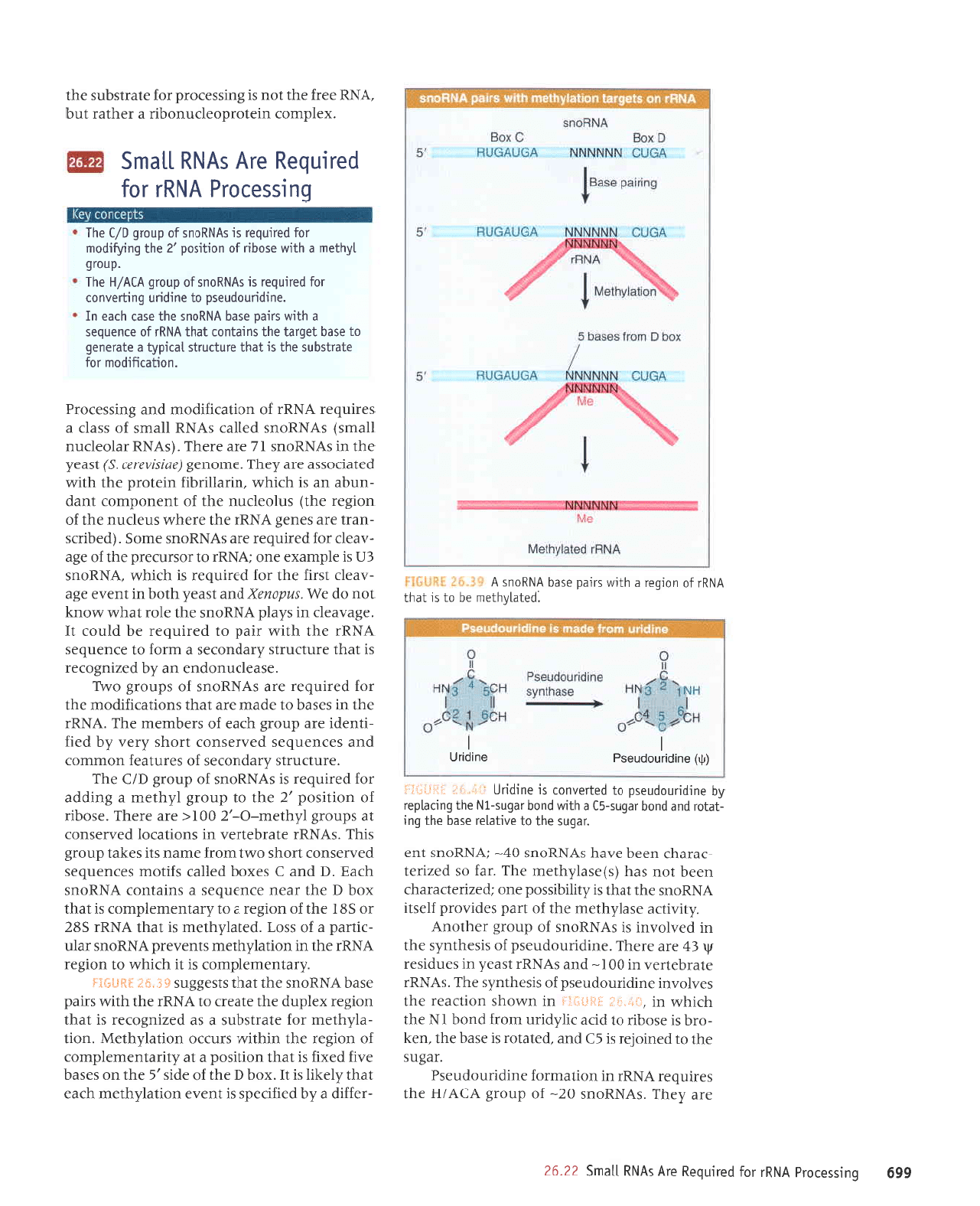
I,r,:.,,!i.
:jijitiii:i
'palelAqlau
aq ol sr
leql
VNUr
lo
uorbat e
q1L,r,r
stLed
aseq
VNUous
V
-reJJIp
e,{q
par;oads
sr
tuelJ
uorleldqlaur
qrea
teqt
llr>lll sl
U'xoq
( rqt
lo
rpls,E rqt
uo sespq
eArJ
pJXrJ
sr
lerl1
uorlrsod e
le
^,i.lrreluauralduror
yo
uo6ar aql ulqrlm sJn))o uoqe1.{qlayrg
'uo1l
-elLqraru
JoJ JleJtsqns e se
pezruSorar
sr
leql
uorSar
xaldnp
aql Jtearf, ot
VNUr
aqt
qtpr,t
srred
aseq
VNUous
aql
teql
stsaSSns
#s-',+i s*fi*Ijj
'.{reluarualdruoJ
sr
1l
q)rqM
o1 uor8ar
VNUJ
Jqt ul uorlel.dqtaru stua,rard
y5lUous
Jpln
-rrued
e
Jo
sso'I
'pate1,{.qtaru
sr
1eql
vNUr SgZ
ro
58I
aqt;o uo€ar e ot dretuarualdruor sr
teql
xoq
11
aql Jeeu eJuanbas e sureluof,
VNUous
qleg
'11
pue
)
sJxoq
pJIIeJ
sJrtou saruanbas
pe^JasuoJ
uoqs
o^\l ruoJJ erueu slr sa>lel dnorS
srqJ
'svNuJ
eleJqaue^ ur suorlPJol
pe^JasuoJ
te
sdnor8
ldqraru-O-,z
00I<
rrp rrrqJ
'rsoqrr
;o
uorlrsod
,Z
erft ot dnorS
ldqiaru
e Surppe
ro1
parrnbar
sr svNgous
;o
dnor8
11/f,
eql
'eJnlJnJls
.drepuoras
Jo
sJJnlpeJ uoururoJ
pue
seJuenbas
paarasuoJ
uoqs
dran ,,(q
par;
-lluapr
are dnor8
qJeJ
Jo
sJJqruJru aqJ
'VNUJ
eql ul sasPq 01 Jppru aJP
leql
suoqe)rJrpou eql
roy
parrnbar
JJe syNgous
yo
sdnor8 o.t41
'JseJlJnuopua
ue
,{q
pazruSorar
sr
lpql
JJnlJnrls ^drepuoras e ruJoJ o1 aruanbas
y5{r
ar{l
qlprzr
rred ol
parrnbar
rq
plnoJ
1I
'a8e.Leap
ur s,{.e1d
vNgous
eql alor
leqm
.tt.ou>I
lou
op a7,4'sndouay
pue
lsear(
qtoq
ur
1ua,ra
a8e
-ApelJ
tsJrJ
Jql ro;
parrnba;
sr
qJrqm
'VNUous
gn
sr aldruexe Juo
lyNgr
o1 rosrnrard aqt
1o
a8e
-^eell
Jo]
parrnbar
are
svN5ous JruoS
'(paqrns
-ueJt
JJe
saua8
yggr
Jql JJeqM snJlrnu Jr{1
Jo
uor8ar aqt) snloalrnu aql
;o
tuauodruo) tuep
-unqe
ue sr
qlrqm
'urJpllrrqrJ
ulalord Jql
qlrM
pJlerrossp
are Laql'aruoua8
(aatsutant
S)
rsead
Jqt ul sVNUous
I/
rre JrJqJ'(sVNU rploJpnu
11erus)
sVNUous
palle)
sVNU
Ilprus Jo
sselr e
sarrnbar
VNUJ
Jo
uorleJrJrpou
pue
Surssaro;4
'uorlPllJr.pour
l0J
olpilsqns aql sr
lpql
elnllnrls
lerrdr\1
e alerauab
o1 aseq
1able1
oql sureluol
leql
VNUr;o
aruenbas
P
qlrM
srreo aseq
vNuous
aql asPl
qrea
uI
'aurpunopnasd
ol aurpun 6uqlanuor
ro;
perrnbar
sr sVNUous;o dnorb
VIV/H
aql
'dnorb
1Aq1eu
e
qlrm
asoqu
lo
uoqLsod
,7
aq1 6urAlrpour
rol
parLnbar
sr sVNUous;o dnorb
Ah
aqL
parrnbau
arv
svNu
lleus
'xaldruor
uralordoalrnuoqrr
e rJr{tpJ
lnq
'VNU
JaJJ aql
tou
sr Surssarord JoJ eteJlsqns aql
(rf)
eurpunopnes6
outpun
6urssarord
VNUr
roJ
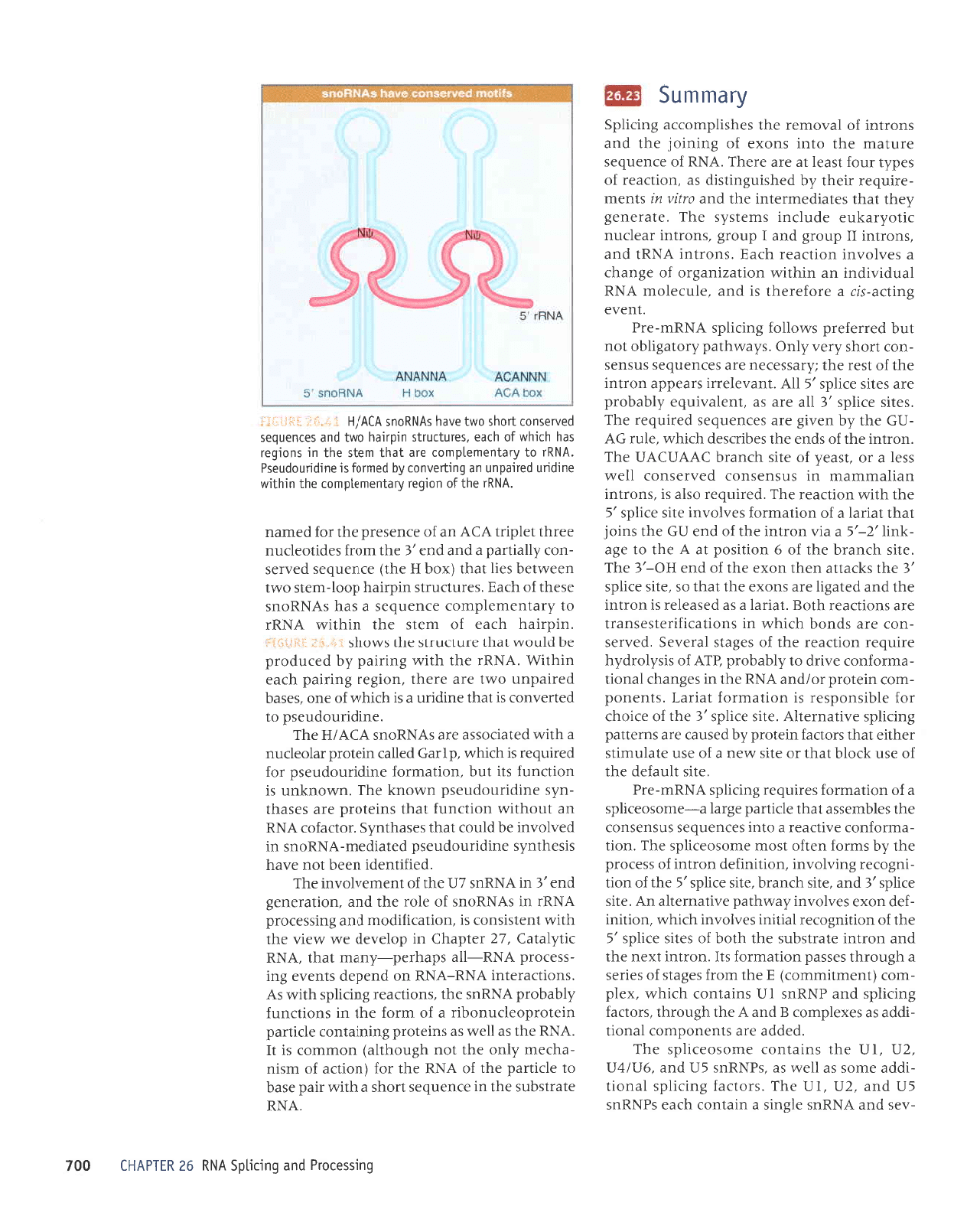
6urssaro:6
pue
6uL:qd5
VNU
9Z
Ull_dVHl
'VNU
Jterlsqns eql ur e)uJnbas
goqs
p
qlIM
rted aseq
o1 aprtred eqt
Jo
VNU
eqt JoJ
(uotpe
Jo
ruslu
-pqJeu
,{1uo aqr
1ou
q8noqrle) uoruuo)
sl
U
'yNd
Jq1 sE
IIaM
sB
sutatord SututBluor
aprped
urato.rdoalrnuoqrr
e
Jo
uJoJ
aql uI suoltJunl
z(lqeqord
VNUUs
Jqt
'suortJper
3uo11ds
qllM
sV
'suortJeJJtul
VNu-VNu
uo
puadap
stua.ta
8ut
-ssarord
VNU-1e
sdeqrad-Lueu
leql
'VNU
lr1L1ete3
'77
nldeqS
ur dola,rap
JM MaIA Jqt
qllM
luJlsrsuof,
sr
'uollpJlllpotu
pup
Sutssarord
vNur
ul svNuous
Jo
JIoJ aql
pue
'uorlerauaS
pua,€
ur
YNuus
4n
aql
Jo
luJrue^Io^ul
JqI
'pelJlluepl
ueeq
lou
e^eq
srsaqtuf,s JurprrnopnJsd
pateparu-VNUous
uI
pe^lonur
aq
plnoJ
teqt
srspqlu,{g
'rone;or
y51g
ue
tnoqtrM
uortJunJ
lpql
sulatord
are saseql
-u^ds
aurprrnopnasd u.trou>l
eqJ
'u,rtou>lun
sI
uorlf,unl sll
lnq
'uorlpruJoJ
autptrnopnesd
ro;
palnbar
sr
qJrqm
'd1re9
pJIIer
ulelord
reloapnu
P
qlr^^
pJlerJosse
JJe
svNuous
v)v/H
rqJ
'aurpunopnasd
ol
peua^uoJ
sl
lPql
auplrn P sI
q)Iq^\
Jo
auo
'seseq
parredun
o.trl JJp a:aqt
'uor8ar
Sutrted
qrea
ulqtl6
'VNUr
rql
qtlm
Surrted
.{q
parnpord
aq
plnoz\A
leql
eJnlf,nJls
JI{] sMoqs
l r'r'l:ir-.
::.llilil;Ii
'urdrreq
qJea
Jo
ruJts
aqt urqlrM
VNUJ
ol
.dretuarualduor aruanbas e spq
syN5ous
Jsaql
Jo
qJpg
'sJrnpnrls
utdrteq
dool-ruals o.ttl
ueJMleq sarl
teql
(xoq
g
Jr{l) eruJnbJs
paArJS
-uor
L11er1red e
pue puJ,€
Jqt ruoJJ sJpltoepnu
aarqt
taldut
V)V
up;o
aruasard eqt JoJ
pJrupu
'VNUr
eqllo uoLbat
fileluaura'1duor aq1 utqltm
euLpun
parLedun
ue 6uLlanuor
[q
peurol
sr aurpunopnasd
'VNUI
01
rtreluauelduol a.lP
lPql
uals
aql uL suotbal
seq
qlrqM
J0
qrPe
'selnlrnrls
urdrreq oMl
pue
saluanDas
pa^iosuol
]Joqs
oMl
aneq sypgous
y3y/g
:
i;1j::.i
00/
-Aas
pup
VNUUs
a13urs
e uretuoJ
qJea
sdN6us
En
pue
'Zn'In
JqI
'srot)e1
Surrrlds
Ipuorl
-rppe
Juos se
IIaM
se
'sdNuus
En
puP
'gnlvn
'Zn'ln
Jql surpluoJ aurosoa:r1ds a,{I
'pJppP
arp sluauoduo)
Ieuorl
-rype
se saxaldruor
g
pup
V
aqt
q8norqt'srolJeJ
Suoqds
pue
dNUus
III
surptuo)
qrrq.,rn
'xa1d
-ruor
(tuarutlruruor)
A
Jqt ruoJJ sa8els
yo
sarras
e
q8norql
sassed uorteuroJ sll
'uoJlur
lxau
eql
pue
uoJtur eleJlsqns Jql qtoq
Jo
sJlrs allds
,g
Jql
Io
uorlruSorar
lerlrur
sJAIoAur
q)lqu
'uorlrur
-Jap
uoxJ sellonur Lernqled JlrlpurJllp uV
'alrs
arrlds,g
pue
'etrs
q)ueJq'a1rs
er11ds,E Jql
Io
uou
-ruSorar
Surnlo,rur'uorlrurJap uoJlur
;o
ssarord
aqt
r(q
suJoJ uJlJo
Jsoru
aurosoa)Tds aql
'uorl
-eurJoJuoJ
JATJJeJT
p
olul
sJJuJnbas snsuasuol
eqt sJIquJSSe
teqt
Jllrued a8rel e-auosoarrlds
p
Io
uorleruJo; salnbar
Suorlds
VNUrU-erd
'alrs
llneJrp
aql
JO
esn
>llolq
lPr{l
Jo Jlrs MJU
e
}o
asn alelnurls
JJqlre
tpqt
sJotJeJ uratord,{q
pasner
are suralted
Sunqds JlrtpurJtlv
'atrs
arrlds
,€
aqt
Io
alror{)
ro; alqrsuodsJr sr uorte[rJoJ
lerref
'sluauod
-ruor
uratord ro/pue
VNU
eql ur sa8ueqr
leuorl
-pruroJuo)
alrJp 01 ,{lqeqord
AJV
Jo
srs,,{1orp,{q
arrnbar uorlJeJJ Jql
Jo
sa8els
leranJS
'pJAJes
-uo)
eJe spuoq
q)lq1\/t
ur suorlpf,rJrJelsesueJl
eJe
suoll)eJI
qlog
'lelJel
e se
pJSealeJ
sI
uollul
aql
pue pale8rl
JJp
suoxJ Jql
teql
os
'a1rs
arrlds
/€
Jql s>lJe]le uaql uoxe rql
Jo
pur
Ho-,€
JqJ
'Jlls
qJuerq
Jqt
Jo
9
uorlrsod
te
V
Jqt ot a8e
-Iull
,z-,s
P ern uoJlur eql
Io
pue
n9
aqt suroI
lpqt lelJel
e
Jo
uorteruJo;
sJ^lo^ur etrs arrlds,g
eql
qllM
uorlJeeJ aq1
'parrnbal
osle sr
'suoJJur
uPrlPruruPru ur snsuJsuoJ
pJAJJSUOJ
IIJM
ssJI e ro
'tseaL;o
rlls
q)uerq
lyyn)Vn
eqJ
'uortur
Jql
Jo
spue 3q1 sJqu)sJp r{)lq1!1
'JInJ
gv
-nD
Jql ^,i.q uanr8 arB
saruanbas
pa;rnbar
aq;
'salts
arqds
,€
IIe
are se
'tuJlellnba
,{1qeqo'rd
JJe SJIrS arrldS
,5
IIv
'lue^JlJrrr
Sreadde uorlur
erll
Jo
lseJ
aql
1^dressarau
aJe saJuenbas snsuas
-
uo)
uoqs,{ran
.d1u
g'
s^de.,vrqted
.drote8rlqo
1ou
tnq
perreJerd
s,no11o;
Surtrlds
yN5ru-ard
'lUaAe
3urlre-srr e aJoJeJaql
sr
pue
'elnJaloru
VNU
Ienprnrpur
up urqlrM
uortezrue8ro
;o
aSueqr
P saAIoAUr
UOrlJeeJ
qJBg
'SUOJlur
VNUI
pue
'suorlur
1
dnor8
pue
1
dno;8
'suortur
JpJIJnu
rrloLrelna JpnlJur
srualsLs aq1
'aleraua8
.{.aqr
reqr
sJlplpeuJelur
Jql
pue
lJilt Lt7 sluaru
-arrnbar
laql [q paqsrnSurlsrp
sp
'uortf,eeJ
Jo
sadLl
rnoy
lseal te
are
JraqI
'VNU
Io
aruanbas
eJnleru eqt otur
suoxa
yo
Sururof Jqt
pup
suoJlul
Jo
leloruer
aqt saqsrldruo>e Suorldg
Areu
unS
