Lewin Benjamin (ed.) Genes IX
Подождите немного. Документ загружается.

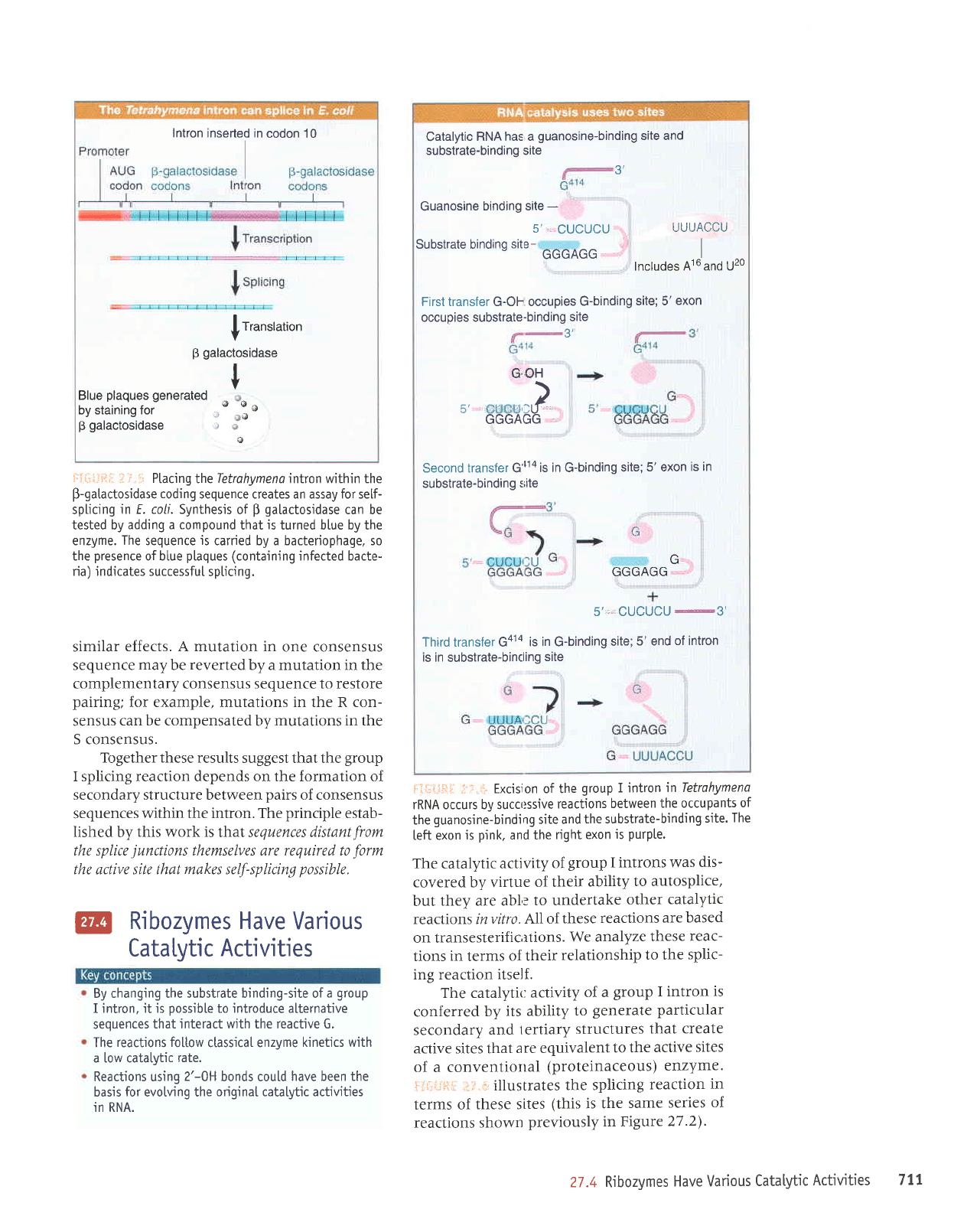
Intron
inserted in
codon 10
I
I
Translation
B
galactosidase
I
V
Blue
plaques
generated
by staining for-
,'
:
*
B
galactosidase o
@
Catalytic
RNA has a
guanosine-binding
site and
substrate-binding site
Guanosine
binding site
-
3',
First transfer G-OF occupies
G-binding
site; 5'
exon
occupies substrate-binding
site
5'
,'
CUCUCU
Substrate bindino
site
-
uuuAuu
q.oH
\
s', eilffiJr/
GGGAGG
UUUACCU
I
lncludes
416 un6
g2o
::
i;.il : il i;
it
.r
.
r;
Ptaci ng
the
Tetrahy
m e na intron within the
B-gatactosidase
coding
sequence creates an assay for se[f-
spticing
in E. coLi. Synthesis
of
B
galactosidase
can be
tested by
adding a compound
that
is
turned blue by the
enzyme.
The
sequence
js
carried by a bacteriophage, so
the
presence
of blue
plaques (containing
infected bacte-
ria) indicates successfuI
splicing.
similar
effects. A mutation in
one consensus
sequence
may
be
reverted
by a mutation in the
complementary consensus sequence to restore
pairing;
for example, mutations in the R con-
sensus
can be compensated
by
mutations in the
S consensus.
Together these results
suggest that the
group
I splicing reaction depends on the formation of
secondary structure between
pairs
of consensus
sequences
within the intron. The
principle
estab-
lished
by
this work is
that sequences distant
from
the splice
junctions
themselves are required to
form
the active site that makes self-splicing
possible.
@
Ribozymes
Have
Various
Catalytic
Activities
By changing the substrate binding-site of
a
group
I
intron. it is
possible
to introduce alternative
sequences
that
jnteract
with the reactive G.
The
reactions foltow
ctassicaI enzyme
kinetics with
a low catalytic
rate.
Reactions using
2'-0H
bonds coutd
have been the
basis
for
evotving the
original catalytic activitjes
in RNA.
Second transfer G't1a
is in G-binding
site;
5' exon
is in
substrate-binding
s;ite
G
/:
GGGAGG
5"..,cucucu
@3'
Third transfer Gala
is in G-binding
site;
5' end
of intron
is
in substrate-bincling
site
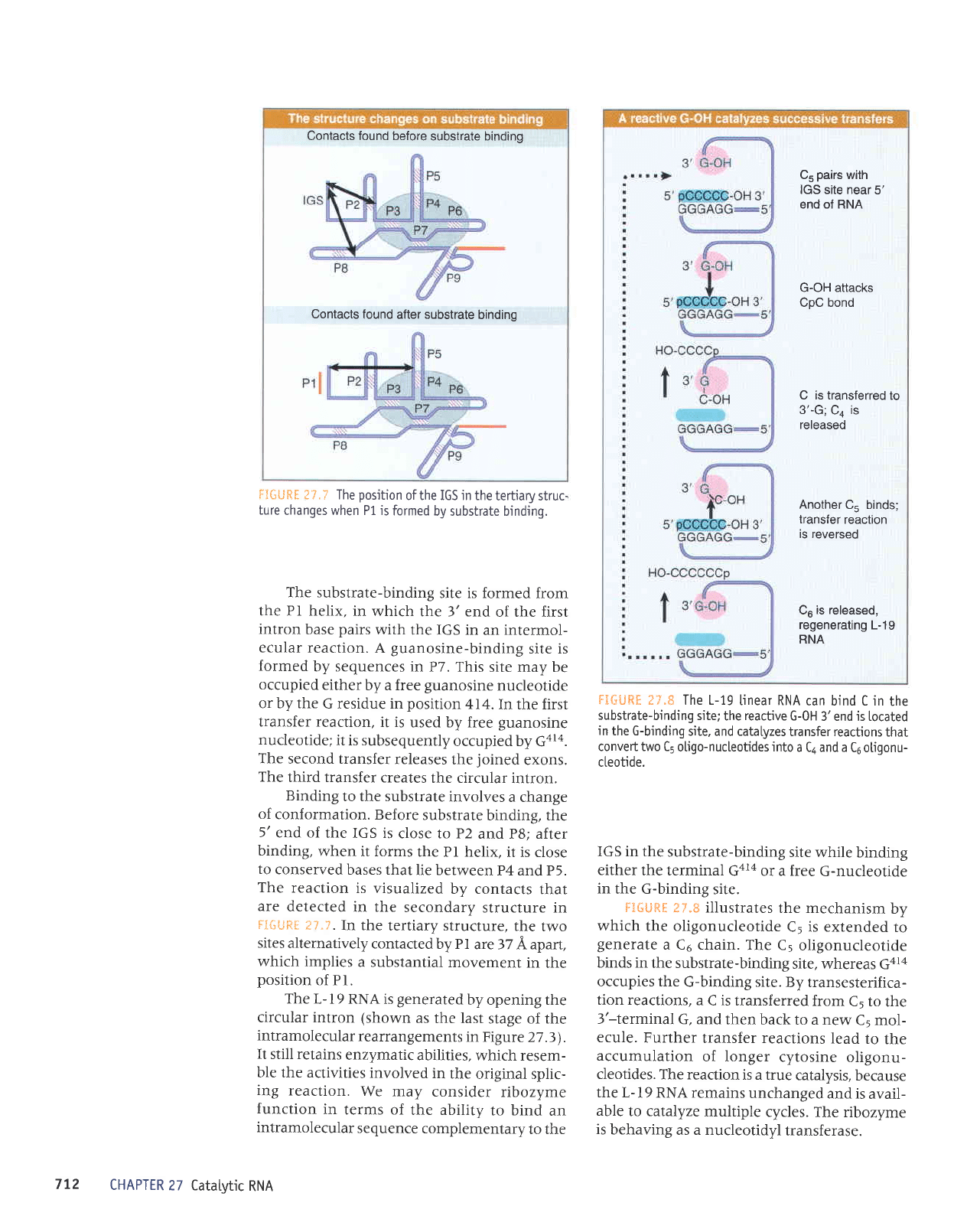
lil{:i.jltt i:,r',ir.
Excision of the
group
I intron
in Tetrahymena
rRNA occurs by successive
reactions
between
the occupants
of
the
guanosine-binding
site
and
the substrate-binding
site.
The
[eft exon
is
pink,
and the
right
exon
is
purpte.
The catalytic
activity
of
group I introns was
dis-
covered by virtue
of their
ability
to autosplice,
but
they are abl,:
to undertake
other
catalytic
reactions invitro.
All of
these
reactions
are based
on transesterific;rtions.
We analyze
these
reac-
tions
in
terms
of their
relationship
to the
splic-
ing reaction
itself.
The catalytic
activity
of a
group I intron
is
conferred
by its
ability
to
generate
particular
secondary
and lertiary
structures
that
create
active sites that
are equivalent
to the
active
sites
of a conventional
(proteinaceous)
enzyme.
l:;i;iji:l{:
l:-i.,:-
illustrates
the splicing
reaction
in
terms of
these sites
(this
is the
same
series
of
reactions
shown
previously
in Figue
27
.2).
u
27.4 Ribozymes
Have Various
Catatytic
Activities
7lt
Contacts found before
substrate bindino
Contacts
tound after substrate
bindino
F:GUftt 27.? The
posjtion
ofthe IGS in the
tertiary struc-
ture
changes when P1 is
formed
by substrate binding.
The substrate-binding
site is formed
from
the PI helix,
in which
the J' end
of the first
intron
base
pairs
with
the IGS in
an
intermol-
ecular reaction.
A
guanosine-binding
site is
formed
by sequences
in P7. This
site may be
occupied
either
by a free
guanosine
nucleotide
or by the
G residue in
position
4l4.In
the first
transfer reaction,
it is
used by free
guanosine
nucleotide;
it is subsequently
occupied by
(+t+.
The
second transfer
releases
the
joined
exons.
The
third transfer
creates
the circular
intron.
Binding
to the substrate
involves
a change
of
conformation.
Before
substrate
binding, the
5'
end of
the IGS is
close to P2
and P8; after
binding,
when
it forms
the Pl helix,
it is close
to
conserved
bases that
lie between
P4 and P5.
The
reaction
is visualized
by contacts
that
are
detected
in the
secondary
structure in
FiSlJftg
t?.7.
In the
tertiary structure,
the
two
sites
alternatively
conracted
by Pl
are 37 A apart,
which implies
a substantial
movement
in
the
position
of Pl.
The
L- l9 RNA
is
generated
by opening the
circular intron
(shown
as
the last
stage of the
intramolecular
rearrangements
in Figure
2 7.
3
)
.
lt still retains
enzymatic
abilities,
which resem-
ble the activities
involved
in
the
original splic-
ing
reaction.
We may
consider
ribozyme
lunction
in
terms of
the ability
to bind
an
intramolecular
sequence
complementary
to the
CHAPTER
27 Catalvtic
RNA
Cs
pairs
with
IGS site near
5'
end of RNA
G-OH attacks
CpC bond
C is transferred
to
3'-G;
Ca
is
released
Another
C, binds;
transfer
reaction
is reversed
C5
is released,
regenerating
L-19
RNA
FtGtlSt
??.& The L-19
linear RNA
can bind C in
tne
substrate-binding
site;
the
reactive
G-0H
3'end
js
located
in
the G-binding
site, and catalyzes
transfer reactions
that
conved two C5
otigo-nucleotides into
a Ca and a
C6 otigonu-
cteotide.
IGS in the
substrate-binding
site while
binding
either
the terminal
G414 or a free
G-nucleotide
in
the G-binding
site.
FI6U*f
?7.8 illustrates
the mechanism
by
which
the oligonucleotide
C5 is extended
to
generate
a C6 chain.
The C5
oligonucleotide
binds in
the substrate-binding
site, whereas
Gala
occupies the
G-binding site.
By transesterifica-
tion reactions,
a
C
is
transferred
from
C5 to
the
3'-terminal
G, and
then back
to a new
C5 mol-
ecule.
Further
transfer reactions
lead
to
the
accumulation
of longer
cytosine
oligonu-
cleotides.
The reaction
is a true
catalysis,
because
the
L- l9 RNA remains
unchanged
and is
avail-
able
to catalyze
multiple
cycles.
The ribozyme
is
behaving
as a nucleotidyl
transferase.
772
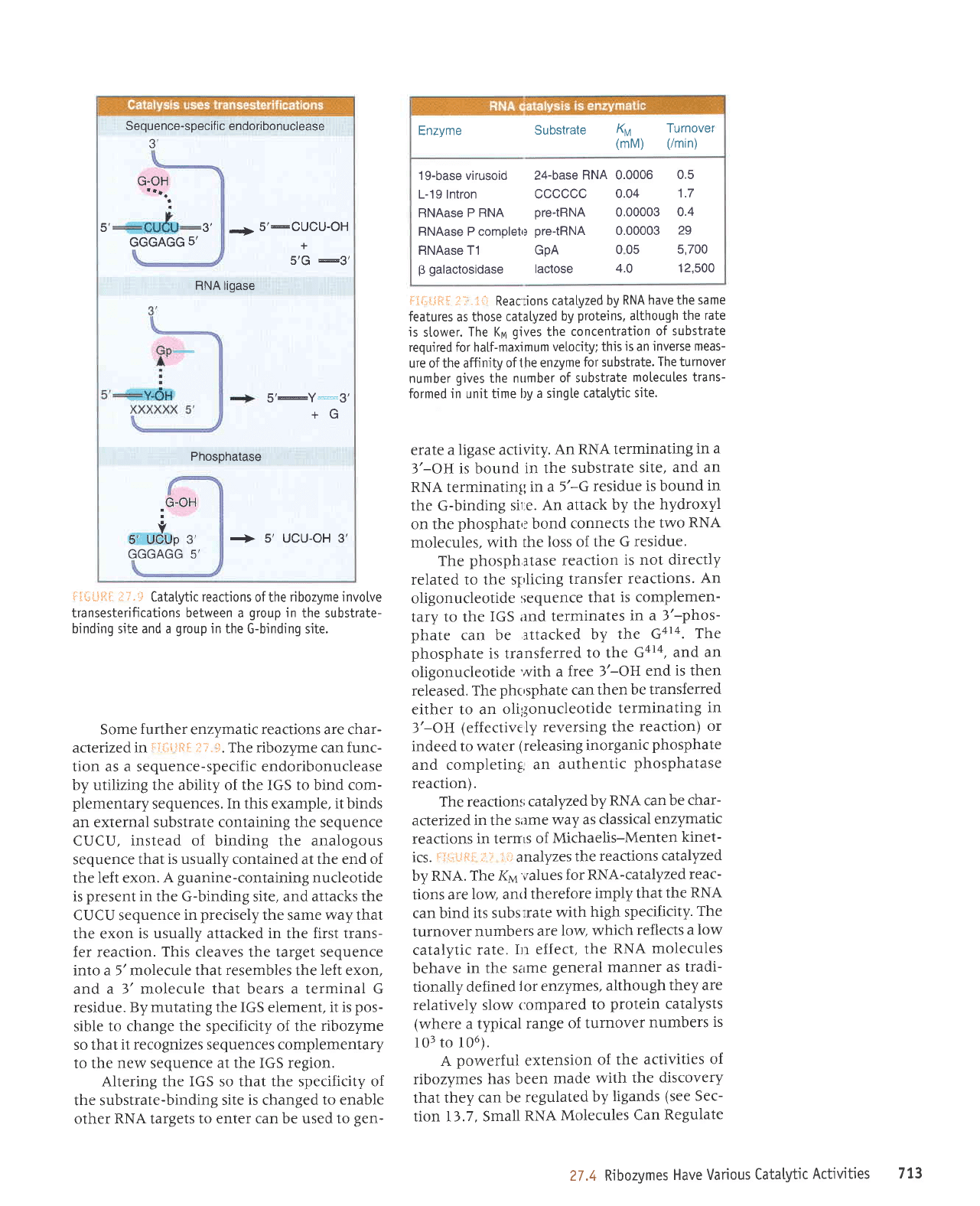
Sequence-specific endoribonuclease
3',
G-OH
5' 3',
-.;5'*QgCU-OH
5'G
@3'
GGGAGG
5'
RNA ligase
5'@Y*"3'
+G
Phosphatase
\r-un
v
5' UCUp
3'
uuuAuu 3
+ s', UCU-OH 3',
il,:+LlitL
:i1".:.1
Catatytic reactions
of the
ribozyme involve
transesterifications
between
a
group
in
the substrate-
binding site and a
group
in the
G-binding site.
Some
further
enzymatic reactions are char-
acterized in
:rTi:,qiiti
-::,!.t.
The ribozyme can func-
tion as a sequence-specific endoribonuclease
by utilizing the ability of the IGS to bind com-
plementary
sequences.
In this
example,
it binds
an external substrate containing the sequence
CUCU, instead of binding the analogous
sequence
that is usually contained
at
the end of
the left exon. A
guanine-containing
nucleotide
is
present
in
the G-binding site, and attacks
the
CUCU sequence
in
precisely
the
same
way that
the exon
is usually attacked in the first trans-
fer reaction.
This
cleaves the target sequence
into a 5'molecule that
resembles
the
left
exon,
and a l' molecule that bears a terminal G
residue.
By mutating the IGS element, it is
pos-
sible
to change the specificity of the
ribozyme
so that
it recognizes sequences complementary
to the
new
sequence
at the IGS region.
Altering
the IGS so that the specificity of
the substrate-binding
site
is
changed
to enable
other RNA targets
to enter can be used to
gen-
19-base virusoid
L-19 lntron
RNAase
P RNA
RNAase P completr:
RNAase T1
B
galactosidase
24-base
RNA 0.0006
0.5
cccccc
0.04
1.7
pre-tRNA
0.00003
0.4
pre{RNA
0.00003
29
GpA
0 05
5,700
lactose
4.0 12,500
+rISLiiqil
,ill:.1i]
Reac.:ions
catatyzed
by
RNA have the
same
features as those
catatyzed
by
proteins,
although
the rate
is
stower.
The KM
gives
the
concentration
of substrate
required for half-maximum
vetocity;
this
is
an
inverse meas-
ure of the affinity
of t he enzyme
for substrate.
The turnover
number
gives
the number
of substrate
molecules trans-
formed in unit time lly
a sing[e catatytic
site.
erate
a ligase activity.
An
RNA terminating
in a
l'-OH is bound
in the
substrate
site,
and an
RNA terminating
in a 5'-G
residue
is bound
in
the G-binding
sit.e.
An attack
by the
hydroxyl
on the
phosphat,:
bond
connects
the
two RNA
molecules, with
the
loss of the
G residue.
The
phosphltase reaction
is not directly
related to the str'licing
transfer
reactions.
An
oligonucleotide
sequence
that
is complemen-
tary to the
IGS and
terminates
in
a 3'-phos-
phate
can be
lttacked
by
the G4r4.
The
phosphate
is transferred
to the
Gaia, and
an
oligonucleotide
',,vith
a free
3'-OH
end
is
then
released. The
phc,sphate can then
be transferred
either to an
oli;lonucleotide
terminating
in
3'-OH
(effective
ly
reversing
the
reaction)
or
indeed to water
(releasing
inorganic
phosphate
and completing,
an
authentic
phosphatase
reaction).
The
reactions catalyzed
by
RNA can be
char-
acterized in the
sirme
way as
classical
enzymatic
reactions in terrrLs
of
Michaelis-Menten
kinet-
ics.
trli.;Lil'dt: ;f
,:
i
ii
analyzes
the
reactions
catalyzed
by RNA. The
I(y'ralues
for
RNA-catalyzed
reac-
tions are
low, anrl
therefore
imply that
the
RNA
can bind
its subs:rate
with
high specificity.
The
turnover
numbers
are
low, which
reflects a
low
catalytic
rate.
Lt effect,
the
RNA
molecules
behave
in the s,rme
general manner
as tradi-
tionally defined
Ior enzymes,
although
they are
relatively slow
<:ompared
to
protein
catalysts
(where
a
typical
range
of turnover
numbers
is
103 ro 106).
A
powerful
extension
of the
activities
of
ribozymes
has been
made
with
the
discovery
that
they can be
regulated
by
ligands
(see
Sec-
tion 13.7, Small
RNA
Molecules
Can
Regulate
Enzyme
Substrate
Ky
Turnover
(mM)
(/min)
27.4 Ribozymes
Have Various
Catalytic
Activities
773
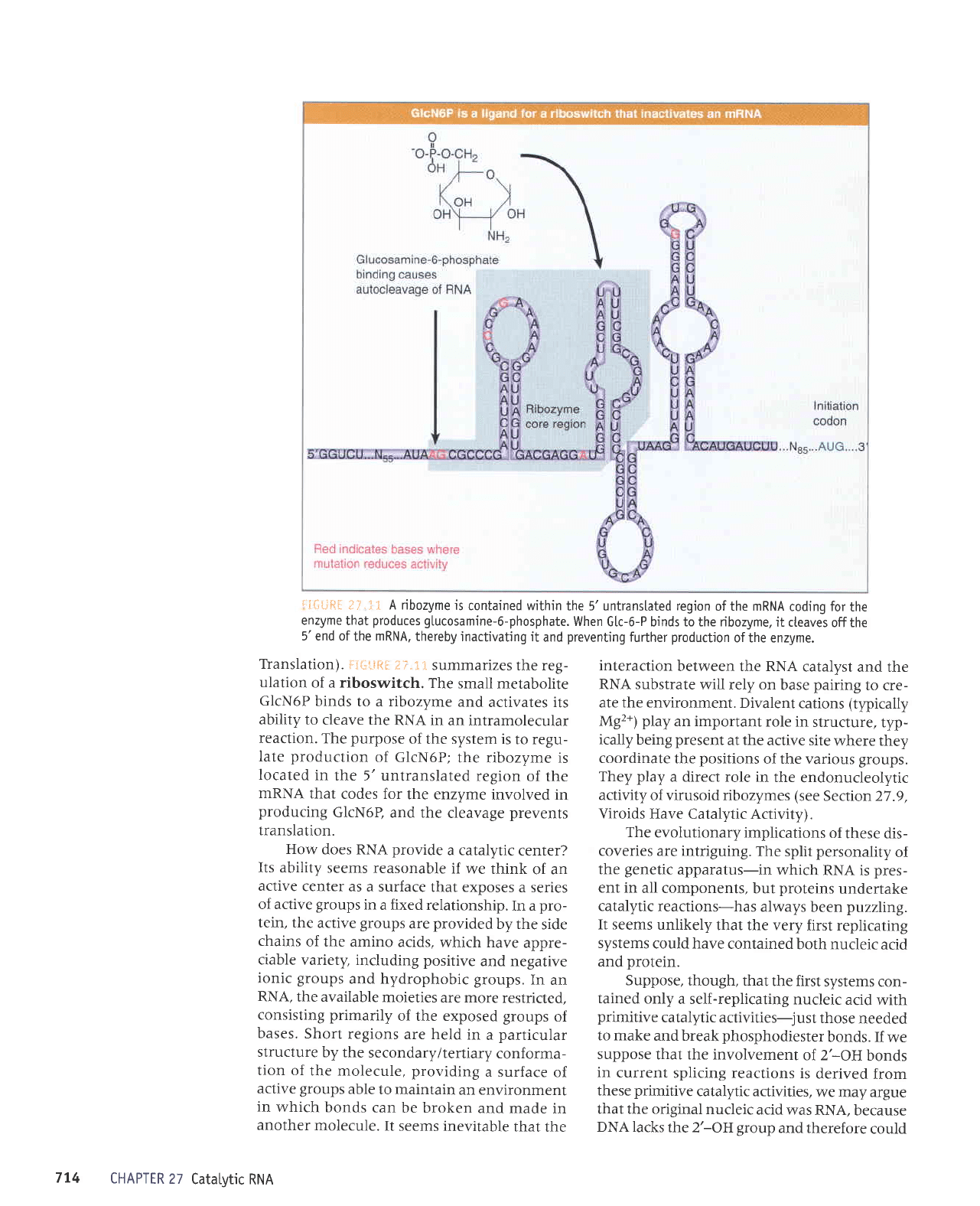
::iti;&i
i:..1
t
A ribozyme
is
contained within
the 5'untranstated region
of the mRNA
coding for
the
enzyme
that
produces
gtucosamine-6-phosphate.
When
G[c-6-P binds to the ribozyme,
it cteaves
offthe
5'end ofthe mRNA,
thereby
inactivating it
and
preventing
further
production
ofthe enzyme.
Translation).
ij'L:ij$ii
i:,::
summarizes
the reg-
ulation
of a riboswitch.
The
small
metabolite
GlcN6P
binds
to a ribozyme
and
activates its
ability to
cleave the
RNA in
an intramolecular
reaction.
The
purpose
of the system is
to regu-
late
production
of
GlcN6P;
the ribozyme is
located
in
the 5' untranslated
region
of the
nRNA
that
codes for
the enzyme
involved in
producing
GlcN6P,
and the
cleavage
prevents
translation.
How
does RNA
provide
a catalytic
center?
Its ability
seems
reasonable
if
we think
of an
active
center
as a surface
that exposes
a series
of active
groups
in
a fixed relationship.
In a
pro-
tein, the
active
groups
are
provided
by the
side
chains
of the amino
acids,
which have
appre-
ciable
variety, including positive
and negative
ionic groups
and hydrophobic groups.
In
an
RNA,
the available
moieties
are more
restricted,
consisting primarily
of the exposed groups
of
bases.
Short
regions
are held
in
a
particular
**.1.'dffitril*,':1"i{ii;:rrur
in
which
bonds
can
be broken
and made
in
another
molecule.
It seems
inevitable
that
the
CHAPTER
27
CataLytic
RNA
interaction
between the RNA
catalyst
and the
RNA substrate
will rely on
base
pairing
to cre-
ate the environment.
Divalent
cations
(typically
Mg2+)
play
an important
role in
structure,
typ-
ically
being
present
at the
active site
where they
coordinate
the
positions
of the various groups.
They
play
a direct role in
the
endonucleolytic
activity
of virusoid ribozymes
(see
Section 27.9,
Viroids Have
Catalytic Activity).
The
evolutionary
implications
of
these dis-
coveries are intriguing.
The
split
personality
of
the
genetic
apparatus-in
which
RNA is
pres-
ent in
all components.
but
proteins
undertake
catalytic reactions-has
always
been
puzzling.
It seems
unlikely
that the very
first replicating
systems
could have
contained
both nucleic
acid
and
protein.
Suppose,
though, that
the first
systems
con-
tained
only
a self-replicating
nucleic
acid
with
primitive
catalytic activities-just
those
needed
to make
and break
phosphodiesrer
bonds.
If we
suppose
that the involvement
of.2'-OH
bonds
in
current
splicing reactions
is
derived
from
these
primitive
catalytic activities,
we may argue
that the original
nucleic
acid
was RNA,
because
DNA lacks
the
2'-OH
group
and therefore
could
714
not undertake such reactions.
Proteins
could
have
been added for their ability
to stabilize the
RNA
structure.
The
greater
versatility
of
pro-
teins then could have
allowed them to take
over
catalytic reactions, leading
eventually
to the
complex and sophisticated apparatus
of modern
gene
expression.
Some Group I Introns
Code
for Endonucleases
That
Sponsor
MobiLity
o
Mobite
introns
are able to insert themselves into
new
sites.
.
Mobile
group
I introns code for
an endonuctease
that makes a doubte-strand break
at a target site.
o
The intron
transposes
into
the site of the doubte-
strand break by a
DNA-mediated
replicative
mechanism.
Certain introns of both the
group
I
and
group
II
classes contain open reading frames that are
translated into
proteins.
Expression
of the
pro-
teins allows the
intron
(either
in its original
DNA form or as a DNA copy
of the
RNA)
to be
mobile:It is able to insert itself
into a new
genomic
site.
Introns
of both
groups
I and II are
extremely widespread,
being
found in
both
prokaryotes
and eukaryotes. Group I in-
trons migrate by DNA-mediated mechanisms,
whereas
group
II introns migrate
by
RNA-medi-
ated
mechanisms.
Intron mobility was first detected by crosses
in which the alleles
for
the relevant
gene
differ
with
regard to their
possession
of the
intron.
Polymorphisms
for
the
presence
or absence of
introns are common in fungal mitochondria.
This is
consistent with the
view
that these
introns originated by
insertion
into the
gene.
Some
light on the
process
that
could be
involved
is cast by an analysis of
recombination in
crosses
involving the
large
rRNA
gene
of the
yeast
mitochondrion.
This
gene
has a
group
I intron
that
contains
a coding sequence.
The intron is
present
in
some
strains
of
yeast
(called
ol+) but absent in others
(trl-).
Genetic crosses between ro+ and
al- are
polar:The progeny
are usually <rl+.
If we think of the cl+ strain as a donor and
the ol- strain
as a recipient, we form the
view
that
in rrl+
x
o- crosses a new copy of the intron
is
generated
in the rrl-
genome.
As a result, the
progeny
are
all rrr+.
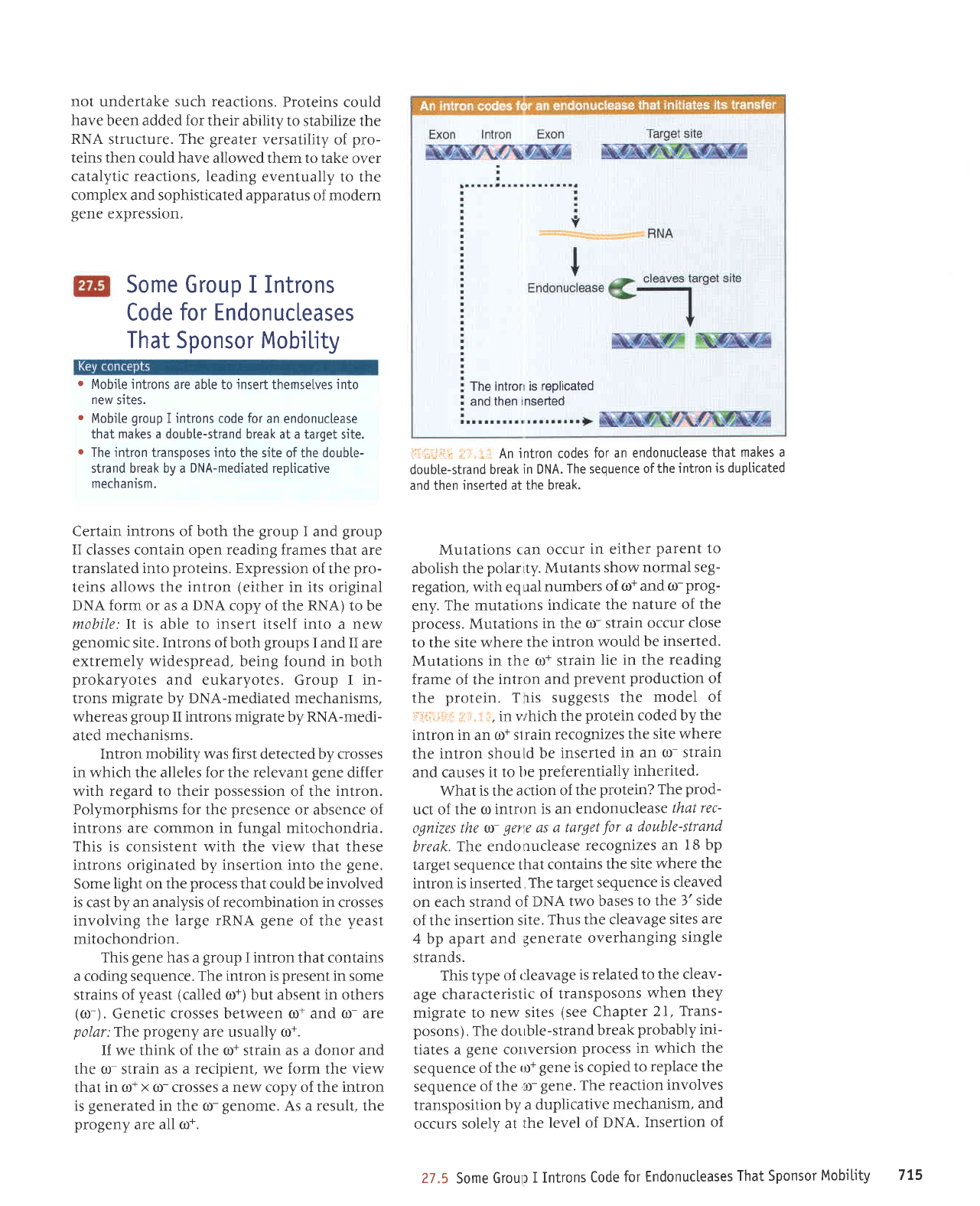
1111i;i:ir li
.;'.:f An
intron codes
for an endonuctease
that
makes a
double-strand
break
in DNA.
The sequence ofthe
intron
is duplicated
and then inserted at
the break.
Mutations
can occur
in either
parent
to
abolish the
polarLty. Mutants
show
normal seg-
regation, with eq
ual numbers
of
o+ and o-
prog-
eny.
The mutations
indicate
the
nature of the
process.
Mutations
in the
ro- strain occur
close
to the site where
the
intron would
be
inserted.
Mutations in the
ot+ strain
lie
in the reading
frame of the
intron and
prevent
production
of
the
protein. Tris suggests
the model
of
]'iii.riti:,r
i'. i"r,
in
v,zhich the
protein
coded by
the
intron in
an
o+ sl
rain recognizes
the
site where
the intron should
be
inserted
in an
co- strain
and causes
it to be
preferentially
inherited.
What is the
action of
the
protein? The
prod-
uct of the
o intron
is an endonu
clease that
rec-
ognizes the o-
ger'e
as
a target
for
a
double-strand
break.
The endonuclease
recognizes
an
18 bp
target sequence
that
contains
the
site where
the
intron is inserted
The
target sequence
is cleaved
on
each strand of
DNA
two bases
to the
l' side
of the
insertion site.
Thus
the cleavage
sites
are
4
bp
apart and
generate overhanging
single
strands.
This type
of
rleavage
is related
to the cleav-
age characteristic
of transposons
when
they
migrate to
new sites
(see
Chapter
21,
Trans-
posons).
The
dorLble-strand
break
probably ini-
tiates a
gene
colrversion
process in which
the
sequence
of the
rrt+
gene
is copied
to
replace the
sequence
of the
o-
gene.
The
reaction
involves
transposition
by
a duplicative
mechanism,
and
occurs solely
at
the
level of
DNA.
Insertion
of
27.5
Some
Group
I Introns
Code
for
Endonucleases
That sponsor
Mobil.ity
775
The
introrr is replicated
and then
inserted
the intron interrupts
the
sequence recognized
by the
endonuclease,
thus ensuring stability.
Many group
I introns
code for
endonucle-
ases that
make them mobile.
Several different
families
of endonucleases
are found;
one com-
mon
feature is
the
presence
of the
amino acid
sequence LAGLIDADG
near the
active site. Sim-
ilar introns
often carry
quite
different
endonu-
cleases.
There
are differences in
the details of
insertion; for
example,
the endonuclease
coded
by the
phage
T4
td
intron
cleaves
a target site
that is 24
bp upstream of the
site at which the
intron
is itself
inserted. The
dissociation between
the intron
sequence
and the
endonuclease
sequence is
emphasized by
the fact that the
same
endonuclease
sequences are found in
inteins
(sequences
that code for
sel{-splicing
proteins;
see
Section 27.12, Protein
Splicing Is
Autocatalytic).
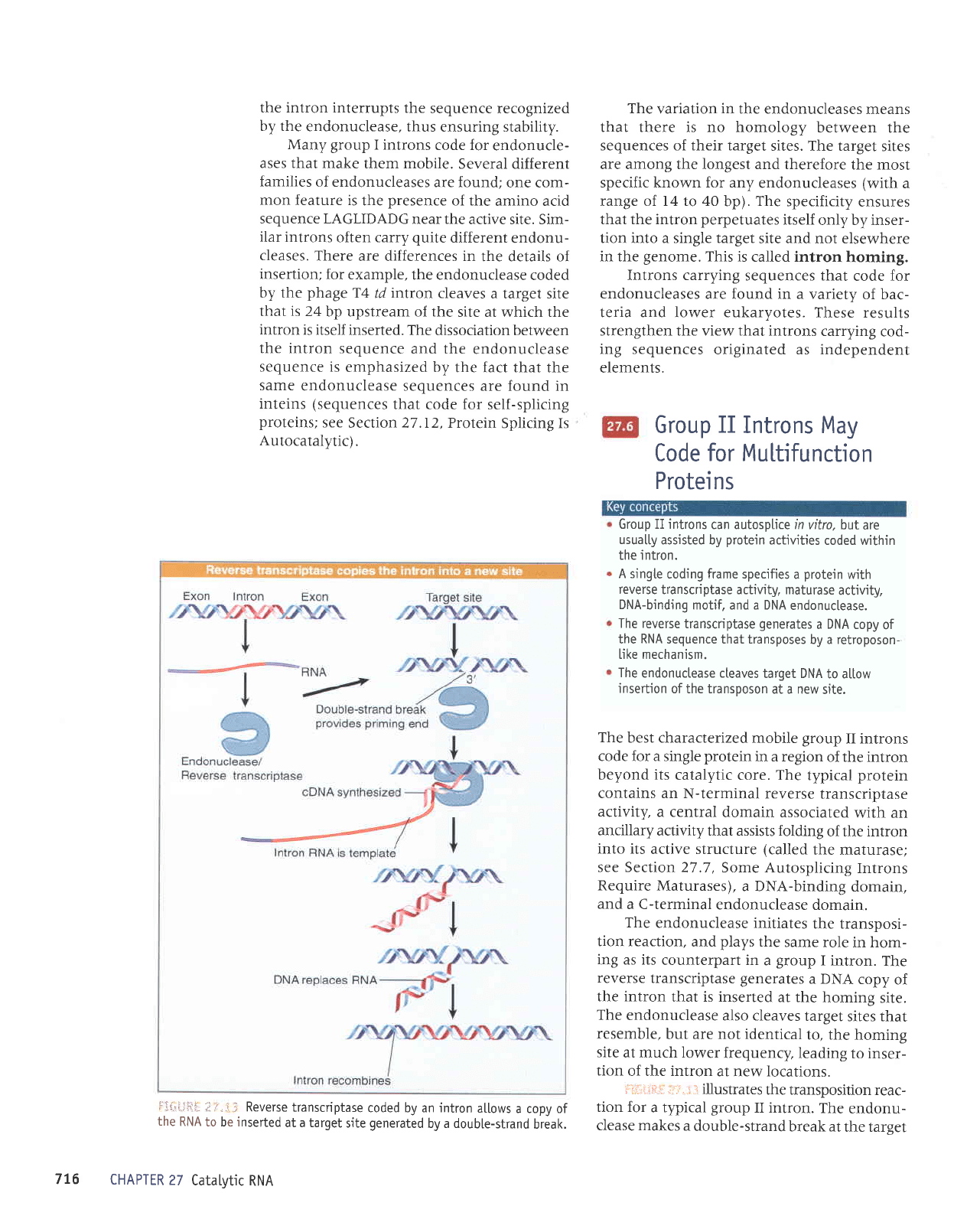
i-1.i.:i.tl.li:
;ir'.
ii:
Reverse
transcriptase
coded by an intron
allows
a copy of
the
RNA
to be inserted
at a target
site
generated
by a doubte-strand
break.
CHAPTER
27
Catatytic
RNA
The
variation
in
the endonucleases
means
that there is no homology
between
the
sequences of their target
sites. The target
sites
are among the Iongest and therefore
the most
specific known for any
endonucleases
(with
a
range of 14to 40
bp).
The
specificity
ensures
that the intron
perpetuates
itself
only by inser-
tion into a single target
site and not elsewhere
in the
genome.
This
is called intron
homing.
Introns carrying
sequences that
code for
endonucleases
are found in a
variety of bac-
teria and lower eukaryotes.
These results
strengthen the view that introns
carrying
cod-
ing sequences
originated as independent
elements.
Group
II
Introns May
Code
for Multifunction
Proteins
Group II introns
can autosptice in
vitro, but
are
usua[[y
assisted by
protein
activities
coded within
the intron.
A single
coding frame specifies
a
protein
with
reverse
tra
nscri
ptase
a ctivity, maturase
activity,
DNA-binding
motif, and a DNA
endonuctease.
The reverse
transcriptase
generates
a DNA copy
of
the RNA sequence that transposes
by a retroposon-
[ike mechanism.
The
endonuctease cteaves
target DNA
to altow
insertion
of the transposon
at a new
site.
The
best characterized
mobile
group
II introns
code for a single
protein
in a region
of the intron
beyond its
catalytic core. The
typical
protein
contains an
N-terminal reverse
transcriptase
activity,
a central
domain associated
with an
ancillary
activity that assists folding
of the intron
into its
active structure
(called
the maturase;
see Section
27 .7
,
Some Autosplicing
Introns
Require
Maturases),
a DNA-binding
domain,
and
a C-terminal
endonuclease
domain.
The
endonuclease initiates
the transposi-
tion reaction,
and
plays
the
same role
in hom-
ing as its
counterpart in
a
group
I
intron.
The
reverse
transcriptase generates
a DNA
copy
of
the intron
that is inserted
at the
homing
site.
The
endonuclease
also cleaves
target
sites that
resemble,
but are not identical
to, the
homing
site
at much lower
frequency,
leading
to inser-
tion
of the intron
at new locations.
t:; ii:
i.i
l;:
h ir
;i . :i :1
illustrates
the transposition
reac-
tion for
a typical
group
II intron.
The
endonu-
clease makes
a
double-strand
break at
the target
776

site.
The
endonuclease activity
requires both
the
domain of the
protein
and the intron
RNA. The
protein
domain cleaves
the antisense
strand of
DNA,
and the intron RNA
actually cleaves the
sense strand. This reaction
directly inserts
the
intron into the DNA
target site.
The intron RNA
provides
the
template
for
the synthesis of cDNA. Almost
all
group
II
introns have a reverse
transcriptase activity
that
is
specific
for
the
intron.
The reverse
transcrip-
tase
generates
a DNA
copy of the intron. the
result being the insertion of
the
intron
into the
target site as a duplex DNA. The
mechanism
resembles the transposition
of retroviruses, in
which the
RNA is
an obligatory intermediate
(see
Section 22.2, Tlrre Retrovirus
Life Cycle
Involves Transposition-Like
Events). The type
of retrotransposition involved in
this case resem-
bles that of a
group
of retroposons that lack
LIRs, and which
generate
the 3'-OH needed
for
priming
by making
a
nick
in the target
(see
Figure 22.20 in
Section 22.I2, LINES Use an
Endonuclease to Generate
a
Priming
End).
Some
AutospLicing
Introns Require Maturases
.
Autospticing introns may require maturase
activities encoded within the intron
to assist
fotding
into
the active catalytic structure.
Although
group
I
and
group
II introns both have
the capacity to autosplice in vitro,
under
physi-
ological conditions they usually require assis-
tance
from
proteins.
Both
types of intron may
code for maturase activities that are required
to assist the splicing
reaction.
The maturase activity is
part
of the single
open reading frame coded by the intron. In the
example
of introns that
code
for homing
endonucleases,
the
single
protein product
has
both endonuclease and maturase activity. Muta-
tional
analysis shows that the two activities are
independent.
Structural analysis shows that the endonu-
clease
and maturase activities are
provided
by
different
active sites in the
protein,
each
coded
by a separate domain.
The
endonuclease site
binds
to DNA, but the maturase site binds to
the intron
RNA. ,'..r,.'.
:
:
:
,'
shows the struc-
ture of one such
protein
bound to DNA.
A
char-
acteristic
feature
of the endonuclease
is the
presence
of
parallel
cr
helices,
which
contain
the hallmark
LAGLIDADG
sequences,
Ieading
to the two cataly.ic
amino
acids.
The maturase
activity is locaterl some
distance
away on the
surface of the
protein.
Introns that code
for maturases
may be
unable to splice
themselves
effectively
in the
absence of the
prctein activity.
The maturase
is
in effect a splicinl;
Iactor that
is required
specif
-
ically for splicing of the
sequence
that codes
for
it. It functions tc, assist
the folding
of the cat-
alytic core to
forrn an active
site.
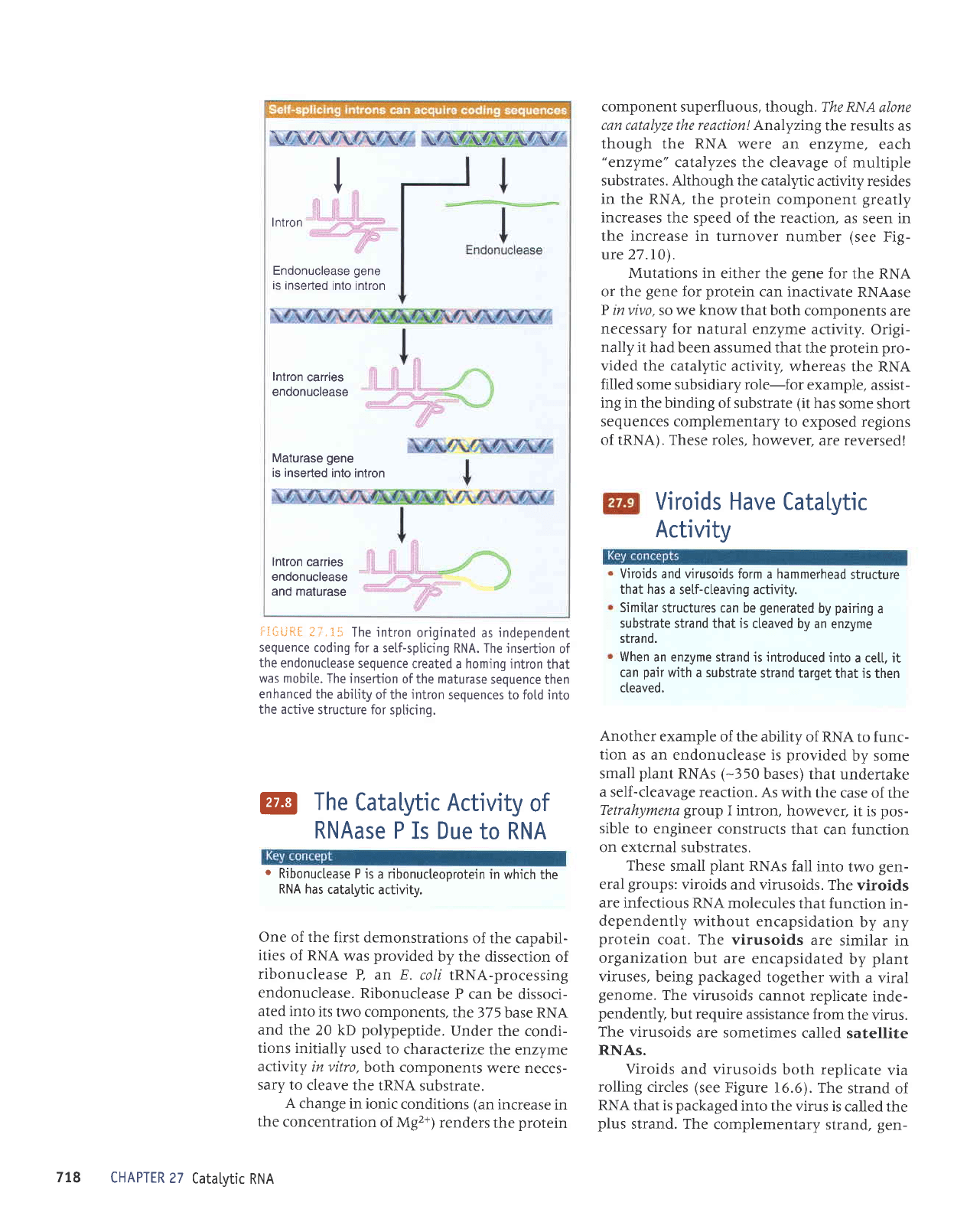
The
coexistence
of
endonuclease
and
mat-
urase activities
in the same
protein
suggests
a route for tht:
evolution
of the
intron.
irirli:iir
,.;' I
suggests
that the
intron originated
in
an
independent autosplicing
element.
The
insertion into th
s element
of a sequence
cod-
ing for
an endonuclease
gave
it mobility.
The
insertion,
howe ver,
might well
disrupt the
ability
of
the R|trA sequence
to
fold into the
active structure,
This would
create
pressure
for assistance
from
proteins
that
could restore
folding ability.
'fhe
incorporation
of such a
sequence
into tlLe
intron would
maintain
its
independence.
Some
group
II introns
that
do not code
for
maturase activit es
may
use comparable
pro-
teins that are
coled by
sequences
in the host
genome.
This sul;gests
a
possible route for the
evolution of
gent:ral
splicing
factors. The
factor
may
have
originirted
as a
maturase that
specif-
ically assisted
the splicing
of
a
particular intron.
The coding sequcnce
became
isolated
from the
intron in the
host
genome, and
then
it
evolved
to
function with ir wider
range of substrates
that
the original
intron sequence.
The catalytic
core
of the
intron could
have evolved
into an snRNA.
:
i';.1rt'1 .
'.
i A homing
intron
codes
for an endonucte-
ase ofthe
LAGLIDADTi
famity that
atso
has
maturase activ-
ity. The LAGLIDADG
sequences
are
paft
ofthe
two a
helices
that terminate
in
tl-e
catalytic
amjno
acids close
to the
DNA duplex.
The maturase
active
site'is
identified
by an
arginine residue
etstrwhere
on
the surface
of the
protein.
27.7
Some
Autospticing
Introns
Require
Maturases
777
\/\t/\t\f\r'
\/\/\t\/\t\/
I
I
v
InIron
Endonuclease
gene
is
inserted
into intron
Intron
carries
endonuclease
Maturase
gene
is inserted
into intron
lntron
carries
endonuclease
and maturase
component
superfluous, though. The RNA
alone
can catalyze the reaction! Analyzing
the results as
though the RNA
were an enzyme,
each
"enzyme"
catalyzes the cleavage
of multiple
substrates. Although the catalytic
activity resides
in the RNA, the
protein
component
greatly
increases
the speed of the reaction,
as
seen in
the increase in turnover
number
(see
Fig-
ure 27.I0).
Mutations in
either the
gene
for
the RNA
or the
gene
for
protein
can inactivate
RNAase
P invivo,
so we know that both
components
are
necessary for natural
enzyme
activity. Odgi-
nally
it had been assumed
that the
protein
pro-
vided the catalytic
activity, whereas
the RNA
filled some
subsidiary
role-for
example,
assist-
ing in the
binding of substrate
(it
has some
short
sequences complementary
to exposed
regions
of IRNA). These
roles, however,
are reversed!
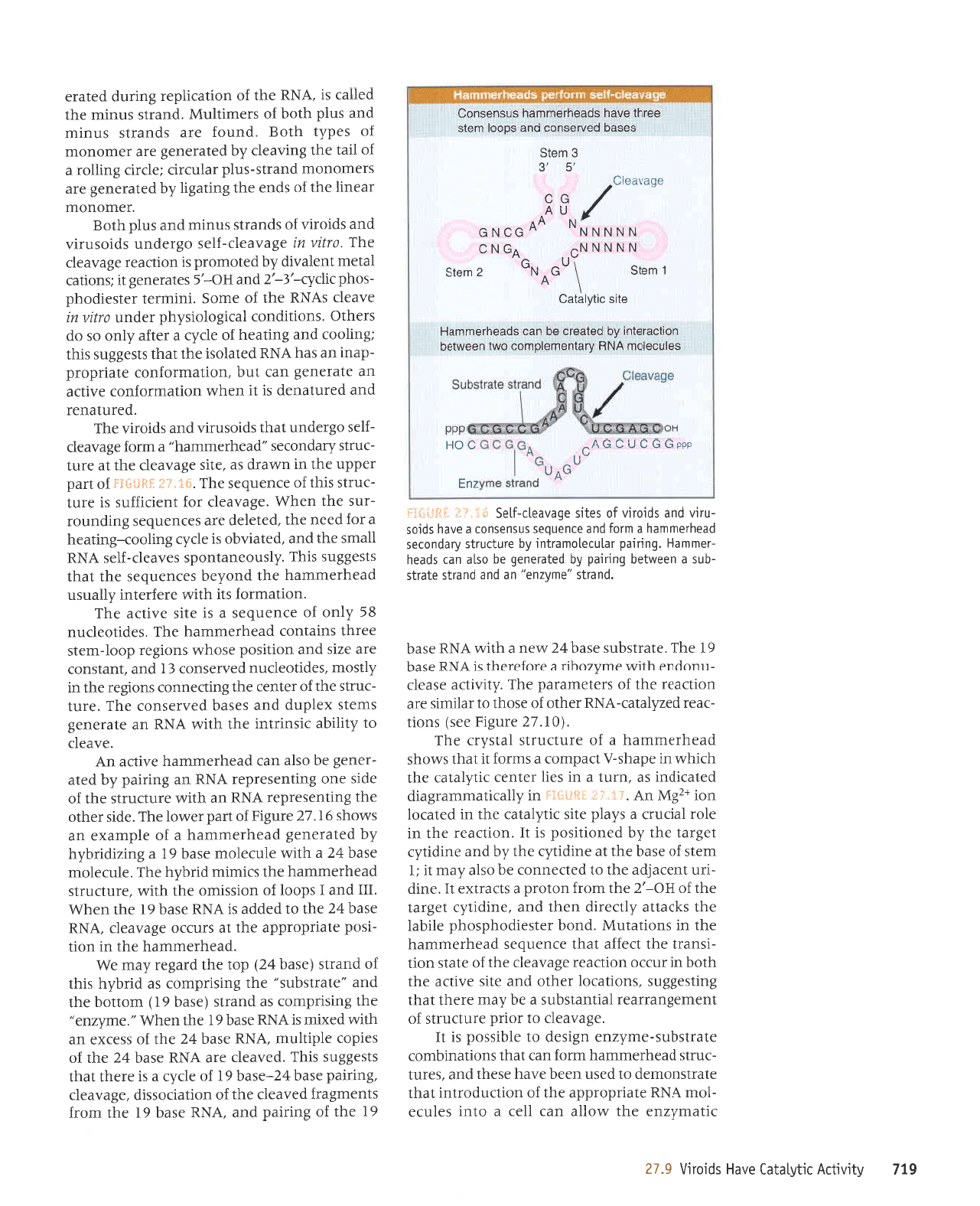
Viroids Have
Catalytic
Activity
Viroids and virusoids
form a hammerhead
structure
that has
a self-cleaving activity.
Simitar
structures can be
generated
by
pairing
a
substrate strand that is
cteaved by an
enzyme
stra nd.
When an enzyme strand is introduced
into
a cet[, it
can
pair
with a substrate
strand target
that is then
cteaved.
Another
example
of the ability
of RNA ro func-
tion as
an endonuclease is
provided
by some
small
plant
RNAs
(-350
bases)
that undeftake
a
self-cleavage reaction.
As with
the case of
the
Tetrahymena
group
I intron,
however,
it is
pos-
sible to
engineer constructs
that
can function
on external substrates.
These
small
plant
RNAs
fall into
two
gen-
eral
groups:
viroids and
virusoids. The
viroids
are infectious
RNA molecules
that function
in-
dependently
without encapsidation
by any
protein
coat. The virusoids
are
similar
in
organizalion
but
are encapsidated
by
plant
viruses,
being
packaged
together
with
a viral
genome.
The
virusoids cannot
replicate
inde-
pendently.
but require assistance
from
the
virus.
The
virusoids
are sometimes
called
satellite
RNAs.
Viroids
and virusoids
both
replicate
via
rolling
circles
(see
Figure
I6.6). The
srrand
of
RNA
that is
packaged
into
the virus is
called the
plus
strand. The
complementary
strand,
gen-
ii';U*[
ti.15
The intron
originated
as
independent
sequence coding
for a self-splicing
RNA. The insertjon
of
the
endonuclease
sequence
created a homing intron
that
was mobi[e.
The insertion
of the maturase
sequence
then
enhanced
the abitity
of the
intron
sequences to fold into
the active
structure for splicing.
@
The
CataLytic Activity
of
RNAase P
Is Due
to RNA
o
Ribonuctease
P is
a ribonucteoprote'in
in
which the
RNA has
catalytic
activity.
One of the
first demonstrations
of the
capabil-
ities
of RNA
was
provided
by the dissection
of
ribonuclease
P, an E.
coli
tRNA-processing
endonuclease.
Ribonuclease
P can
be dissoci-
ated into
its
two components
,
the
37 5 base RNA
and
the 20 kD
polypeptide.
Under
the condi-
tions initially
used to
characterize
the enzyme
activity
in vitro,
both components
were neces-
sary to
cleave the
IRNA
substrate.
A
change in ionic
conditions
(an
increase
in
the
concentration
of Mg2+) renders
the
protein
CHAPTER
27
Catatytic
RNA
778
6r.L
flrnrpy
rLy{1e1e3 o^eH sproltA
6'12
rrleru.dzua
Jqt
,tt.olle
uer
IIJI
e olul salnJe
-loru
VNU
alerrdordde Jql
Jo
uorpnpoJtur
leql
alerlsuoruJp 01
pJSn
uJJq J^Pq JSJqI
pue
'sJrnl
-JnJls
peeqJJrurueq
ruJoJ ue)
]eq1
suollPulqluof,
eleJlsqns-eruz(zua u8rsap o1 alqrssod
s1
11
'a8errealr
ol
rotrd Jrnl)nrls
Jo
luarua8uerrpJJ
Iertuplsqns
e eq
Leur araql
teqt
8uusa83ns
'suorleJol
Jer{lo
pue
alrs elulp
eql
qloq
uI JnJf,o uollf,par a8erreelr
Jqt
Jo
Jlpls
uop
-rsueJl
eql
peJJp
leql
a;uanbJs
ppJqrJurueq
aql
q
suorlelnw
'puoq
rrlserpoqdsoqd
apqel
aql s>lJelle
,{1na,np
uaql
pue
'autptl.{l
p8rcI
Jqr
Jo
HO-,z
Jqt ruorJ uolord e slJpJlxJ
1I'JuIp
-lJn
IUJJP[pe
eql 01
pJl]JuuoJ
aq
osle
,leu
lt
11
ruets
Jo
aseq eql
1e
aurprldr aqt l.q
pue
autptil.t
la8rel
aqt
,(q
pauorlrsod
sl
tI
'uolpeeJ
Jr{l ul
aloJ
IprJnrJ
e s.{e1d alrs rr1^,i.leler eqt
ut
pateJol
uot
*.3141
uV
':i
i""r.i1
]iJi-11:!i.+
ut
Llprrleuruer8etp
paleJrpu
se
'uJn1
p
ur serl JJ]UJJ
ru.dlerer
aql
qJIqM
ur adeqs-n
pedruor
p
sruJoJ
tl tpql
sMoqs
peaqrJurreq
e
Jo
Jrnl)nJls
letsArr
aq1
'hytZ
arn8rg aas)
suot]
-rear
paz.dpleJ-VNu
JJqlo
Jo
Jsoql ot
relIuIS
eJe
uorlJpeJ Jql
Jo
sJeleuered aq1
',{.lrnrlre
aspap
-nuopue
qt4n
aruLzoqrr
p
aJoJeJaql
sr
VNU
Jspq
6I
rql'rlerlsqns eseqvz Mru e
qll^1.
vNu
esEq
'puells
,,eu[zue,.
up
pup
puerls
alPlls
-qns
p
uoeMleq
6uuLed [q
palerauab
aq oqe
up] spPaq
-rourureH
'6uured
lplnlalouerlur
fiq
ernpnrls
fi.repuoras
peeqlourupq
p
ulo;
pue
eruanbes snsuasuol
p
a^Pq
sptos
-nrr^
pue
sptolt^
Jo
salts abeneelr-;1a5
..
'
rr.,,':-,tri.i
6I
eql;o
Sutrted
pue
'VNu
eseq
6I
Jql IUoJJ
sluau8e4
paleell
eqt
Jo
uopelrosstp
'a8eneap
'8urrrcd
aspq
iz-aseq
61
;o
altzb e sI
JrJql
leql
slsaSSns
srql
'pa^eall
rre
VNU
rseq
7Z
eqt
Jo
sardor a1dr11nru
'yNu
aseq
VZ
erp
Jo
ssaJXJ ue
r{tpr
pJXrrJ
sl
VNu
JSeq
6l
Jql ueq6
,,'au(zua,,
aq1 SursudruoJ
s€
puerls
(aseq
61)
uroltoq
aql
pup
,,alensqns,,
aql
Sutstrdruor
se
puqLq
stqr
Jo
puerls
(aseq
77)
dor
ael
pre8ar
Leru a6
'peaqJJrrrueq
eql uI uoll
-rsod
aletrdordde
aqr
1e
sJnlJo
a8errealr
'y55
rseqvz
rql 01
peppe
sl
YNU
aseq
6I
eql uaqM
'11I
pup
1
sdool
Jo
uoISSIuro
Jql
qllr\
'eJnlJnJls
peaqrJruueq eq1 s)Iurru
plJq^q
JqI'JInJJlotu
JSeqVZ
e
qtIM
elnJelou
Jseq
6I
e Surzptrq,{q
.dq
paleraua8
peaqraruueq
e
Jo
aldurexa
ue
sMoqs
9y
77
anfug;o
tred
ra,Lrol eq1,
'apIS
Jeqlo
aqt
Surluasardar
ygg
ue
qlIM
eJnpnrts
Jql
Jo
aprs
Juo Surluasardar
vNu
ue
SuutBd
Lq
pare
-raua8
Jq os1e
ueJ
peaqJeruueq JAI1;e uY
'eAPJIJ
ot,{fluqe
lIsuIJluI
Jq1
qUl!|
VNU
ue aleraua8
suats
xaldnp
pue
saseq
paAJJSuoJ
eqJ
'JJnl
-)nJls
eql
Jo
Jaluat
aql SuIDJuuoJ
suot8ar
aql ut
u(psoru
'saplloJlJnu
pelJesuoJ
€ I
pue
'luelsuoJ
eJe ezrs
pue
uotlrsod
asoqm
suorEar
dool-rua1s
JJJqI
sulPluoJ
pPJqJauueq eqJ
'sJplloapnu
gg
,{1uo
yo
aruanbas
p
sI J]IS
elll)e
JqJ
'uoIlPIuJoJ
sll
qlIM
JrJJrJluI
z(11ensn
peJqJeruupq aqr
puou(aq
saruanbas
eq1
leql
s1sa33ns
srq;',{lsnoauetuods
seleelJ-JIas
YNU
Ilelus
Jql
pue
'palel^qo
st apzb
Suqoo>Suqeaq
e JoJ
peeu
Jql
'pelJlap
are saruanbas
Sutpunor
-Jns
aqt
ueq6
'a8e.teap
JoJ
IUJIJIJJnS
sI
aJnl
-)nrls
srql;o
aruanbas
ee;,
'';"i=ifr
5uii*{i
yo
ired
raddn
Jql uI
uMeJp se
'ells
aSenealr
Jql
1e
ernl
-rnr1s
fuepuores
,,peeqJeruueq,,
e rrrJoJ a8e,teap
-;1as
o8rapun
leql
spIosnJIA
pue
spIoJIA aqJ
'paJnleueJ
pue
paJnleuep sl
1l
uaqM
uolleluJoJuo)
aAIlJe
ue aleraua8
upJ
lnq
'uolleruroJuol
aletrdo;d
-deur
ue seq
VNu
palelosl eql
teql
s1sa33ns srql
lSuqoor
pue
Suueaq
yo
apzb
P Ja{P
Lpo os op
sJeqro
'suolllpuo)
prrSolorsz(qd
rapun o4n
ul
eneep
syNU
aql
Jo
Juros
'IuIIuJel
ralsarpoqd
-soqd
r1p,{r-,
€-,Zpue
Ho-,s
sateraua8
1l
lsuolleJ
Ieteu
luJIeAp
,{.q
parourord sr uorDear
a8eaeap
JrtrI'
lJlt^ ut aEe
nealt
-;1as
o8rapun
spIosnJIA
pue
sprorr^
Jo
spuerls
snurur
pue
snld
qtog
'Jaruououl
reauq
eqt
Jo
spua
aqr Supe8q
z(q
paleraua8 are
srJruouoru
puerls-sn1d Jelnf,JIJ
1a1lrn 3uq1or e
Jo
Irel
Jql 3uu,ea1r
,{q
pateraua8 are ratuouoru
yo
sadr{1
qlog
'punoJ
aJe
spueJls
snulur
pue
snld
qtoq
Jo
sJaulllnw
'puPrls
snulru
Jql
peilel
sr
'VNU
eql
;o
uotterqdar
Suunp
palerJ
..
zu=
sun
^n
ddcr
SOCnCOV"
COH
selncelour
y1r1g
fueluaue;duroc ornl
uaoMloq
uortcEraiur
Iq
paleetc
oq ueo speoLuar!ueH
alrs cry{1e1e3
\v
t
LUoIS
\
ne
'
ruO
Z
urals
NNNNNC
VEruC
NNNNNNT
..veCNe
-t\
./
nv'
^ /
cc
oDEAEOIS
,9
,e
e
Luels
saseq
peruesuoc pue
soool Luals
ooJr,.ll o^eq speaqreuruJPLl
snsuosuoc
s', 3',
GC
Stem3!
A
t/u
GA
UA
.
,'.:i..
:
-'
-.
i
:
A
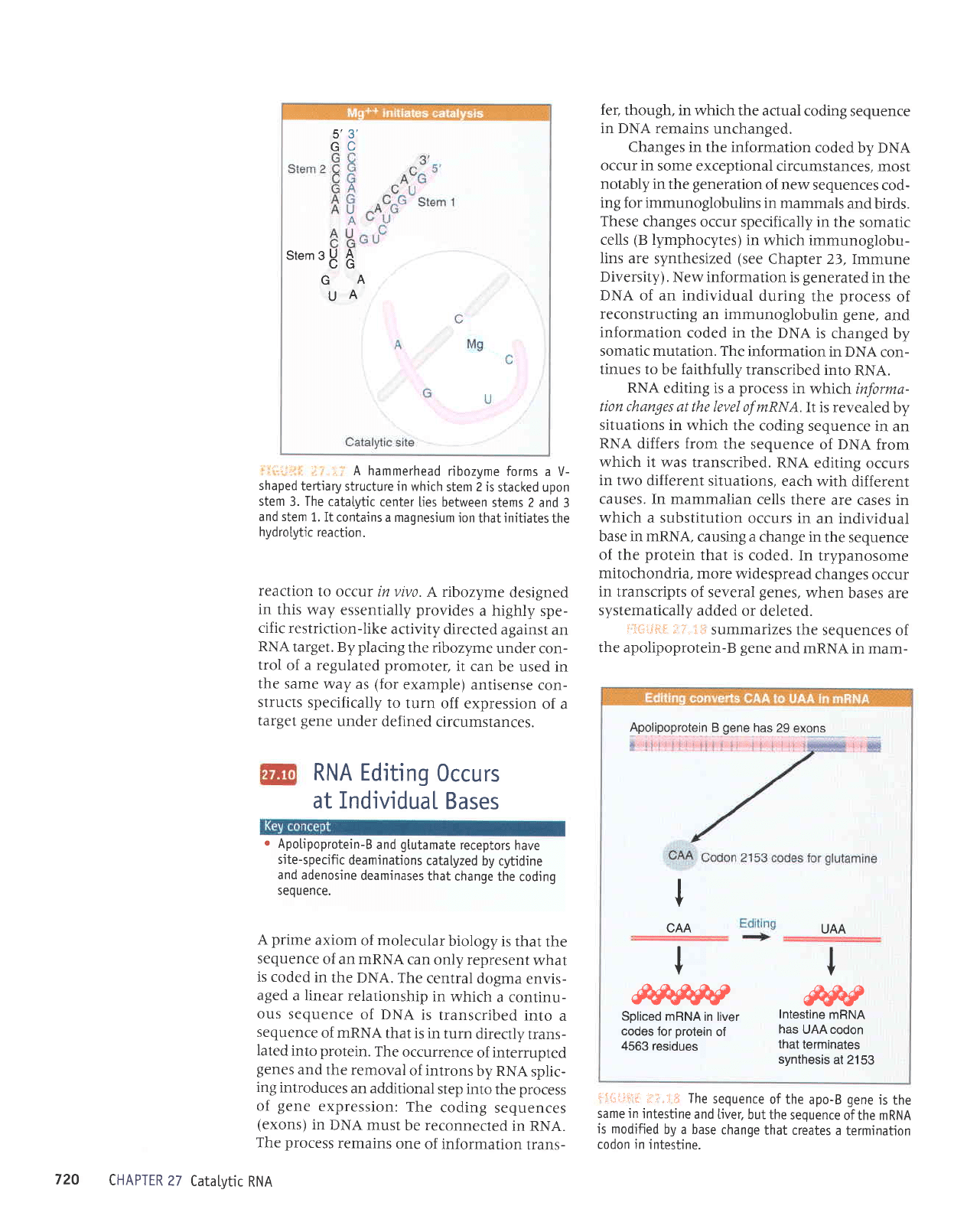
hammerhead
ribozyme
forms a V-
shaped
tertiary
structure in which
stem 2 is
stacked upon
stem
3.
The
catatytic
center
lies between
stems 2 and
3
and stem
1. It
contains a maqnesium
ion
that
initiates
the
hydroLytic
reaction.
reaction
to occur in vivo.
A ribozyme
designed
in
this
way essentially provides
a
highly
spe-
cific
restriction-like
activity
directed against
an
RNA
target.
By
placing
the ribozyme
under con-
trol
of a regulated promoter,
it can
be used in
the same
way as
(for
example)
antisense
con-
structs
specifically
to
turn off
expression
of a
target
gene
under
defined
circumstances.
RNA Editing
Occurs
at IndividuaL
Bases
.
Apotipoprotein-B
and
glutamate
receptors
have
site-specific
deamjnations
catalyzed
by cytidine
and
adenosine
deaminases
that
change the
coding
sequence.
A
prime
axiom
of molecular
biology
is that
the
sequence
of an nRNA
can
only represent
what
is coded
in
the DNA.
The
central
dogma
envis-
aged
a linear
relationship
in
which
a continu-
ous
sequence
of DNA
is
transcribed
into
a
sequence
of mRNA
that is
in turn
directly
trans-
lated
into protein.
The
occurrence
of
interrupted
genes
and
the removal
of introns
by RNA
splic-
ing
introduces
an additional
step into
the
process
of
gene
expression:
The
coding
sequences
(exons)
in
DNA
must
be
reconnected
in
RNA.
The
process
remains
one
of information
trans-
CHAPTER
27
Catatytic
RNA
fer,
though, in which
the actual
coding sequence
in DNA
remains unchanged.
Changes in the information
coded by DNA
occur in
some exceptional
circumstances,
most
notably
in the
generation
of new
sequences
cod-
ing for immunoglobulins
in
mammals
and birds.
These
changes
occur specifically in
the
somatic
cells
(B
lymphocytes)
in which
immunoglobu-
Iins are
synthesized
(see
Chapter 23,
Immune
Diversity).
New
information
is
generated
in
the
DNA of an individual
during
the
process
of
reconstructing
an immunoglobulin gene,
and
information
coded in the DNA
is changed
by
somatic mutation. The
information
in DNA
con-
tinues to
be
faithfully
transcribed
into
RNA.
RNA
editing is
a
process
in
which informa-
tion changes
at the level of
mRN,4. It is revealed
by
situations
in which
the coding
sequence
in an
RNA differs from
the sequence
of
DNA
from
which it
was transcribed.
RNA
editing occurs
in two
different situations,
each
with different
causes. In mammalian
cells
there
are cases in
which
a substitution
occurs in
an individual
base in mRNA,
causing a change
in
the sequence
of the
protein
that is
coded. In
trypanosome
mitochondria,
more
widespread
changes
occur
in
transcripts
of several
genes.
when
bases are
systematically
added
or deleted.
t t;;;i3 i:S ;'
r-. i ij
summarizes
the
sequences
of
the apolipoprotein-B gene
and mRNA
in mam-
i:l{i,i!'ii.'
r i.li
i* The
sequence
of the
apo-B
gene
is the
same in
intestine
and Uver,
but the
sequence
ofthe mRNA
is
modjfied
by a base
change
that creates
a terminatjon
codon in
intestine.
Apolipoprotein
B
gene
has 29
exons
UAA
I
v
oftry
Intestine
mRNA
has
UAA codon
that
terminates
synthesis
at
2153
I
Y
CAA
I
V
ofrfufury
Spliced mRNA
in liver
codes
tor
protein
of
4563
residues
720
