Lewin Benjamin (ed.) Genes IX
Подождите немного. Документ загружается.

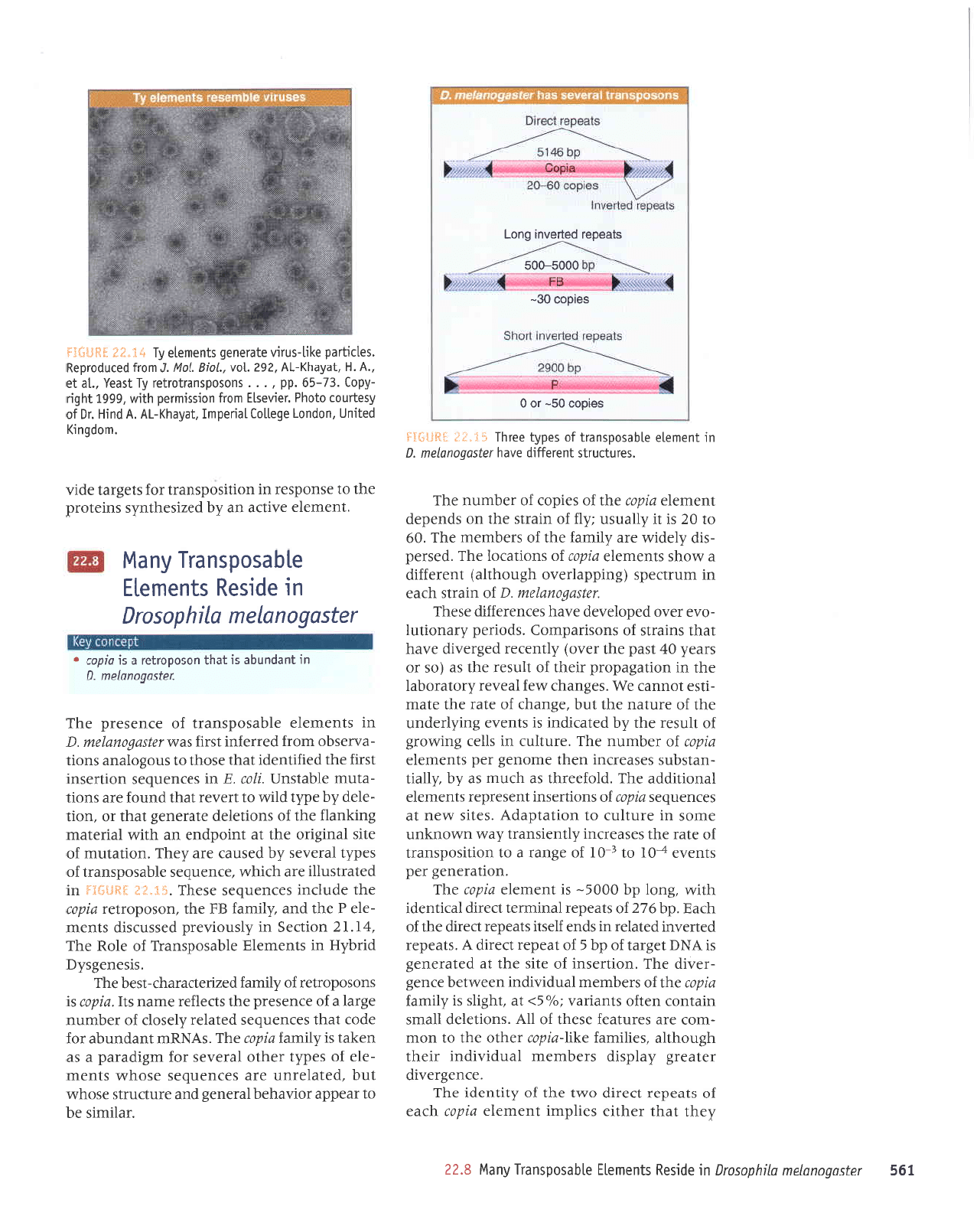
I99
talsz1ouDlaw qLqdosot1
ul aprsa5 slueuell
alqesodsupll Aueyl
g'77
,{aqr
reqr
rJqlrJ
sarldrur
luaruJla
adot
qtea
;o
sleadar
lJeJrp
omt
Jql
Jo
,(tlruapr aqJ
'aJuabJeAIp
ralear8 deldsrp
sJJqruJru
lenprlrpur
Jreql
q8noqrle
'serlrrupJ
anq-adot raqlo aqt ot
uoru
-uroJ
eJP saJntPeJ
esaql
Io IIy'suorlJlJp IIPrus
ureluo)
uelJo sluerre L
'.o/os>
le
'rq8ls
sr Lpruey
udot
aql
Jo
sJJqrueu
pnprnrpur
uaaa,rlaq arua8
-Jelrp
aqJ
'uoruJsur
Jo
alrs
Jql
]e
pale:aua3
sl
VNq
la8rel
Jo
dq
s
Jo
leadar
prrlp
V
'sleadar
pJuJAur pateler
ur spuJ
Jlrsrl
sleadar
Drrrp
Jql
Jo
qreg'dq
gLZlo
steadar
Ipururel
paJrp
Ielrtuapl
qtyvt
'3uo1
dq
OOOE-
sr
luJrrelJ
adoc at71
'uorleraua8
rad
stuJle
r0I
ol
e-0I
Jo
a8uer e o1 uottrsodsuerl
Jo
JlpJ
aql saspaJJur.dpuarsuerl Le.,r,r u.uoulun
Jtuos ur JrntlnJ
ot uorletdepV
'sJlrs
MJU
le
saruanbas atdot
1o
suoruesur
luasardar
sluJuala
Ieuoqppe
eqJ
'ploJaeJqt
sP
q)nru
se
,{q
'z(1er1
-uelsqns
saspeJJur uJql Jruoue8 rad sluauala
atdot
1o
Jaqunu JqJ
'eJnlln)
ur slla) 3ur.uor8
Jo
llnsal
aql '(q
pelPJrpur
sr sluJAJ Surrllrapun
eql
Jo
JrntPu eqt
lnq
'a8ueqr
Jo
Jler aql etelu
-rlsJ
touuef,
a14'sa8ueqr ,lral
IpeAaJ,{.ro1e;oqe1
rql
ul uorle8edord
Jreqt
Jo
llnsJJ
eqt se
(os
:o
sread
97
lsed
aqt rano) Llluarar
pa8ranrp
aneq
lpql
sureJls
yo
suosrreduro3
'sporlad
rlreuorlnl
-ola
JJAo
padola,rap
alpq sef,uJJJJJIp JsJqJ
ulsaSoualatu'C.
Jo
urens
qJee
ur runrlrads
(3urddega,l,o
q8noqlle)
tueJJJJrp
p
Moqs
sluauJlJ adot
1o
suorte)ol
aq1
'pasrad
-srp
.{1apwr are Lpuey
eql
Jo
sraquaru eqJ
'09
ot
0Z
sl
U
^llensn
1L1;
Jo
urens aqt
uo spuadap
lueruJlJ
atdoc
ar4t;o sardor
Jo
Jaqrunu aqJ
'sarnpnlls
luaroJJrp
atteq nlsooouopw
'g
ur
tueuele
alqesodsuerl
;o
sadfl aa.rql
;{ I"If; lA{ilSgr{
serdoc
gg-
ro
g
soloos
0e-
0009-{09
sleeder
pepenur
6uo1
'Jelruns
aq
ot readde JorlpqJq
IpJauaB
pue
eJnttnrts
asoqM
lnq
'pJtelJJun
eJp saluanbes
JSoqM sluJlu
-ele
Jo
saddl
raqlo
IpJeAas
ro; ru8tpered e se
ue>let
sr Lpue;
atdoc arqT
'syNgru
luepunqe
roJ
epo)
]eql
saruanbas
pJrplJJ
.{1aso1r
Jo
rJqunu
a8rel e;o aruasa.rd
aql sDelJeJ
arupu s1I
'atdot
sr
suosodorlar
;o
z(lrue;
pazualJpJeq)-lsaq
aq1
'srsauaBsdq
plrq,{H
ur slurrualg
alqesodsue4l
Jo
alog aqJ
'VI'IT.
uortJJS
ur z(lsnornard
passnrsp
sluelu
-rle
d
eql
pue
'Lpue;
g{
aqt
'uosodoilat
ardoc
aql
JpnlJuI saruanbas
eseqJ
'.$E.ar
*e{fi$li
uI
pelpJlsnllr
aJe
qJIqM'aruanbas alqesodsuerl
;o
sadl.1
leraaas
dq
pasner
are
daql
'uollelnrx
Jo
atrs
leur8rro
aql
le lurodpue
up
qq,lr
IeIJJlpIu
3ur1ue1;
Jqt
Jo
suopalap
aleraua8
1eql
ro
'uoll
-a1ap
,{q ad,{1
ppr,l
ot
uelJr 1pql
punoJ
eJe suoll
-plnur
elqplsun
'1n 'E
uI saJuJnbJs
uollJesul
lsJrJ
erp
perJrluapl
teqt
asoql o1 snoSoleue
suotl
-enJesqo
uorJ
paJJeJul
IsJIJ
serlr Jrlsadoualaut
g
ur
slueruale alqesodsuen
Jo
JJuasard aq1
'JalsoDouDlaw '0
ur
luepunqp
sr
1eq1
uosodorlar
p
st
prdot
c
ralso
6 o u oP
w ol ul dosor
0
ur
aprsau sluauall
alqesodsuPrl
Aue6
'luaurela
alrlJe
up
dq
pazrsaqlu^,{.s
sutalord
aqt ot
asuodseJ uI uolllsodsuerl
ro; sla8rel apn
'tuopDu
L;
polrun
'uopuol
a6a1o1
leuedurl
'1e[eq;-'1y 'V
pulH
t0
Jo
r\salnor oloLld laneql
uorJ uolsslulad
qluvt
'6661
lqbu
-fido3
'E1-99 'dd
' ' ' '
suosodsuetlotlat
[1
1sea1
'1e
1a
"y
'1.1 '1e{eq;-N
'Z6z'lo^'lolg
lory
't
wu!
parnpordaS
'sa1rr1ed
altt-sntr,l
elerauab slueuola
fit
tl'*g
$HiltSI*

suosodorlau
puP
sasnlnollau
'suor+lunJ
6uLpor eneq
lou
op
leql
sartLueJladns
lplr^uou
aql
pup
SINII
ro alrt-snlno.lJa.l
arp
lpql
sartrrup;ladns
le.lr^
oql olur
pepurp
aq upt suosodollau
S?.-frfr
HEtlflilj
slHO
(ypgtu
cruoueOqns
paldnralurun
ur
penouret)
suorlur
oN
o/v\l lo ouo
suotlur ureluoc
Ieyl
uorlezrueojo
(slcnpord
uosodsuerl
rol
osealcnuopuo/ esetbalur.roTpue sotlrntlce
Outpoc auou ro)
euop eselducsuetl
oslo^au eseldlrcsuetl astanag
eu.rIzu3
dq
rc-L
dq
rc-L
dq
9-t
sleeder
1a6re1
sleadeJ
oN
sleedet op sleedat
;eurultal
6uo1 tutrrllol
slducsuerl
111 lod 1o
(esnou)
seue6opnes6
tg
'Cl
ZS
'tE
Qa7se6oue1ew 6)
erdoc
(sleuureu) g3plg
(ueunq)
11
@e1qaercc'g)
A1
sad,{1 uouruo3
AlrLueyedng
lel^uoN
Sf Nll
I;rrue;.redng
;errl
zz
ulldvHl 299
-qf,aru
luJreJJrp
p
asn
pue
sUJl >pe1 daqt
lnq
'(^lrueJradns
1err,r
aqt
Jo
srJqrueu
tuelslp
atotu asrrdruoJ
01
pJrJprsuo)
Jq JJoJeJer{l Leur
pue)
.{trnrDe
asetdrJJs
-uert
JSJaAJT
JApq osle
(SENff)
saruanb
-JS
peleedal
pJsJJdslalur
Suol JeJ
.
'sJrTJ
pue
lseaL
lo
stuJtuJle
atdot
pue
[1
Jql ur
pJZrJJlJpJeq)
tsJq
Jre,{aq1
'ui.ro1
snorl)JJur
tuapuadapur
ue
qSnorql
8ur
-sspd
lou
ur ruJql
ruoJJ
IJJIIp
lnq
sasnJrl
-ortJJ
se lJuupru
arues Jqt
ur a:npo;dar
z(aqt
'suosodoJler
rJqlo
a>lrl
'sJrlrlrl)e
eseJ8elur Jolpue
aseldrnsueJl
JSJaAaJ roJ
apor Ilnrreyadns pr.ra
eel
jo
srJQrueli\f
o
:Sg'fi*
*Hf1?ll
ur
paqsrn8urlsrp
Jre suosodortJl
Jo
sJSSelf,
eaJqJ
'VN(
olul
VNU
;o
uorldrnsuerl
esJe^er eAIoAur
leql
uortrsodsuen
roJ sursrup
-q)aru
Jo
asn reql
^q
pJurJJp
are suosodo:1ag
'auouab
upunq aql
Jo Jlprl
lsorllp
elnlrlsuor
suosodollal
pue
suosodsuell
o
'
uotlrsodsuell
azfleler
uel
lpql
seuAzua
loJ
apol so^lasuoql
lou
op
Aeql
1nq
'1uene
uoqrsodsuell polprpout-y11X
ue
fq
peleraueb
olam
lptll
punoJ
oq upl
sluauala te{}Q
o
'sull
a^eq
lou
op sloLllo
spataqM
'suL'l
Jo
osn lreql
ur sasnlrnollal
olquosal f11rat1p
suosodorlar
eur0g
o
'elrryed
snoqloJut up
rujoJ
lou
seop
lPql
vNU
uP Pr azurqou
leq}
suosodsupll
are flrueyadns
lplrn
aq]
Jo
suosodorlaS
r
sassPll
aarql
olut
llPl
suosodorlax
@
'rlllreder
snorpaJur ,{.ue
ro;
eJuaprla
ou sr
alJqt
(sluaurala
[1q11z,z'
JSeJ
eql
sr se)
lnq
'sasodsueJt
tl
leql
'JSJnoJ
Jo
'^rou>l
eM'sassJssod
1r
suortrunJ
Ierr^orlJJ
Lueru anoq
z{es ol
preq
sl
tI
'sJSnJIAoJlJr
Jql ot
patelar
ur8
-rJo
uP e^eq
lsnlu
adot
Teql
alqelr^aur surees
1r
lPql
UOIIEZIUESJO
IPJIAOJ1JI
qll^t
SAJUPIqIUESAJ
Lueur
os eJp eJaql
'llnseJ
e se
luorlrsodsueJl
Jo
Jporu s,atdot
JoJ a)ueprlJ
DJJrp
>lJel
eM
'suralord,{.1od
;o
a8eneap
pue
VNU
Jo
Sunrlds se
qJns
sluala Surnlorrur .,{.1q
-eqord'parnpord
aJe suraloJd
lpJa^aS'slBadar
IeurruJJl
eqi
Jo
auo
Jo
elpplru arll ur uorle
-l1lur
ruoJJ stlnser
q)lql!\
'snurr[Ja],E
uoururof,
e eleq
sVNUru aq1
'sldrnsuerl
qfual-ued
pue
qfual-llnJ
qloq
Sunursardar
'sy111gru
*(y)L1od
tuppunqp
se
punol
arc
ptdot;o
stdursue4l
'salrtued
J>lll-snJIA alPJaueB
01 JIqe
aq o1
.{1a1qun
sr atdoc
leql
sueeu
qJIqM
'snJln
eql
Jo
adolazrua
aqt
roJ
paJrnbar
saruanbas
Aua
letrlorlal
qlrM
drqsuorlelar .r{.ue sr sar8
-olouoq
aql uoJJ J)uJsqe Jlqplou
v
'sasnJr^
-oJlJJ
Jo
s3)uanbas
pd pue
6a6
aql
pue
aureJJ
Sutpear u)do otdn
Jqr
Jo
strBd uaa,lrrlaq sar8
-oloruoq
aJe JJaTIJ
'dq
rcZV
Jo
eurprJ Surpear
-3uo1
a18urs
p
suretuof, aruanbas atdoc
aq7
'petou
uaeq eAPr{
yyl6atdot
Surureluor sellrued
'auo,{1uo
seq uJoJ Jeuoqs
aql
pup
steadar
Ieuruuel
o,lt.l spq ruroy ra8uol
Jqt
'saprlJ
VNe
Ierr^oJlJr
a>lrl
1yN(
JelnJJrJ
aeJJ
Jo
ruroJ Jqt
q punoJ
Jre sluJrxJIJ atdot sau4l
tV
'(lplJaleru
Suruaaratur
aqt
palelap
uorleu
-rquo)JJ
g paterauaS
aq 01 ruaql
DedxJ
plnom
am
q8noqrle) paparap
uaJq
lou
a,leq sleadar
IeurruJat
aqt;o
sardor
1enprnrput
itretur
sLe.up
are aruoua8
eql ur sluaruJle atdot
er47
'sesnJrloJteJ
qrlm
drqsuollpleJ
p;o
arrqsa8
-8ns
sr srql
'sluerue;p
4L
lo
eseJ rplrrurs
Jr{l ur
sy
'uorlrsodsuerl
Surrnp
tuauele
rolruaSord e
yo
sleadar
1rerrp
aql
Jo
Juo tuoJJ
palereueS
erp
qloq
leql
Jo
'sluala
uoIlJJrJoJ
tlurrad
o]
lJeJJlur

anism from
retroviruses
to
prime
the
reverse
transcription
reaction.
They are
derived
from
RNA
polymerase
II
tran-
scripts. A minority
of the
elements in
the
genome
are fully functional
and can
transpose autonomously;
others have
mutations,
and thus
can only transpose
as the result
of the action
of. a
trans-
acting autonomous
element.
.
Members
of the nonviral
superfamily
are identified
by external
and internal
features that
suggest that they originated
in RNA sequences.
In
these cases,
though.
we can only
speculate on how
a DNA copy was
generated.
We
assume
that they were
targets for
a transposi-
tion event by an enzyme
system coded
elsewhere, that is, they
are always
nonautonomous. They
originated in
cel-
lular transcripts.
They do not
code
for
proteins
that have transposition func-
tions.
The
most
prominent
component
of this family is
called short interspersed
repeated sequences
(SINES).
These com-
ponents
are
derived from RNA
poly-
merase III
transcripts.
i:i{,,rif.i.r
r.:.i..r
shows the
organization and
sequence relationships
of elements that code for
reverse
transcriptase. Like retroviruses,
the LIR-
containing retroposons
can be
classified
into
groups
according to the number
of independent
reading frames for
gag, pol,
and int, and the order
of
the
genes.
In
spite of these superficial
differ-
ences of organization, the common feature is
the
presence
of reverse
transcriptase and integrase
activities.
\pical
mammalian LINES
elements
have two reading frames;
one codes for a nucleic
acid-binding
protein
and the other codes for
reverse transcriptase and endonuclease
activity.
LTR-containing
elements can
vary
from
integrated retroviruses to retroposons
that
have
lost
the capacity to
generate
infectious
parti-
cles. Yeast and fly
genomes
have
the
Ty
and copia
elements that cannot
generate
infectious
par-
ticles. Mammalian
genomes
have endogenous
retroviruses that, when active, can
generate
infectious
particles.
The mouse
genome
has
sev-
eral active endogenous retroviruses, which are
able to
generate particles
that
propagate
hori-
zontal infections.
By
contrast, almost all endoge-
nous retroviruses lost their activity
some
50
million
years
ago
in
the human lineage, and
the
genome
now has
mostly inactive remnants
of the endogenous retroviruses.
LINES and SINES comprise a major
part
of
the animal
genome.
They were defined origi-
nally by the existence of
a large
number of rel-
atively
short sequences
that
are related
to one
another
(comprising
the moderately
repetitive
DNA
described
in Section
4.6,
Eukaryotic
Genomes Contain
Both Nonrepetitive
and
Repetitive DNA Sequences).
The
LINES com-
prise
long interspersed
sequences,
and the SINES
comprise short interspersed
sequences.
(They
are described as interspersed
sequences
or inter-
spersed repeats
because
of their common
occurrence
and widespread
distribution.)
Plants contain
another
type of small
mobile
element, called
MITE
(for
miniature inverted-
repeat transposable
element).
Such elements
terminate
in inverted
repeats,
have a 2 or 3 bp
target sequence, do
not have
coding sequences,
and are
200 to 500 bp
long. At
least nine such
families exist in
(for
example)
the rice
genome.
They
are often
found
in the
regions flanking
protein-coding genes. They have
no relation-
ship to SINES or
LINES.
LINES and SINES
comprise
a significant
part
of the repetitive
DNA
of animal
genomes. In
many higher eukaryotic
genomes,
they occupy
-50%
of the
total DNA.
i:iil"tfli:
;i:.,l'* summarizes
the distribution
of the
different
types of trans-
posons
that constitute
almost
half of
the human
genome.
Except for the
SINES,
which are
always
nonfunctional, the
other
types of
elements all
consist of both
functional
elements
and
ele-
ments that have suffered
deletions
that elimi-
nated
parts
of the
reading
frames
that code
for
the
protein(s)
needed
for transposition.
The rel-
ative
proportions
of these
types
of transposons
are
generally
similar
in the
mouse
genome.
A common
LINES
in mammalian
genomes
is called Ll.
The typical
member
is
-6500
bp
long and terminates
in an
A-rich tract.
The two
open
reading frames
of a
full-length
element
i:r::iigi: ;:;i. t
i'
Retroposons
that
are ctosely
related to
retrovjruses have a simjlar
organization,
but
LINES share
onty the
reverse transcriptase
activ'ity.
LTR
Homologywith:,i;:,,9a9
$SSPol
mtnt
22.9
Retroposons
Fa[[
into Three Classes
563

Element
Organization
Length
(Kb) Human
genome
LINES
(autonomous),
e.9., L1
AFIFI
.{po|,
.
..,.,
SINES
(nonautonomous),
e.9., Alu
DNAtransposon
)',.'Tr*nspoeaee'r.
d
Number Fraction
1-11 450,000
8%
6-8 850,000 17%
<0.3
1,500,000
15"/"
2-3
300,000 30/o
FIGURE22.1B
Fourtypesoftransposableetementsconstituteatmosthatfofthehumangenome.
are called
ORFI and ORF2. The number
of full-
length
elements is usually
small
(-50),
and the
remainder
of the
copies are truncated. TIan-
scripts
can be found.
As implied
by
its
presence
in
repetitive
DNA, the LINES family
shows
sequence
variation
among individual members.
The
members
of the family
within a species,
however,
are relatively homogeneous
compared
to the variation
shown
between species. LI is
the
only member
of the LINES family
that has
been active in
either the mouse
or human lin-
eages. It
seems
to
have
remained highly
active
in
the mouse,
but has
declined in the human
lineage.
Only
one SINES has
been active in
the
human
lineage:
the common Alu
element. The
mouse genome
has a counterpart
to this ele-
ment
(Bl),
and also
other SINES
(B2,ID,B4\
that have
been active. Human
Alu and mouse
Bl
SINES
are
probably
derived
from the 7SL
RNA
(see
Section
22.10, The
AIu Family Has
Many
Widely
Dispersed Members).
The
other
mouse
SINES appear
to have originated
from
reverse
transcripts
of tRNAs. The
transposition
of the
SINES
probably
results from
their recog-
nition
as substrates
by an active
Ll element.
individual members
of the
family
are dispersed
around
the
genome
instead of being
confined
to tandem clusters. Again, there is
significant
similarity between the members
within a species
compared
with variation between
species.
In the human
genome,
a
large
part
of the
moderately repetitive
DNA exists
as sequences
of
-300
bp that are interspersed
with nonrepet-
itive
DNA. At Ieast half of the renatured
duplex
material is
cleaved by the restriction
enzyme
AluI
at a single site located 170
bp along the
sequence. The
cleaved sequences
all are mem-
bers of a single family known
as the Alu fam-
ily, after the means
of
its
identification. There
are
-100,000
members in
the haploid
genome
(equivalent
to one member
per
6 kb of DNA).
The
individual Alu sequences
are
widely dis-
persed.
A related
sequence family is
present
in
the mouse
(where
the 50,000 members
are
called the Bl family), in
the Chinese
hamster
(where
it is called the Alu-equivalent
family),
and in other mammals.
The individual
members
of the Alu family
are related rather
than identical. The
human
family
seems to have
originated
by
means
of a
130
bp tandem
duplication, with
an unrelated
sequence of 3I
bp
inserted
in the right
half of
the dimer. The
two
repeats
are
sometimes
called
the
"left
half"
and the
"right
half"
of the Alu
sequence. The individual
members
of the Alu
family have
an average identity
with
the con-
sensus sequence
of.87'/". The mouse
Bl repeat-
ing
unit is 130
bp long and
corresponds
to a
monomer
of the human
unit. IthasT0o/o-80o/o
homology
with the human
sequence.
The Alu
sequence is related
to 7SL RNA,
a
component
of the signal recognition particle
(see
Section
I0.9, The
SRP Interacts
with the
SRP Receptor). The
7SL RNA
corresponds
ro
the left
half of
an
Alu
sequence
with an inser-
tion
in the middle.
Thus the ninety
5'terminal
bases
of 7SL RNA
are homologous
to
the left
end
of Alu, the
central 160
bases of 7SL
RNA
The
ALu Family
Has
Many
WideLy
Dispersed
Members
.
A major
part
of repetitive
DNA in mammatian
genomes
consists
of repeats
of a single famity
organized
[ike
transposons and
derived from RNA
potymerase
III transcri
pts.
Ihe most
prominent
SINES
comprises
mem-
bers
of a single family.
Its
short length
and high
liifi
:['lJ',"'::1fi
:B'iT,Tii:xT;rfi
CHAPTER 22
Retroviruses
and Retroposons
564
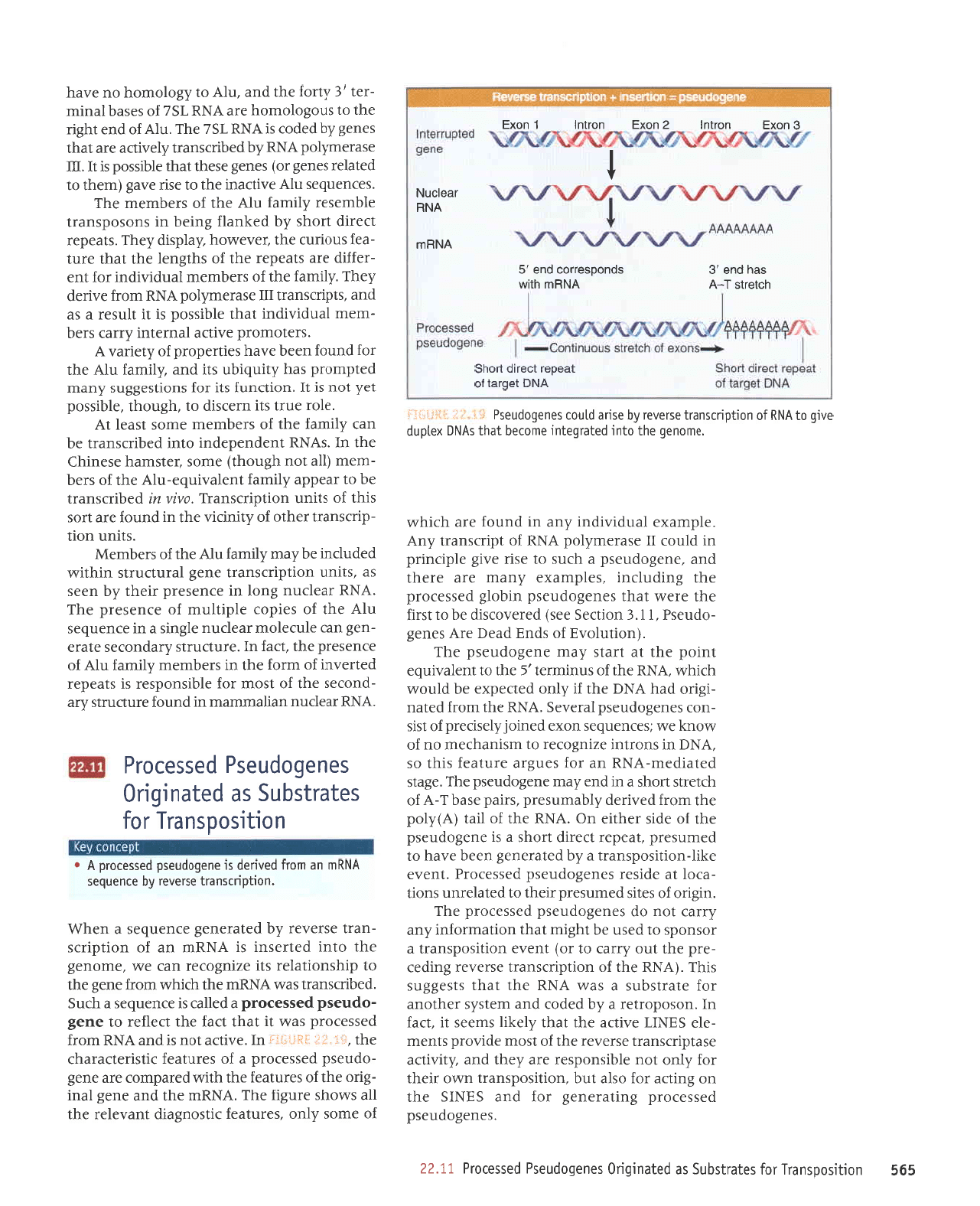
999
uol]rsodsuerl
roJ saleltsqns
sp
patpul6u0
seua6opnasd
passalold
II'zz
'sauJSopnasd
pessa)oJd
Surleraua8
JoJ
pue
SSNIS
aq1
uo Surlre
JoJ oslp
1nq
'uorlrsodsueJl
u,rl.o JrJq]
ro; ^d1uo
1ou
alqrsuodsar
JJe daqt
pue
,{tr,trlre
aseldrnsuerl
esJaAJJ Jr{l
Jo
lsoru
apt.ord sluaru
-JIe
SENIT
anrtre
eql
leqt
z(1a>g1
sruJas
tl
'tJeJ
u1
'uosodorlar
e
dq
pepoJ pue
ruatsz(s JJqloup
roJ
JteJtsqns e spm
VNU
eql
leqt
slsaSSns
slql'(t/NU aql
Jo
uortdrJf,suert JsraAJJ Surpar
-ard
aql
lno
,{.r.rer o1 ro)
1ua,ra
uoltlsodsuerl
e
.rosuods
ol
pJSn
aq
tq8ru leqt
uolleuroyur
due
zi.uer
lou
op sauaSopnasd
passarord
aq1
'ut3uo;o
sJtrs
pJrxnserd:raql
01
peleleJun
suoll
-eJol
lp
JprseJ sauaSopnasd
passaror4'luale
a1t1-uorlrsodsueJt
e
,{q
palerauaB
uaaq
aleq 01
parunsard'teadar
DJJrp uoqs
e sr auaSopnasd
aql
Jo
aprs raqur uO
'VNU
rqr
Jo Iret
(y)L1od
aq1 ruoJJ
pJArJap
[lqerunsard
'srred
aseq
1-y;o
I{JlJrls
uoqs
p
ur
pua
^deru auaSopnasd aq1
'a8ers
pelptpeu-yN5
up
ro; san8re ernleal
srql os
'VN11
ul suoJlur azruSolar ol rusruer{Jaru ou
Jo
/!\ou>l JAl. lsaruanbas
uoxa
pauroIAlasnard yo
lsts
-uor
sauaSopnasd
prana5
'VNU
aqt
ruoJJ
pateu
-t8rro
peq yNCI
rql;r,{po
paDadxa
rq
plnolror
WIqM
'vNu
Jqt
Jo
snururJJl
,E
eql ot
tualenrnba
lurod
aqt
te
uels
z(eru auaSopnasd aq1
'(uo4n1o,Lg
Jo
spug
peaq
ary saua8
-opnJsd
'I
I'€
uorpes aas)
paranorsrp
Jq ot
lsJrJ
eql eraM
leql
sauaSopnasd urqo13
passarord
aql
Surpnlrur
'saldruexa
.,{.ueru aJe
JJJqI
pue
'auaSopnasd
e
qJns
ol asrr a,rr8 aldoutrd
ur
plnor
1
aseraru^d1od
VNU
;o
ldrnsuerl
duy
'aldruexa
lenprlrpur
z{ue ur
punoJ
eJe
qJrqM
'auouab
eql olur.
palerbalur
aulolaq
1eq1
sy1r16
xaldnp
anL6o1
y1151ouor1dursuet1
astenarr{qasueplnotsaua0opnas6
,:i
lil ,iiilllti;r
Jo
Jruos
^,{.Iuo
'sarnleay
rrlsouSetp
luelelal
aql
IIe
sMoqs arn8r;
aq1
'VNUru
eql
pue
aue8
leut
-3rro
aql
yo
saJntpal Jq1
qll,lr pJJBduror
are aua8
-opnasd
passarord
p
Jo
seJnlpeJ
JIlsIrJlJeJeqJ
eqr
'+
i
't;i
;;ilt!:,t.ti
uI
'aAIlJe
1ou
sI
puP
vNu
luoJJ
passarord
spM
tr 1eql
pe;
aqt
DalJar
ot
auaE
-opnasd
passaro.rd
p
palleJ
sr aruanbas
p
q)ns
'paqu)suerl
spM
VNlIru
Jqt
qJrqM
lrror; auaS aql
o1 drqsuorlelar
s1t aztuSolJJ
ueJ am
'aruoua8
Jqt olul
peuesul
sl
vNgtu
ue
yo
uotldtrts
-uer1
JsJeAer
,(q
pale:aua8
aruanbas
e uJqM
'uoqdursuerl
asranal
fiq
aruanbas
VNUur
up urolJ
paluap
st
eueDopnasd
passerotd
y
r
uorlrsodsuPtl
loJ
salellsqns
se
pelPu r6u0
VNO
labrel
lo
leeder tca:rp uoqs
qcla.rls
I-v
vNHur
r.lllM
sprl
puo
/g
spuoossJJoc
pua
,9
VNHUI
VNH
JeolcnN
t
I
I
e
uoxf
uorlul
z
uoxf uorlul
I
uoxf
saua6opnesd
passalold
'1trNU
Jpallnu
ueqelurueru
ur
punoJ
arnDmls
fue
-puo)as
Jq1
Jo
lsour
roJ alqrsuodsar sr
sleadar
palJa^ur
Jo
ruJoJ
eq1 uI sJeqruaru
,{lrue; nlv
Jo
aruasard
eql
'peJ
uI'eJnDnrts
zhepuoras alera
-ua8
uer elnJJIoIu
Jpapnu
a18uts e ur aruanbas
nlv
aqt
;o
satdor aldttlnut
;o
aruasard
aql
'VNU
Jeepnu
3uo1 ur aruasard
rraql
dq
uaas
se
'strun
uolldrnsuerl
aua8
lprnlJnrls
ulqllm
pJpnlJur
aq z{eru
dpuPJ
nlv
aql
Jo
sraqruJw
'slrun
uorl
-drnsue.rl
Jeqto
Jo
z(ltunrn eq1
ut
punoJ
aJp
uos
srql
Jo
slrun
uorldlJ)sue{
'lttn
ut
paqlrJsueJl
aq ot
readde
,{puey
luale.ttnba-nry
eq1
}o
sJaq
-ruaru
(1e
1ou
q8noql)
etuos talsueq
JsJuIq)
eql uI
'sVNu
luapuadapul
olut
peqlrJsueJl
aq
uer
z{prue; eql
Jo
sJeqlueu
auos
lseel ]v
'JIor
JnJl slr
uraJSIp or
'q8noql
'alqrssod
taz( lou
sl
lI
'uolpunJ
sll
roJ suorlsaSSns
LuBru
paldruord
seq
dlrnbrqn stl
pue
fpuey nlv eql
JoJ
punoJ
uJaq aneq sauradord;o
t(1atrea.
y
'sraloruord
all1re
IeuJalur
drrer sreq
-ruJru
lpnprlrpur
teqt
alqtssod
sI
ll
tlnsar
p
se
pue
'sldursuerl
11
aseraru[1od
y51g
uroJJ
aAIJJp
.{aq1
'.,(pue1
Jql
Jo
srJqlueur
IpnpIAIpuI
JoJ
lua
-reIIIp
are
sleadar aqt
yo
sqrBuJI
eql
lpql
eJnt
-eal
snorJnl
Jql tJ^eMor{',{.e1dstp
^daq1
'sleada:
tJaJrp
uoqs
z(q
pa1ue11
Sutaq
ut suosodsuerl
alqruesal
z(pue;
nlv
Jq1
Jo
sJaqurJru
JqI
'sa)uanbJs
nly enrDeur aql
01 JsIJ aaeS
(uraqt
ol
peleleJ
saua8
ro) saua8
asaql
teqt
alqrssod
q
tI
'm
aserarudlod
y1.1g
z(q
paqrnsuerl z(larrBre ere
1eq1
saua8
,{q
papor
sr
VNU fSZ
aqJ
'nlv
Io
pua
tq8tr
aql
ot snoSolouoq
eJe
VNU
f Sl,
Io
seseq
IPUIU
-JJt
ft,{1roy
aql
pue
'nlv
o1
,(8o1oruoq ou alpq
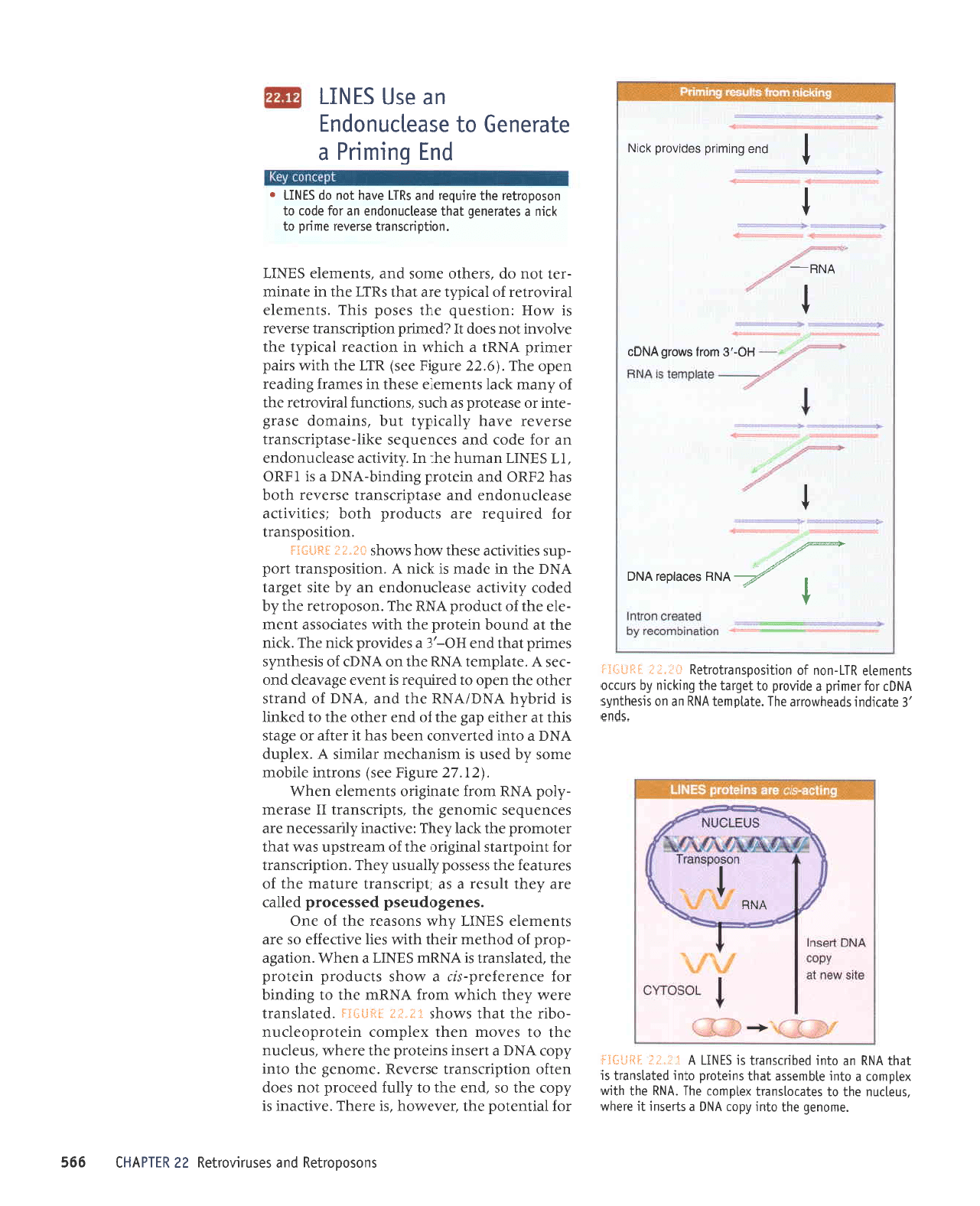
'auroua6
aql olur fdor
yp6
e silesur
lt
alaq/v\
'snallnu
aql ol selprolsuetl
xalduor
aql
'VNU
aql
qlyv\
xalouol
e olur
olquassp
lpql
suralojcl olur
pelelsupll
sr
lPrl]
vNU
uP olur
paqulsuerl
sr.
slNIl
v
i;_1'I
j
l}ji:*ij
'sDua
,€
alelrpur speeqmol.lp
aql
'e1e1dua1
VNU
ue
uo srsaqlur{s
VNqr
loJ laLuud
e aprnotd
o1
1abte1
eq1
burlrru
fq
srnrro
sluaualo
UII-uou
;o
uorlLsodsuellollou
i.rr"l:
:+*s-:ij
suosodorlau
pue
sasnrnortau
zz
ulldvHl
roy
1er1ua1od
Jql ?JAaMoq
'sr
JJeqI'J^rlJeur sr
,{dor
aqt os
'puJ
rql ot ,{gny
paarord
tou
saop
uauo
uorldrJ)suerl esra^au
'JruouaS
eql 01uI
,{dor
VN(
e
uJsur
suraloJd Jq1 aJeqm'snJIJnu
eql ot sJloru
uJqt
xalduoJ
urelordoalJnu
-oqIJ
eql
lPql
sMoqs
ix.;1I
;$fl*ij
'palelsuerl
araaa.daqt
q)lqM
rrroJJ
VNUIU
Jql ol Surpurq
JoJ JJueJalard-srr
p
Moqs
slrnpord uralord
aqt
'palelsuerl
sr
vN5ru
SUNIf
p
uaqM'uorteEp
-dord;o
por{lJru
neqt
qtru
sJrl
eArlJeJJJ os JJe
slueruJla
sIINIT
.,{.qu.
suoseal eql
Io
euo
'seuatopnasd
passarord
paler
are .{aqt
llnseJ
p
se
lldrnsuerl
eJnleu aqr
Jo
sarnleeJ aqt ssassod dflpnsn daql
'uorldlJJSupJ]
ro}
lutodtrets
leut8uo
eql
Jo
uperlsdn se^tzr
1eql
raloruord
aql
pe1
,i.aq1 :a,rrlreurzllrJpssaJJu
JJe
saruanbas JrruouJ8 Jql
'slduJsueJl
II
aspJaru
-,{1od
ypg
uro:; aleur8rJo sluauJlJ uJqM
'ky
tZ
arnSrg aas) suorlur elrqou
aruos Lq
pesn
sr ursrueqf,eru Jelrurs
y
'xaldnp
yN11
e olur
pauenuoJ
uJJq spq
1r
JJtJp ro aSels
slql
lp
JJrIlrJ deE aqr
Jo
pua
raqto
Jql ot
pe>luu
sr
prrq,{q
VNO/VNU
eqt
pue
'vNO
Jo
puprls
JJqlo Jql uado
ot
parmbar
sl
tuJ^a
a8e,reap
puo
-ras
y
'ateldurJl
vNu
eq1 uo
yNqJ
yo
srsaqluds
saruud
teql
puJ
HO-,€
e sapnord
pru
aqJ
'>ptu
Jqt
tp
punoq
uralord aql
qlrm
sJlprf,ossp
lueru
-ela
eql
Jo
lrnpord
yNg
aql
'uosodorlat
aqt z(q
papo)
dlrnrpe
JspJlJnuopua
ue
dq
atls
la8rel
yNCI
aqt ur eppru sr >lJru
y'uorlrsodsuerl
trod
-dns
satltntDe Jsaql ,lroq smoqs
*9"fri *lii]*lj
'uolllsoosuPrl
ro;
pannbar
eJp sl)npord
qroq
lserlrnrlJp
JspelJnuopua
pue
asetdrJJsuerl JSJJAJJ
qloq
seq
Z{UO
pue
uralord Surpurq-y1qq
p
sl
IdUO
'If
SINIf
uerunq
aqt uJ'.{rrarttp JSpJIJnuopuJ
up JoJ Jpo)
pup
saruanbas a>1q-asetdrrtsuerl
asraAeJ aneq l.11errd,{l
rnq
'suretuop
aser8
-Jlur
ro
asBalord se
qJns
/suorlf,unJ
IpJrAoJleJ
Jql
yo
,{.ueru
>lJel sluJrualJ
Jsaql ur sJrupJJ Supear
uado aq1
'g'77
an8q aas)
911
aql
qr1,lr
srred
rarutrd
vNUt
e
qJIqM
ur uorlJpar
lerrd,i(t
aqr
eAIoAur
1ou
seop
lJ
2paruud
uo4dursuerl asrJAaJ
sI
.ttoH :uorlsanb
aq1 sasod srqJ
'stueualJ
IprrAoJlJJ;o
1errd,{r
aJe
leql
sUJl Jqt ur Jteurur
-Ie1
lou
op
'sraqlo
aruos
pue
'sluJruala
sgNIT
'uoqdursuerl
aslanel aurud o1
llru
e salpraua6
1eq1
asealrnuopua up loJ apor ol
uosodorlaj
aq1 arrnbar
pue
sUIl a^pq
lou
op
SINII
.
pul
6urrulrd P
aleraua9
ol osPallnuopul
999
+
VNU
t
+
,
/tops
secepderyp6
*s
t
HO-,0
urorl srvrorb
ypgc
t
I
t
I
t
or"
bururrd
saprnord >1cr1
I
uP asn
slNIl
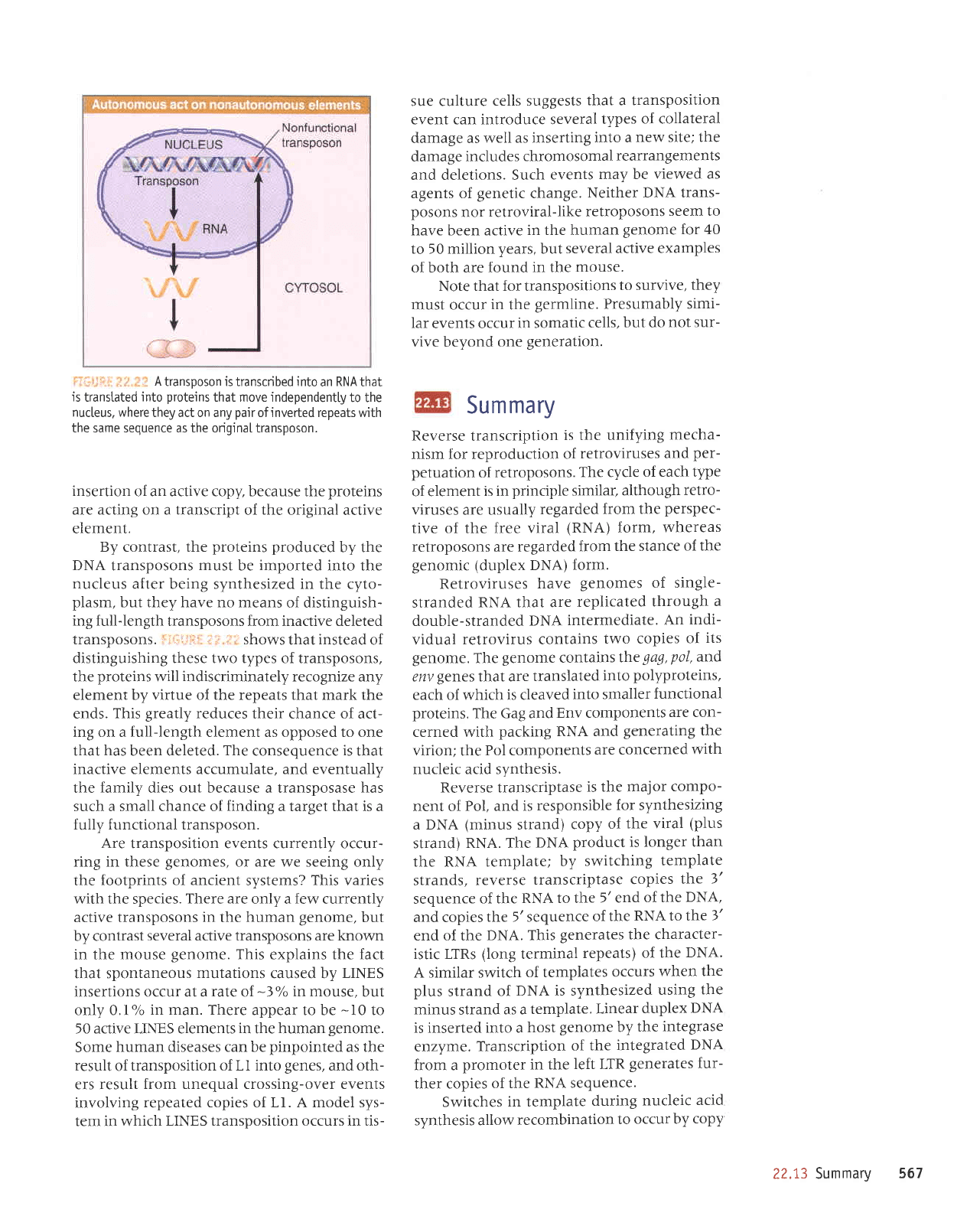
ri,,,:r,.,'
Atransposonistranscribedintoan
RNAthat
is
transtated
into
proteins
that move independently
to the
nucleus, where
they act on any
pair
ofinverted repeats with
the same sequence as the originaI transposon.
insertion
of an active copy, because the
proteins
are acting on a transcript
of
the
original active
element.
By contrast, the
proteins produced
by the
DNA
transposons
must
be
imported
into the
nucleus after being synthesized in the cyto-
plasm,
but they have no means
of distinguish-
ing full-length transposons from inactive deleted
transposons.
r-r;,r:ril
,r
:t .r
i
shows
that
instead
of
distinguishing these two types of transposons,
the
proteins
will
indiscriminately
recognize any
element by virtue of the repeats that mark the
ends.
This
greatly
reduces
their chance of act-
ing on a full-length element as opposed to one
that
has
been deleted.
The
consequence
is
that
inactive elements accumulate, and eventually
the
family
dies
out
because a transposase
has
such a small chance of finding a target that is a
fully functional transposon.
Are
transposition events currently occur-
ring in these
genomes,
or are we seeing only
the
footprints of ancient
systems?
This
varies
with the species.
There are
only a
few
currently
active transposons
in
the human
genome,
but
by contrast
several active transposons are known
in the mouse
genome.
This
explains the fact
that spontaneous
mutations
caused by
LINES
insertions occur at a
rate
of
-3o/"
in mouse,
but
only 0.17o
in man. There
appear to be
-10
to
50 active LINES elements
in
the human
genome.
Some
human diseases can be
pinpointed
as the
result
of
transposition of LI into
genes,
and oth-
ers result
from unequal crossing-over events
involving
repeated copies
of
LI. A model sys-
tem in which
LINES transposition occurs in tis-
sue culture
cells suggests
that a transposition
event can introduce several
types
of collateral
damage as
well as inserting
into a
new site; the
damage
includes chromosomal
rearrangements
and deletions. Such
events
may be
viewed as
agents of
genetic
change.
Neither
DNA trans-
posons
nor
retroviral-like
retroposons
seem to
have been active
in the human
genome for 40
to 50 million
years,
but
several active
examples
of both are found
in the
mouse.
Note
that for transpositions
to survive,
they
must occur
in
the
germline. Presumably
simi-
Iar
events occur
in somatic
cells,
but do
not sur-
vive beyond one
generation.
Sum
mary
Reverse
transcription
is the unifying
mecha-
nism for reproduction
of
retroviruses
and
per-
petuation
of
retroposons.
The cycle
of each type
of element
is in
principle
similar,
although
retro-
viruses are usually
regarded
from the
perspec-
tive of the free
viral
(RNA)
form,
whereas
retroposons
are regarded
from
the stance
of the
genomic
(duplex
DNA)
form.
Retroviruses
have
genomes of single-
stranded
RNA that are
replicated
through
a
double-stranded
DNA
intermediate.
An
indi-
vidual
retrovirus contains
two copies
of
its
genome.
The
genome
contains
the
gag, pol,
and
env
ge\es
that are
translated
into
polyproteins,
each of which is cleaved
into smaller
functional
proteins.
The Gag and
Env components
are con-
cerned with
packing RNA and
generating the
virion;
the
Pol components
are concerned
with
nucleic acid synthesis.
Reverse transcriptase
is the major
compo-
nent of
Pol,
and
is
responsible
for synthesizing
a DNA
(minus
strand)
copy
of
the viral
(plus
strand)
RNA. The
DNA
product is longer than
the RNA template;
by
switching
template
strands,
reverse
transcriptase
copies
the
3'
sequence
of the
RNA to the
5'end
of the
DNA,
and copies
the 5'sequence
of
the
RNA to the
l'
end of the
DNA. This
generates the character-
istic LTRs
(long
terminal
repeats)
of the
DNA.
A similar switch
of templates
occurs
when the
plus
strand
of
DNA is synthesized
using
the
minus
strand
as a
template.
Linear
duplex
DNA
is inserted into a
host
genome
by
the
integrase
enzyme.
Transcription
of
the integrated
DNA
from a
promoter
in the
left LTR
generates
fur-
ther copies
of the
RNA sequence.
Switches
in template
during
nucleic
acid
synthesis allow
recombination
to
occur by
copy
22.13 Summary
567

choice. During
an infective
cycle, a
retrovirus
may exchange
part
of its
usual sequence for a
cellular
sequence; the resulting
virus is
usually
replication-defective,
but can
be
perpetuated
in
the course
of a
joint
infection
with a helper
virus.
Many
of the defective
viruses have
gained
an RNA
version
(v-onc)
of a cellular
gene (c-onc)
The
onc sequence
may be any
one of a number
of
genes
whose expression
inv-oncform
causes
the
cell to
be transformed into
a tumorisenic
phenotype.
The integration
event
generates
direct tar-
get
repeats
(like
transposons that mobilize via
DNA).
An inserted
provirus
therefore has direct
terminal
repeats
of the LTRs, flanked
by short
repeats
of target
DNA. Mammalian
and avian
genomes
have
endogenous
(inactive)
proviruses
with
such
structures.
Other elements
with
this
organization
have
been found in
a variety of
genomes,
most
notably in
S. cerevisiae
arrd
D. melanogaster.
p
elements
of
yeast
and copia ele-
ments
of flies
have coding
sequences with
homology
to reverse
transcriptase and mobi-
Iize via
an RNA form.
They may
generate par-
ticles
resembling
viruses,
but do not have
infectious
capability. The
LINES sequences
of
mammalian genomes
are further removed
from
the retroviruses,
but retain enough
similarities
to
suggest a
common origin.
They use a differ-
ent type
of
priming
event to initiate reverse
transcription,
in
which an endonuclease
activ-
ity
associated
with the reverse
transcriptase
makes
a nick that
provides
a 3'-OH
end for
priming
synthesis
on an RNA template.
The fre-
quency
of LINES
transposition is
increased
because
its
protein products
are cls-acting;
they
associate
with
the mRNA from
which
they were
translated
to form
a ribonucleoprotein
complex
that is
transported
into the
nucleus.
The
members
of another
class
of
retro-
posons
have
the hallmarks
of transposition
via
RNA,
but
have no
coding sequences
(or
at
least
none
resembling
retroviral
functions).
They
may have
originated
as
passengers
in a
retroviral-like
transposition
event, in which
an RNA
was a target
for a reverse
transcrip-
tase.
Processed pseudogenes
arise by such
events.
A
particularly
prominent
family
that
appears
to have
originated
from a
processing
event
is the
mammalian
SINES;
it includes
the
human
Alu
family.
Some
snRNAs, including
7SL
snRNA
(a
component
of the SRP),
are
related
to this
family.
References
The
Retrovirus
Life
Cvcle Involves
Transposition-Like
Events
Review
Varmus,
H. E.
and
Brown,
P. O.
(1989).
Retro-
viruses. In Howe,
M. M. and Berg,
D. E., eds.,
Mobile DNA Washington,
DC: American
Soci-
ety for Microbiology,
pp.
53-108.
Resea rc h
Baltimore,
D.
(1970).
RNA-dependent
DNA
poly-
merase in
virions
of RNA
tumor
viruses.
Nature 226, 1209-l2ll.
Temin, H. M.
and
Mizutani,
S.
(1970).
RNA-
dependent DNA
polymerase
in
virions
of
Rous
sarcoma
virlus. Nature 226, l2ll-1213.
Virat DNA is
Generated bv Reverse
Transcription
Reviews
I(atz, R. A.
and Skalka, A. M.
(1994).
The retrovi-
ral
enzymes. Annu. Rev. Biochem.
63,
B3-17J.
Lai, M. M.
C.
(
1992). RNA recombination
in
ani-
mal and
plant
viruses.
Microbiol
Rev.
56, 6l-79.
Negroni, M. and Buc, H.
(2001).
Mechanisms
of
retroviral recombination.
Annu.
Rev.
G
enet.
3 5.
275-302.
Resea rc h
Hu,
W. S. and Temin, H. M.
(1990).
Retroviral
recombination
and
reverse
transcription.
Sci
ence 250, 1227-12)).
Negroni,
M.
and Buc, H.
(2000).
Copy-choice
recombination
by reverse
transcriptases:
reshuffling
of
genetic
markers
mediated
by
RNA
chaperones. Proc
Natl. Acad
Sci. USA 97
6385-6390.
Viral. DNA
Integrates into
the Chromosome
Review
Goff, S. P.
(1992).
Genetics of retroviral
integra-
Iion. Annu. Rev.
Genet.26, 527-544.
Resea rc h
Craigie, R., Fujiwara,
T., and Bushman,
F.
(1990).
The IN
protein
of Moloney murine
leukemia
virus
processes
the
viral DNA
ends and
accomplishes
their integration
in vitro
Cell 62,
829-837.
CHAPTER
22
Retroviruses
and Retroposons

699
salualaJau
'8f.€-LZ{-
'}il
ltD
or11^ ut
(L\Iqelsur
JrleueE
qlrM
pelP
-rrosse
sr uorllsodsuprlorlrr
I
I
uprunH
'
(ZOOZI
'q 't'r>lrog
pup
''C 'gerSruue4 ''f 'g 'tso:)
''W'g'otnde)'J'S'IezS''3'zi.1auuo3''g'g
tau.dg
'
Le6- LI6'
L8
Ua)'sllar
uellPluluelu
pernllnr
ur uorlrsodsuprtortor
,{.ruanbary
qBU
'(gOOt
)
'H
H'uevezex
pue
''o
f
'e{eog ''f
'u 'slu
-rprPregeq
'a 'I'SPPN
"g
's 'seuloH 'A
'f 'uProw
'
SO9-S6E' Z L
I
p)'
uor
lrsodsup:ro.r
lo.t
ull-uou
roJ trsrueq)JuI
p
:Jlrs
ta8.ret
leuos
-oruorq)
eqt
lp
>l)ru e ,{q
parurrd
sl
VNU
urgzu
;o
uorldrnsuerl asralag'
(tOOt)'lp
le'('O'uen'I
'rs86-1V86 ',r6
VSn
'DS'pwv'lloN
)ord'uontsodsuertorlar
1,{tr
Eurrnp
pelrrequr
lou
sr aruanbes
VNU1
-rarurrd
rqJ
(766I
)
'C 'I 'a>lrog
pup
.n 'uuplurene'I
'
9Z€.-S t €' OI I
t I
D'
uor
lrsodsue:rorlrr
I
-:lNll
uodn
palearc
suorlalap )ruoua5r
'(ZOOZI
A
'f 'uerow
pue "S
'aE8rr4-21n1
''N 'uaqllD
'9
I 6-s06' Lg
il
D'uorlrsodsuello:ta.t
ro;
palnber
JspJIJnuopuJ
palrasuoJ
e sapoJue
uosodsuerlorlrr
I'I
uerunH
'lgOOt)'(
f
'a{eog
pue
''H 'H'ue\zeze>I'1 't'uelow ''o
'8uad
q
lr Pasau
'8€S-I0E'E€'puaD
^aY
'nuuy'suosodsuprloJleJ
11
uerleruureu
;o
.{3o
-lols'(I002)'H'H'ueveze>I
pup'W'E'6et:etso
Mar^au
pu3
6urtuu6
e
alelaua! o]
esPallnuopul
uP
asn
slNIl
'299-0zE'jzv
a]nt'N
'atuoua8
Jsnour aql;o srsLyeue alrleredruol
pue
Sunuanbrs
Iertrul
'(ZOOZ) 'lp
la
uolsralpM
'€€6-826'6OV
arnlttN
'dnorg
3urryo6
deW
aNS
Ipuorlpuralul
eqJ
'susrqdroru.dlod
aprtoalrnu
a18urs uolllnu
Zt'
I
Eurureluol uorlplJel aruanbas
aruouaE
upunq
yo
deru
v
'(I002)
Ip
ta
'U 'ruppueueplq)es
'z8l-89
l'9'
lotg tlal
pw'suosod
-sueJloJleJ
ur
punoJ
sarnleeJ
lera^as
puP
pue
,E
petpedrr,{1ruapue1
e slpeler
tuourale
py{I'I
a8rel e;o aruanbes aqJ
'(986I) 'le
ta
'C 'O
'qao'I
'v6zl-t8zr 'v
ila)
7ua14
'sauaB
azrpru
qtrM pelprJossp .{lruanbar;
sluerurle
tpedar
pJuaAuI
llerus;o
,{1uue; a8rel
e
:lsunoJ,
'keat)
'u 's
ialsseM
puP
'a 'I 'nPerng
'
6ZS8-VZS8't6
VSn
DS'pnv'UoN
'cot4 'saua?
aru edLl-p1u
ur sluaruala
leadar
-petrJAUr
IIptus Jo
JJueurruopeld
aqt
sleona.r
,{arrrns rrlBruats.{s
paseq-ralndruo)
V'
(
966 I )
'U 'S
telsseM
pup
'')
a
'plPuou
''g 'J 'nPerng
Ll llPasau
'I99-I€9
'gS
'waLpotg
'^a[ 'nuuv
'uorleruJoJul
ctlauaS
Jo
.lrolJ asrazrer
aql {q
paleraueE slueurela
alqesodsuerl
pue
'saua8
-opnasd'saua8
:suosodorla.r
1er1,ruoN'(9861
)
'Y 'slpellPrlsJf,
puP
'"I
4
taSururaq "w
'v
teuleM
'8€S-I0E 'S€'puaD'^aa
nuuy
'suosodsuerloJler
11
uellerurueu;o
,{6o
-lolS'
(
I O0z)'H'
H'
ue\zeze>I
pue'W'
g'3egets6
'LIg-f
65'dd
',{5o1orqonrry
ro;
.{1a
-r)os
upJrJJruy
:1q
'uol8urqseM'VNO
altqow
''spa
''g 'q '8rag
pue
'W'W
'e/!{oH
u1
'aruoua8
)rlo^re>lna
aqt uI
seJuonbas
pateadar
pasrads
-ra1ur
3uo1
:suosodortal
prtelar
pue
SgNI'I
'(OgOt)
'H'I/{
'lleEpg
pue
''u .1'1'eaqrqs
"o
'('reo'I '')
's 'salpreH
"Y'1
') 'uoslqrlnH
'
9€9-619'dd,lEo1otqonq41
ro;
[1anog ueJrratuv
:3q
'uol8urqseM'VNQ
afqow
''spe
''g'q 'Erag
puP'W'I
l'a,lloH
uI
'seto.fuplna
raq8rq
uI stuauala
y51q paleedar
pasradsralur
troqs
:sgNIS
'(OgOt)
'f
a
te8ulurac
sMa[Aau
sessell
aarql
olut
llPl
suosooollau
'8€9I-0€
9I
'
t
tolg Ip)
7o74'suralord
Iprllorter
pue
erdor
ueaMlaq,{Eoloruoq
:erdor
luaruala
alqesod
-sue[
pqtldosote
erlt
;o
aruanbas
apltoelJnu
arelduro3
'(SeOt)
'W 'D 'ulqng
pue
'W'S 'lunory
q
ll
eesau
raJsDDOuDlaw
'0
ur aplsau
slueuroll
alqesodsuell
nuPW
'00s-r6v
'ov
il:D'alPrpeuralul
vNu
uP
qEnorql
asodsuBrl
sluatuela,lr'(SSOt)'lp
ta'o'f
'a>laog
qllPaseu
sosnrnorlau
elquaseu
sluouell
f1
1sea1
'uotlrsoosup.l]
ur slnllo
lpt{l
uorllpal
uorlnlosar
alrl-esptauosrodol
oql salqulasot
uorJlpat oLll
r
'laulPloaq
aql
le
sa^ealt
pue
lgvu
01 spurq
zgvu
'uorleurqulo)al
l0J satuanbas
snsuasuol
taulpuou eql
sozlu6olel
lgvu
.
'uorlleel
e6P
-^Palr
eql
lo]
luaDlJJns
pue
^ipssalou
olp
suralold
gvu
aHl
r
uolunou pue
abplpalg
azflplpl
suralold
gvu
eql
'(pebuerear
flentlrnpold
sr urpql Aneaq
auo)
sureqt
A^paq
o1
pue
(pabueuea-r
flanrlrnpotd
aq
r{eu
epqurel
to eddel
euo fluo)
sureqr
lq6q
ol nlalelpdas
sarlddp uorsnllxa
lrlallv
.
'10u
saop
lueur
-aEueuear
anrlrnporduou
e seataqM
'6uutnlro
uolJ
luoul
-abuej.lpol
raqlnJ l{ue
sluanatd
Juauo6uellpa.l
a^rllnpold
V
.
'uLelord
a^rlle
up
1o
uorssardxa
01
speal
1L;r
anLlrnpord
sr
auab
uLuqolbounuur
llelur
ue eletaua6
o] uor]€utQr!O)a!
o
luaue6uPlleau
a^qlnpold
fq
para66u1
sI uorsnllxl
lrtallv
'alllD
pesDxa
uP
J0
uorlalap
Jo
ppalsur
uotsla^ur
up sr
alaql
,uorlelue
-u0
lra.lrp
J0
ppalsur
pa1.talut
alp saua6 bururquorat
eHl
JI
o
'(ureqr
g)
l-C-0
ot
A
pue
l-t
o1
6
ro,(ureqr
1)
l-f
ol
A
urof
o]
palurt
fi11ua1enor
olp spua Eurpor
eq1
e
'luaubptJ
tpinl.lo pasoxa
up elelauab
ol urol
Allensn
sleatq
aql uaemlaq
luaLubetl
oql
Jo
spua
leu6rs
aq1
o
'saruanbas
snsuasuol
oMl
Jo
slaupl
-deq
eq1
lp
s)potq
puplls-alqnop
Aq
srnrro uorJputQruoJO!
r
su0r.sla^UI
lo suoqala0
salelauag
uollpurquolau
'sbuDeds
luera#rp
a^pq
leql
saluenbas
snsuasuol
oMl
uaaMleq
slntlo uorleutquoJO!
o
'lauPuou
e
urolJ s.lrpd
aspq
€Z
lo
ZI
leqlta r{q paletedas
tauel
-daq p
sr uorlpurquolol
toJ
pasn
aluonbos snsuosuot
all
o
aluanbas
snsuosuol
Jo
sad^t o,1
I
sasn uotleurquolau
aunuul
'sluauEes
C
inoJ
pup
'sluaubas
6
97
,sauab
1
ggg
bururquor fq
suLeqr
gOOt<
atnpold uel
snlol
H
uV
r
'sauo6
I
a^q ol lno1
qluvr
seua6
1
ggg
6uLurquor fq
suLeqr
gggl<
ernpotd
upl snlol
urptll
lqbrl
V
.
AlrsranLg
ontsualxl
salelauag
uotJpurquioleu
oL9
'su0x0
lPlaAos Jo
slstsuol
luaubas
I
e{l
r
'l-C-q
ol
A
suroI uorlpurQruo]et
puolas
alf
r
'l-t
ol
0
surol uo4eurquo]o.l
lst!
e{l
r
'lueu6es
auab
l-t
p
pup
']ueuDos
0
p
'eue6
A
e
arp uorlpurquolal urpql {neaq rol
slrun all
r
suoqeurquroleu oml riq
palquassy
alv surpql
n^eaH
'aueb
3
aq1 6urpar
-erd
slueubes
t
a^!JJo auo
pue
luaubas
auab
A
e uoamlaq
uorleurquolal
al6urs
p
fq
palqruasse
sr urpql
lq6rt
edde)
v
.
'uorbel
6urpol-l
pup
'uorlur 'uoxa
6uLpor-1
iloqs
p
spq
]uauEes
euob
l-C
aql
.
'uor6et
6urpor
-alqeue^
pue
'uorlur'uoxa
loppel
p
spq
luau6os
aue6
4
eq1
r
'lueubas
auab
l-f
e
pue
auab
A
p
uoamloq
uoqeurquola.l a16uLs
e
fiq
palquasse
sr urpqt
lq6rl
ppqupl
V
.
uoqeulquorou albuls
e
Aq
palqurassv
arv sureql
lq6rl
'luaurbesauaEIpqluv\
luaubes
euo6
A
e uLo[ o1 uorleurquo]el
tqpuos [q
pale
-rauob
sr uLeqt
urtnqol6ounululr
llplur
ue loJ 6urpor auab
y
r
'sluaubas
aua6
J
pue
sluaubas
auab
A
fq papor
r\leleredas
ate suleuop
I
pup
surputop
fi
o
'asuo0s0r
lolroJJe
aql
saprnord urpurop
I
eql
pue
ua6rlup sezru6orar
ureuop
A
eql
r
'(1)
uorbar
luelsuol
lPurulal-l
p
pue
(A)
uor6ar alqeuea
leurulal-N
up sptl urpLll
r]tpl
o
'AlruPJ
albur.s P ut.loJ
sulPql
fireeq lseqruel
eddel
pue
ppquel
aql olul
lleJ
suleqr
1q611
o
'suleql
A^Poq oMl
pup
surpql
lqbq
om1
1o
taupllal
p
sr urlnqolboufiutulr
u!
r
salfloqdu^l
ur
sued rlaql
uolJ
pelqutassv
alv sauag urtnqol6ounuul
'llar
oql
Jo
uorlerqdrllnu
lpuoll
sle66ul
loldalor
llel
I
ro
urlnqolEounuulI
ue ol 6utpu[q uabrluy
o
'sroldalor
ller I
pue
sutlnqolbounuutlo fi1auen
ebtel r{ren
p
sr o.laLll
o
'roldarer
llal f
olEurs
p
sassaldxa afroqdur{1
1
qree
pup
urtnqolbounuur
alburs e
sassaldxa a$roqdrufl
g
qlpl
.
suaDquv
lenpwpul
ol
puodsau
lpql
sailloqdu^l
sa4qdu.ry uolpalas
lpuoll
u0qlnp0lluJ
3NI]J-NO
U3J-dVH3
@
GE'|
fzil
fi11srenlg
eunuul
