Lewin Benjamin (ed.) Genes IX
Подождите немного. Документ загружается.

tz9
abod
jrau
uo
panuLluo)
'luauber;
luluolp euo
pup pua
ualolq
p
pue
arauorluel
e seq
lpql
euosouro.lql
auo seleraue6
>lparQ
a{l
o
,,'sluauala
Du rll0.l1u0r.,
J0
uorl
-rsodsuerl
r{q
palerauab
slparq aurosourorqr oql
Jo
s}laJJa
oql
Jo
osnPlaq
paiaAOlstp
sPM azteu Ut U0tllS005UPJf
r
sluaua6uareau
pue
a6p)palg asnpl azrehJ ur sluaulall 6uq'|or1uo1
'uorlrsodsup.ll
Jo
alp.l aql
llalJp
sursrupqrau a1dr11nj1
o
'euroue6
aql olur
palnpo.llur
alP
uosodsuPrl
eues eql
Jo
serdor raqlo ueqM uosodsuerl
p
Jo
^dol ouo
r\ue
1o
uorlrsodsuerl
Jo
alpr aql selnper uoqrqrqur r\dorq1ny1
o
slolluol aldqlnhl seH
0IUl
Jo
uor.lr.sodsuPrl
'vN0
0q1
01
punoq
Allualenor sr uralo.ld aql
qlrqM
ur olPrpauralur uP
qbnor q1
ssed
qrLqm'suorllsar
uor.lpurqurolel rgoads-e1rs
al!l-aspraurostodol;o
r\1Lue1
lerauab
aq1
o1 sbuoleq
pue
ut010l0
lul
epqulel salquasol ase^losal 0{l
}0
uoL}lP
all
e
'suoltldai
oMl
eql espelar ol asp^losar e
pup
alnlln.lls alelboluror aql uro1
ol
espsodsuprl
p
sa.lrnbar
Vul
Jo
uorlrsodsup.ll o^rlplrldau
o
3se^losau
puP
asPsodsuerl salrnbau uolllsodsuerl
vul
'(orur)
labrel
atll
o1 6uLurof aro;oq
lnr
are spuplls lnoJ
llp
raqtaqM
ro'(9u1)
1nr
are rred
puoras
aql ero;aq
1a6re1
aq1 o1
pauroI
orp spuells
uosodsuerl
1o
rred
lsrg
aq1
reqleqm o1 6ur
-prolle
regrp uorlLsodsupll alrlelrldaruou roJ
sAenqled oml
o
'pa1e6Lt
are uosodsuerl aql
Jo
aprs raqlra uo spuplls
1a6
-rPl
aql
puP
spuPrls louop
Jo
lrPo ualolqun aql uo
pallru
sr
alnpnrls
ronossorl
p
Jr
sllnsel uorlrsodsuell anrlelqderuoll
.
uo!unau
pue
a6eleerg
Aq spaa:or6 uorlrsodsuerl a^qPlrldaluoN
'uosodsue.ll
aq1 r\q
papor
asellosel
e Aq
pazAleler
si uor+leal uorlpurquolar aql
.
'uoso0suerl
aql
Jo
[dor e
peureb
seq
luerdner
aql
lnq
'suorrldar
leut6uo
eq1
salplauabar saldor
uosodsuell
aql uaemlaQ
uorlpurQruola!
o
'suoltldat
leutEuo
aql
uaaMleq
art
qlrqM
'uosodsuerl
eq1
1o
sardor oMl sPq alerbaluLor aq1
0
'suorrtdar
1abre1
pue
rouop eql
Jo
uorsn1
p
sr
qrrqm
'a1er6
-alurol
e salelauab
xalduor
lajsueil
puerls
P;o
uorlerqdax
r
elubalurol e
qbnorql
spealord uor.llsodsuerl a^qeltldou
'parreda.r
sr.lt
Jt
sMolloJ
uoLltsodsuell
e^tlPltldaluou
pue
palerrldar sL
xalduor
aql
Jt
s/v\olloJ
uorlLsodsue.rl
antlerqdeg .
'uolllPal
lojsuPll
puprls
p
uaql
pup
'6ur1rtu
r\q
pamollo;
'VN6
n111
j0
spua
aql 6ursdpur\s
[q xalduor
aql suloJ
osesodsuPll
nlrl a!]
o
'pue
qlea
le
puerls
auo
qbnotql
alLs
lebrel
aql
ol
papeuuor
s1 uosodsuell
aql
qltqM
uL
xalduor
raJsuerl
pueils
e
6ututtol
Aq s1e1s uolllsodsuell .
uotltsodsuPll
loj salPlpaulalul
u0tll txol
'uorsnxo
astlaldLut
to astlald
ol
pPal
,{Pu uosod
-suprl
P;o
sleader
aql uaaMlaq
uotlPutquolal
snoboloulo;-1
o
'vNc
lsoq
Jo
lueuoouelleel
sasnel
uosodsuPll
e;o satdor
a1d41ntu
uaaMlaq
uoqeutquolal
sno6o1otlo11
o
vN0
Jo
]uaua6uelleau
asneJ
suosodsuerl
'arrqertldaluou
lo a^qeltldal
st uotltsodsuell
loqlaqM
autulalap
ylq
1e6le1
pue
uosodsuPll
ueaMlaq
suotllauuol
oql
J0
elnleu
pexa pue
sluaAa
J0
lap.lo
all
r
'pollg
a.rp sdeb
aq1
puP
'spuo
6urpntlord
aq1
o1
pauLof
s! uosodsuprl
aq]'VN6
1a6te1
ur apPul
olP slrtu
palo6
-6e1s
qrLqrvr
ul utstuPqlout
uotutuol
P asn suosodsuet]
ll!
r
sulsruPqlahl
a^qel!ldeluoN
pue
a^qelrldau
qro8 r{q srnr:g uolJtsodsuPll
Alluapu00eput
asodsuetl
osle
r\eu
pua
leqlto
lP luauala
SI
a^qlP
ue
lnq
'lrun
P sP osodsuell
Aeu uosodsupll
allsoduol
v
.
'uotltsodsuPtl
alPilopun
ol alqe aq
feu uosod
-suer1
alLsodtuol
e
Jo
sluauale
SI
aql
Jo
q10q
lo auo la{}tl
e
'pua
qlee
lP
luaualo
sI
ue
[q
pa1ue1;
uot6ar
letluar
e a^Pq
suosodsuPll
allsoduo]
.
'uotltsodsuetl
tol
6ut
-pol
esoql ol
uotltppP
ut sauab
loqlo
Aller uPl
suosodsuell
.
salnpow
sI
a^PH
suosodsuPll
alrsoduol
'uosodsuerl
rPlnrHed
r\ue lo; rLlst
-rellereql
sr
pue
dq
6
ol
9
s!
lPadai
pattp
aq1
1o
q16ua1
aq1
o
'uosodsuell
oql
Jo
spua aql
le
uoqPluauo
paitp
ur
sleadol
oml
utlo; o1
ssaootd uotilosut
aq1
6uunp
paler
-Ltdnp
sL
pailasur s! uosodsuell
P
qltqm
1e
alts
1a6te1
aq1
o
'sleed0l
lPululal
poile^ut
loqs
[q
paluep
uotltsodsuell.lo]
papeeu
(s)auAzua
aql
roJ sapol
leql
uosodsuPll
P sJ
aluanbes
uot|.losu]
u[
r
salnpohl
uorlrsodsuPrl
aldults
alv
saluenbos
uoHasul
uoqlnpollul
3Nr''lrn0
ulrdvHS
'slue^a
qlns
loj
sla6
-rel
apr^old uel
suosodsupll
Jo
satdol
.sluaua6upllpel
sosnPl
soluanbos
palplol
uoaMleq
la^o 6urssojl
lpnbaun
'uorssardxa
aueb
uo saluanbasuot
llalrp
anpq r\eu
srq-1
'olrs
Meu
e o1 uosodsuetl
p
Jo
luoualou
aql
st auouab
e
urqlrm
ebueqr etuenbes
Jo
asnpt
tofeu
y
l"il
ldft*IJ
sacuanDos
peleloJ
uoeMloq
srncco
rano-6urssolc
lenbaul
I
alrs r.uopupl
1e
{doc
rr,reu
seleleue6
uosodsuell
suoso0suerl
Iz
uSIdvHl
zzg
'i''t;
3sfiL1:j
ur
pJzrrpruruns
eJe
sJSneJ
roleu aql;o
o^41
'JruouJB
aql o1
puratur
sassarord
dq
perosuods
Jrp sluJure8-ueJreJU
'(sJJuanDes
relnllrf,
eJnpsue{l
.de141
sasnrr,r o
nep'
g'
ZZ
uorpa
5
aas
)
ap,{.r
aarDa;ur
ue SurJnp
uolleruJoJur
)ltaue8
]JJSUEII
UEJ
,SJSNIIAOITJJ
Jql Z{IqCTOU
'SJSNJIA
auos
'seloIJe>lne
uI
'(erralreg
Jo IerJJtpW
JIlJueg
aql sI
vNo
'z'I
uorllJs aas)
uorleruro;
-sueJl
Jo
sueatu,{q
errJDeq
Jtuos uJJMleq
sJnJJo
VNq1
Jo
rJJSueJt
tf,JJrC
'uotqdar
uMo JrJql r{lrM
3uo1e
saua8
tsoq
JaJsueJl dlleuorserro
sa8eqd
pue
spruspld qrog
'(sar8alerlg
a8eq4
'VI
Dt
-deql
aas)
uorpalur,{q
peards
sa8eqd
searJqm
'(vNo
pJpuerts-JISutg
sra;sue41
uorle8nluo3
'0I '9I
uollJa5
aas)
uotlB8nfuor .dq
anoru spnu
-se1d
'erralrpq
uI
'lerJJteru
trlaua8
yo
sqr8ual
(goqs
raqter,{.11ensn)
Jo
raJsuerl
aqt Surleparu
Lq,'(11e1uozlJotl
uollpluJoJur
Jlour
sluJruala
IeuosoruoJqJerlx!['saruoua3
ueJMleq uorlpur
-ro;ur.{.rrer
ol sJolJaA
Jo
,{l1tep
aql
ruoJJ sllnsJr
saruanbas
zlrau
Jo
uorlJnpoJtur
ueppns
JqJ
'saruanb
-Js
Surlsrxa
Sur8uerrear
,{q
pue
saruanbas
zltau
Surrrnbre
,{.q
qloq
JAIOAJ
saruouJD
uoqlnporlul
Aleuuns
'alrileJ
ureuol
sassoll
0lpuej
d
x
alpu
l^l
fqrvr
sureldxa
tossatdar
eq1
1o
aruasetd
aq1
o
'use1do1r\r
aq1 uL [1eu
-.leleu
paluoqur
sr
qtrqM
'uorlrsodsuell
;o
rosserdat
e sarnpord
olp
luauala
d
aUI
.
'asesodsuerl
aq1
ro; aruanbas
buLpo:
eq1 saletauab qlrqM
,uollut
auo senouro.l
luana
6uoqds
rgoads-anssrl
P asnPlaq
sossoll
alpueJ
l^l
x
aleur
d
J0
euqurab
aql ut
polp^tllp
o.lp
sJuautola
I
r
aurlrulsg
aql ur
palp^rllv
etv
sluauall
d
'alIIoJUr
ssoll
oql
solPul
pue
sauaE fueu
selenrpeur
sasso.lt
aseql
u[ sells
Mou
|P
sluotxola
d
J0
uotilesut
aql
.
'pele^rlte
sI uorlrsodsuPlJ
'aleual
[/\l
ue
qlrM
possoll
sr
olpu
d
e ua![r\
r
'surplls
l^l
ur
lou ]nq
,alsD,ouDlaw
o1rydosot6
Jo
surp.lls
d
ur
pau
-lPl
ele
lPql
suosodsuP.l]
e.lP
slueu0la
I
o
srsaue6sfg puqfp
ur
sluauall
elqesodsuerl
Jo
aloU€ql
'uorlel^qleu
Aq
polp^rlleur
are
sluaLuale
ud5
o
'uosodsuerl
eql
Jo
spua
oql
lp
salrs
la6rel
slr o1
spurq uralord
ydut
eq1 uaqM
'uorilesur
jo
salrs
.rroql
1e
uorssardxe
eue6
lrage
sluauale
Lud5
o
uorssardx3
auag
aluan1;ul
s1uarua13
urd5
'uot1e1r\q1aur
Jo
alpls aq1
ur sabueqr
Jo
llnsal
e se lelle
serladord
rraql
uaqM
'aseqd;o
sa6ueqr
o^eq
slueuala
6ur11ot1uor
snoulouoln!
r
'suLalord
&pssarau
aql sapn
-ord
luauela
snououolne
ue uaqrn asod
-suell
upl faql
inq
'uoLlrsodsuetl
az[1e1er
o1 flLreder
rraql
olpuuurlo
lpql
suorlelnu
elpq
slueurala
6uqlotluor
snOulouolnpu0[
r
'asodsuerl
ol ueql
alqpua
lpql
suralord
loJ epol
quaulala
6uqloiluor
snououolrl!
o
'sluaulola
6uqlorluor
sn0urouolneuou
pup
sn0ul0uolne
q10q
seq
ozrpul uL
suosodsuetl
1o
flrue; qre3
.
suoso0suPjl
J0
saurueJ
ulol
sluaulall 6urllolluol
'uorleEeuen
lqeuos
Jo
aluellnllo
aql loJ alqlsuodsol
sr
elrr{r abpuq-a6e1eatQ-uorsnJ
a{l
r
'uotsnJ
pue
a6eleetq
;o
salrfr
taq1n1
obrapun
qltqM
'seuos
-ouolql
luluelrp
seletauab
oulosouolql
oql
l0
spua ualotq
aql uaaMlaq
uolsrlJ
r
'e1oEr{zorelaq
e ur salelle
lueurulop
1o
arueread
-desrp
aq1 fq
papelep
aq upl srr.ll
jsrs
-o1rur
6uunp
1so1
sr
lueu6erJ
tulualp
orll
r
9'ri?7
w
Oi[Iill

Unequal recombination results from mis-
pairing
by the cellular systems for homologous
recombination. Nonreciprocal recombination
results in
duplication or rearrangement of loci
(see
Section 6.7, Unequal
Crossing-Over
Rearranges Gene Clusters). Duplication of
sequences within a
genome
provides
a
major
source
of new sequences.
One copy of the
sequence can
retain
its original function,
whereas
the other may evolve into
a
new func-
tion. Furthermore, significant
differences
between
individual
genomes
are found at the
molecular level because of
polymorphic
varia-
tions caused
by recombination.
We saw
in
Sec-
tion 6.14,
Minisatellites
Are Useful for Genetic
Mapping, that recombination
between
mini-
satellites adjusts their lengths so that every indi-
vidual
genome
is distinct.
Another major cause of variation is
pro-
vided by transposable elements or trans-
posons:
these are discrete sequences
in
the
genome
that are mobile-they
are able
to trans-
port
themselves
to other locations
within
the
genome.
The mark of a transposon is that
it
does
not utilize an independent form of the element
(such
as
phage
or
plasmid
DNA),
but
moves
directly
from
one site
in
the
genome
to another.
Unlike most otherprocesses involved in
genome
restructuring, transposition does
not rely on
any relationship between the sequences at the
donor and recipient sites. Transposons are
restricted
to moving themselves,
and sometimes
additional sequences, to new sites elsewhere
within the same
genome;
they are, therefore,
an
internal counterpart to the vectors that can
transport sequences
from
one
genome
to
another.
They may
provide
the
major
source
of
mutations in the
genome.
Transposons fall into two
general
classes.
The
groups
of transposons reviewed
in this
chapter exist as sequences of
DNA
coding
for
proteins
that are able directly to
manipulate
DNA so as to
propagate
themselves within
the
genome.
The transposons reviewed in
Chapter 22,
Retroviruses
and
Retroposons,
are
related to retroviruses, and the source
of
their mobility
is
the ability to
make DNA
copies
of their RNA transcripts; the
DNA
copies
then become integrated at
new
sites
in the
genome.
Transposons that
mobilize
via
DNA are
found in both
prokaryotes
and eukaryotes.
Each
bacterial
transposon carries
gene(s)
that code
for the
enzyme activities
required for its own
transposition,
although
it may
also
require
ancillary
functions of the
genome
in
which
it
resides
(such
as
DNA
polymerase
or
DNA
gyrase).
Comparable
systems
exist
in eukary-
otes,
although their
enzymatic
functions are
not so well characterized.
A
genome
may con-
tain both functional
and
nonfunctional
(defec-
tive) elements.
Often
the
majority of
elements
in a eukaryotic
genome are defective,
and
have
Iost the ability
to transpose
independently,
although
they may
still be
recognized
as sub-
strates for transposition
by
the enzymes
pro-
duced by
functional
transposons.
A eukaryotic
genome
contains
a large
number
and variety
of
transposons.
The
fly
genome has
>50
types
of
transposons,
with
a total
of several
hundred
individual elements.
Transposable
elements
can
promote
rearrangements
of the
genome
directly
or
indirectly:
.
The transposition
event
itself
may cause
deletions
or inversions
or lead
to the
movement
of
a host
sequence
to a
new
location.
.
Transposons
serve
as substrates
for
cellular
recombination
systems
by
func-
tioning
as
"portable
regions
of homol-
ogy"; lwo
coPies
of
a transPoson
at
different
locations
(even
on
different
chromosomes)
may
provide
sites
for
reciprocal
recombination.
Such
exchanges
result in deletions,
insertions,
inversions,
or
translocations.
The intermittent
activities'of
a transposon
seem to
provide a somewhat
nebulous
target
f or
natural
selection.
This
concern
has
prompted
suggestions
that
(at
Ieast
some)
transposable
elements
confer
neither advan-
tage
nor disadvantage
on
the
phenotype, but
could constitute
"selfish
DNA"-DNA
con-
cerned only
with
their
own
propagation.
Indeed,
in considering
transposition
as an
event
that is distinct
from
other
cellular
recombina-
tion systems,
we tacitly
accept
the view
that
the transposon
is an
independent
entity
that
resides
in the
genome.
Such a
relationship
of the
transposon
to the
genome
would
resemble
that of
a
parasite
with
its host. Presumably
the
propagation of an ele-
ment
by transposition
is balanced
by
the harm
done
if a transposition
event
inactivates
a nec-
essary
gene,
or
if the
number
of
transposons
becomes
a
burden
on
cellular
systems'
Yet we
must
remember
that
any
transposition
event
conferring
a selective
advantage-for
example,
a
genetic rearrangement-will
lead to
prefer-
ential
survival
of the
genome carrying
the
active
transDoson.
21.1
Introduction
@
Insertion
Sequences
Are
Sim
pLe
Transposition
Modules
.
An insertion
sequence is
a transposon
that codes
for
the enzyme(s)
needed for
transposition
ftanked
by short inverted
terminaI repeats.
r
The
target site at which
a transposon
is inserted is
dupl.icated
during the insertion
process
to form
two repeats in
direct orientation
at
the ends of the
transooson.
o
The
length of the
direct
repeat
is
5 to 9 bp
and
is
characteristic
for
a
ny
particu
[a r
transposo n.
Transposable
elements were
first identified
at
the molecular
level in
the form
of spontaneous
insertions
in
bacterial
operons. Such
an inser-
tion
prevents
transcription
and/or
translation
of
the
gene
in which
it is inserted.
Many
different
types
of transposable
elements
have now
been
characterized.
The
simplest transposons
are called inser-
tion
sequences
(reflecting
the
way in which
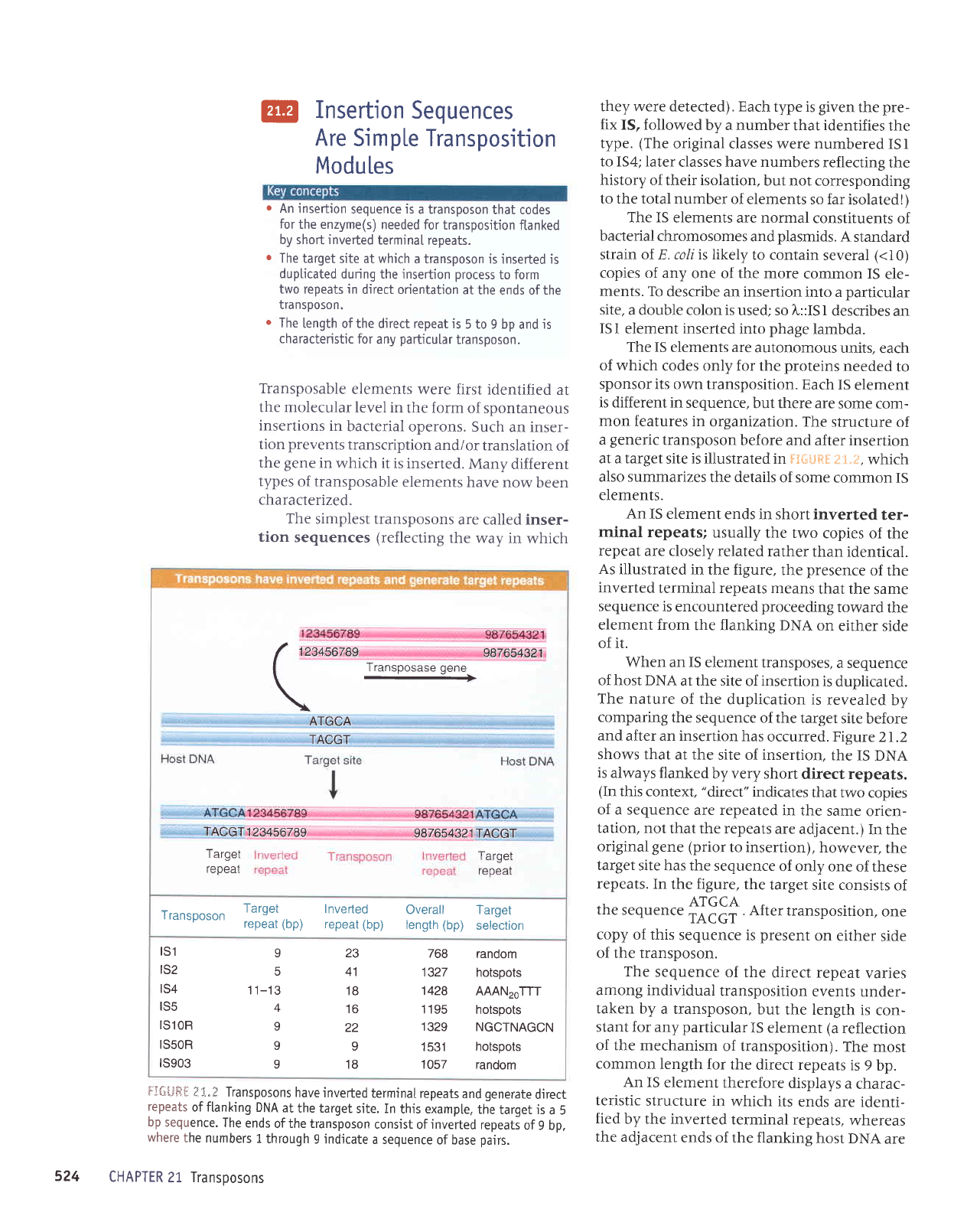
FJ{-;ltFil
I
3.} Transposons
have inverted
terminal repeats
and
generate
direct
repeats
of flanking
DNA
at
the target
site. In
this exampte.
the
target is
a 5
bp
sequence.
The
ends
of the transposon
consist
of
inverted
repeati
of 9 bp,
where
the numbers
1
through
9 indjcate
a sequence
of base
pairs.
CHAPTER
2L Transposons
they were detected). Each
type is
given
the
pre-
fix IS, followed
by a number
that identifies
the
type.
(The
original classes were
numbered IS I
to IS4; later
classes have numbers
reflecting
the
history
of their isolation.
but not corresponding
to the total number
of elements
so far isolated!)
The IS
elements are normal
constituents
of
bacterial
chromosomes
and
plasmids.
A standard
strain of E. coli is likely
to contain
several
(<10)
copies of any
one of the more
common IS
ele-
ments. To
describe an insertion
into a
particular
site,
a double colon is
used; so
7,::IS
I
describes
an
IS
I element inserted
into
phage
lambda.
The IS
elements are
autonomous
units,
each
of which
codes only for
the
proteins
needed
to
sponsor its own
transposition. Each
IS
element
is different in
sequence,
but there are
some com-
mon
features in
organization.
The structure
of
a
generic
transposon
before and
after insertion
at a target
site
is
illustrated in FSGiJftr
? 1-F,
which
also summarizes
the details
of some
common IS
elements.
An
IS element
ends in short inverted
ter-
minal repeats;
usually
the two
copies of
the
repeat
are closely related
rather
than identical.
As
illustrated
in the figure,
the
presence
of
the
inverted
terminal repeats
means
that the
same
sequence
is encountered
proceeding
toward
the
element from
the flankine DNA
on
either side
of it.
When
an
IS
element
transposes,
a sequence
of host DNA
at the site
of
insertion
is
duplicated.
The nature
of the
duplication is
revealed
by
comparing
the
sequence of the
target
site before
and
after an insertion
has
occurred. Figure
2I.2
shows
that at the
site of insertion,
the
IS DNA
is
always flanked
by very short
direct repeats.
(In
this context,
"direct"
indicates
that two
copies
of a sequence
are repeated
in the
same
orien-
tation, not
that the repeats
are
adjacent.)
In the
original
gene
(prior
to insertion),
however,
the
target
site has the
sequence
of only one
of these
repeats.
In
the figure, the
target
site consists
of
,
ATGCA
the
sequenc.
i;CGi.
After
rransposirion,
one
copy
of this
sequence is
present
on either
side
of the transposon.
The
sequence
of the
direct repeat
varies
among individual
transposition
events
under-
taken by
a transposon,
but
the Iength
is
con-
stant for any
particular
IS element
(a
reflection
of the mechanism
of transposition).
The
most
common
length
for the
direct repeats
is
9 bp.
An IS
element
therefore
displays
a
charac-
teristic
structure in
which
its ends
are identi-
fied
by the
inverted
terminal
repeats,
whereas
the adjacent
ends of the
flanking
host DNA
are
ai.
|
'123456789
987654321
I
Transposase gene
\
-.+
\
\,u"o
TACGT
TACGT1234s67B9
987654321TACGT
Target
Target
repeat
repeat
Transooson
Target
Inverted
Overall Target
'
repeat
(bp)
repeat (bp)
length
(bp)
setection
ls1
g
23
768
random
lS2
s
41
1927
hotsoots
ls4
1 1-13
18
1428
AAAN2oTTT
lS5
4
16
1195
horspots
lS10R
9
22
1329
NGCTNAGCN
lS50R
I
I
1531
hotspots
15903
I 18 1057
random
524
identified by
the
short direct repeats. When
observed
in a sequence
of
DNA,
this type of
organization is taken to be diagnostic
of
a trans-
poson,
and suggests that the sequence origi-
nated in a transposition event.
Most IS elements insert at a variety of sites
within
host DNA.
Some, though, show
(vary-
ing
degrees
of
)
preference
for
parlicular
hotspots.
The inverted
repeats
define the ends of a
transposon.
Recognition of
the ends
is common
to transposition events sponsored by all types
of transposon.
Cli-acting mutations that
prevent
transposition are
located in
the ends, which are
recognized by
a
protein(s)
responsible for trans-
position.
The
protein
is
called a transposase.
All
the
IS elements except IS I contain a sin-
gle
long coding
region, which
starts
just
inside
the inverted
repeat
at one end and terminates
just
before
or within the inverted repeat at the
other
end. This codes for the transposase.
IS I has
a more complex organization. with
two sepa-
rate reading frames; the transposase
is
produced
by
making a frameshift during translation to
allow both
reading frames to be used.
The lrequency of lransposition
varies among
different elements.
The overall rate of transpo-
sition
is
-I0-l
to
l0a
per
element
per genera-
tion.
Insertions in individual targets occur at a
level comparable
with the
spontaneous
muta-
tion
rate, usually
-
l0-5
to
l0-7
per generation.
Reversion
(by
precise
excision of the IS ele-
ment)
is usually infrequent, with a
range of
rates of
l0-6 to l0-10
per generation,
which
is
-103
times less
frequent
than
insertion.
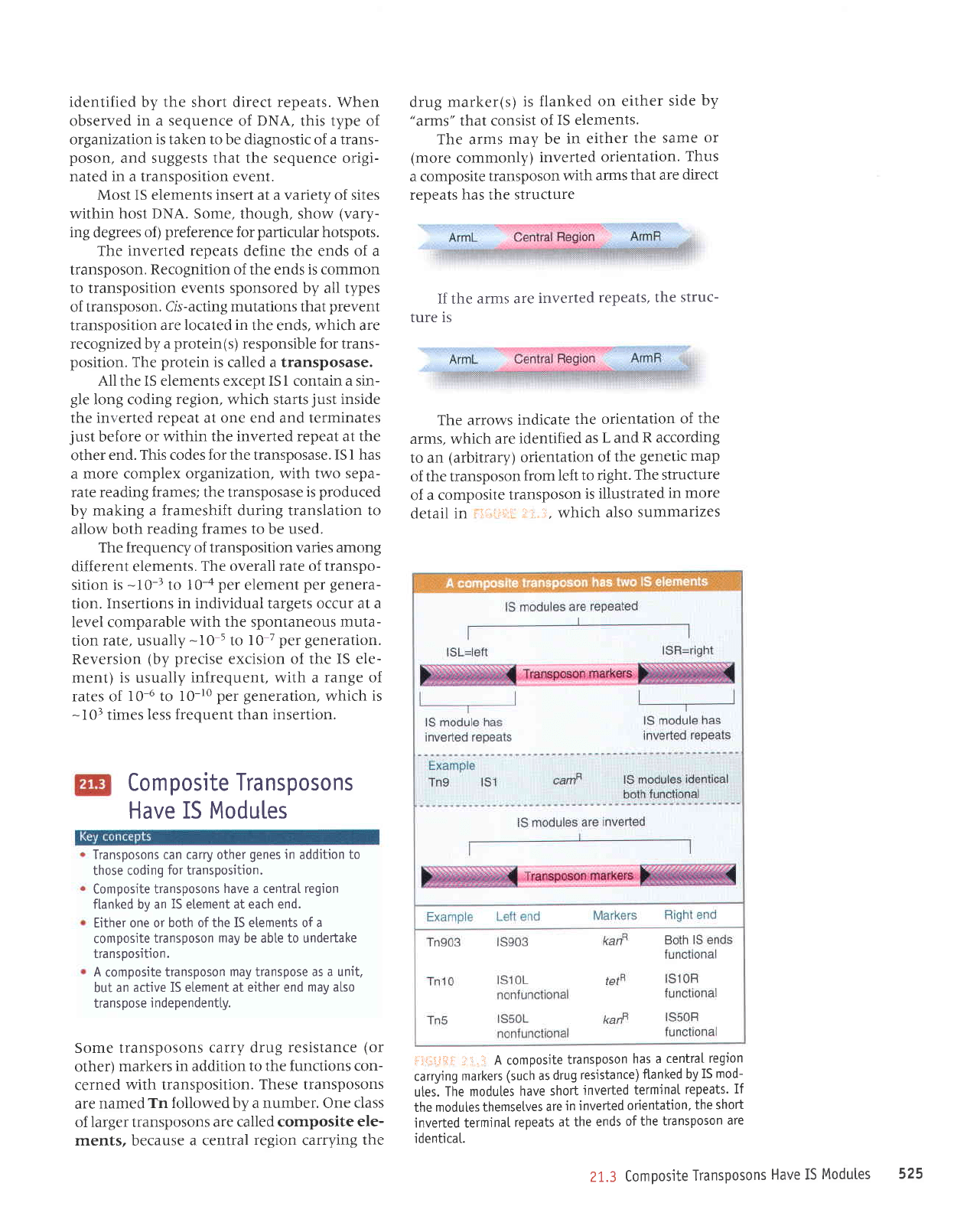
Composite
Transposons
Have IS Modules
Transposons can carry other
genes
in
addition
to
those coding
for transposition.
Composite
transposons
have a central region
flanked by an
IS
etement at each end.
Either one or
both of the IS etements of a
composite
transposon
may
be abte to undertake
transposition.
A
composite
transposon
may
transpose
as a unit.
but an
active IS element at either end
mav also
transpose
i ndependent[y.
some transposons
carry drug
resistance
(or
other)
markers
in
addition
to the functions con-
cerned
with
transposition.
These transposons
are
named
Tn followed by a number. One
class
of larger transposons
are called cornposite
ele-
ments, because
a central
region
carrying
the
drug marker(s)
is
flanked
on either
side
by
"
arms"
that consist
of IS
elements.
The arms
may be
in either
the
same
or
(more
commonly)
inverted
orientation.
Thus
a composite
transposon
with
arms
that are
direct
repeats has the
structure
If
the
arms are
inverted
repeats, the
struc-
ture is
The arrows
indicate
the
orientation
of
the
arms, which
are identified
as
L
and
R according
to an
(arbitrary)
orientation
of
the
genetic map
of the transposon
from
left
to right.
The structure
of a composite
transposon
is illustrated
in more
detail
in
j'ii.l.i!ii
:r':.:i,
which
also
summarizes
+:j{:1-lIil
:l:i
.;:I
A composite
transposon
has a
centraI
region
carrying
markers
(such
as
drug
resistance)
ftanked by
IS
mod-
utes.
The modules
have
short
jnverted
termjnal
repeats.
If
the modules
themsetves
are
in inverted
orientation,
the short
inverted termina[
repeats
at the
ends
of the
transposon
are
identicat.
21.3
Composite
Transposons
Have IS
Modules
525
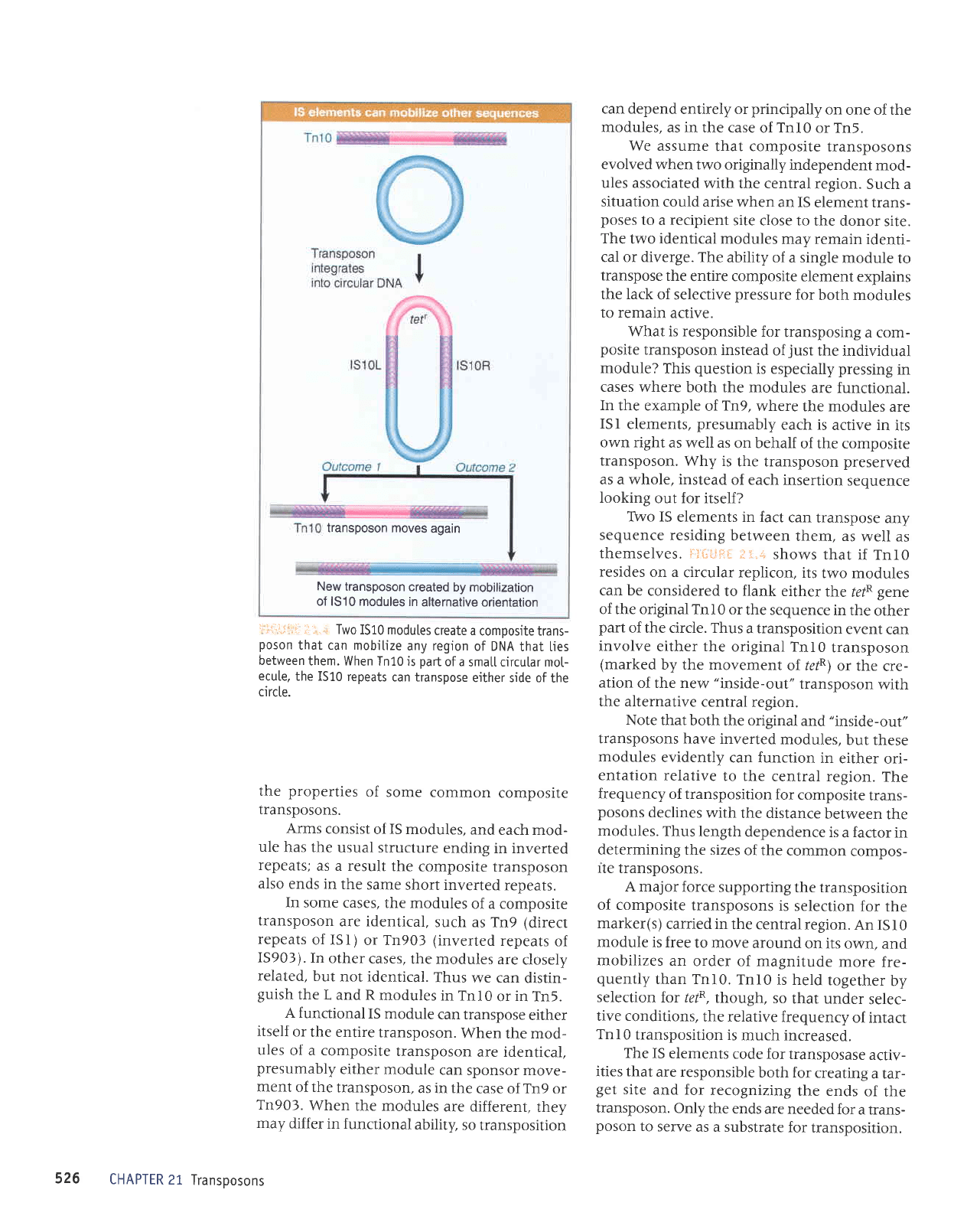
New transposon
created
by mobilization
of lS10
modules in
alternative
orientation
transposon moves
again
r:.r:-;iiri,.
i: I ..:
Two
15L0 modules
create a
composite trans-
poson
that can
mobitize
any region
of DNA
that Lies
between
them. When
Tn10 is
part
of a
smatl circular mot-
ecule,
the IS10
repeats
can transpose
ejther side of
the
circ[e.
the
properties
of
some common
composite
transposons.
Arms
consist
of
IS
modules,
and
each mod-
ule
has
the usual
structure
ending
in inverted
repeats;
as a result
the
composite
transposon
also
ends in
the
same
short inverted
repeats.
In
some cases,
the modules
of a composite
transposon
are identical,
such as
Tn9
(direct
repeats
of ISI)
or Tn903
(inverted
repeats
of
IS90l).In
other
cases,
the modules
are
closely
related,
but
not identical.
Thus
we can
distin-
guish
the
L and R
modules
in Tnl0
or
in
Tn5.
A functional
IS module
can transpose
either
itself
or
the entire
transposon.
When
the mod-
ules
of a composite
transposon
are identical,
presumably
either
module
can
sponsor move-
ment
of the
transposon,
as in the
case
of
Tn9
or
Tn903.
When
the modules
are different.
they
may differ
in functional
ability,
so
transposition
CHAPTER
2L
Transposons
can depend
entirely or
principally
on
one of the
modules,
as in the case
of
Tnl0
or Tn5.
we assume
that composite
transposons
evolved
when two originally
independent
mod-
ules associated
with the central
region.
Such a
situation
could arise
when an IS
element trans-
poses
to a recipient
site close to
the donor
site.
The
two identical modules
may
remain identi-
cal or diverge.
The ability
of a single
module
to
transpose
the
entire composite
element
explains
the lack
of selective
pressure
for
both modules
to remain
active.
What is responsible
for
transposing
a com-
posite
transposon
instead
of
just
the individual
module?
This
question
is especially pressing
in
cases where
both the modules
are functional.
In
the example
of
Tn9,
where
the modules
are
IS
I elements,
presumably
each is
active in
its
own right
as well as
on behalf
of the composite
transposon. Why
is
the transposon preserved
as a whole, instead
of each
insertion
sequence
looking
out for itself?
TWo IS
elements in fact
can
transpose
any
sequence residing
between
them, as
well as
themselves.
i:*i.jitl
;l::.ri
shows
that
if Tnl0
resides
on a
circular replicon,
its
two modules
can be
considered
to
flank
either
the
/e/R
gene
of the original
Tnl0 or
the sequence
in the
other
part
of the circle. Thus
a transposition
event
can
involve
either the
original Tnl0
transposon
(marked
by the movement
of rerR)
or the
cre-
ation
of the new
"inside-out"
transposon
with
the alternative
central
region.
Note thatboth
the original
and
"inside-our"
transposons have
inverted
modules,
but these
modules
evidently
can function
in
either
ori-
entation
relative
to the
central region.
The
frequency
of transposition
for
composite
trans-
posons
declines
with
the distance
between
the
modules.
Thus
length
dependence
is
a
factor
in
determining
the sizes
of the
common
compos-
rte transposons.
A major
force
supporting
the transposition
of composite
transposons
is
selection
for
the
marker(s)
carried in
the central
region.
An ISI0
module
is free
to move
around
on its
own, and
mobilizes
an
order of magnitude
more
fre-
quently
than TnI0.
Tnl0
is held
together
by
selection
for tetR,
though,
so
that under
selec-
tive conditions,
the relative
frequency
of intact
Tnl0
transposition
is
much increased.
The
IS elements
code
for transposase
activ-
ities that
are responsible
both for
creating
a tar-
get
site
and for
recognizing
the ends
of
the
transposon.
Only
the ends
are needed
for
a trans-
poson
to
serve
as a substrate
for
transposition.
526
LZ9
sursruptlla6.e^qplrtdaluoN
pup
e^qellldau
qlog
r{q srnr:6 uorlrsodsuetl
7'17
P JalJe alnJJloru Jouop
Jql 01 suao
-deq
req14'elrs
Jouop aql ruorJ
tsol
pue
alls
taSret
Jql
tp
pJuJsuI
Jq ol
lueuale
eq1 Jsnpl uorlrsodsuerl anrlertldaruou
Jo
srusrueq)Jtu
qlog'(uorlrsodsue;J
JoJ sJlerpeurJJlul uoluruof,
'9'IZ
uoll
-ra
g
aas
)
uortrsodsuerl
a,r.r1err1dar
q11.la.
sdats aruos seJeqs
pue
saruanbes
VN11
la3:et
pue
Jouop
Jo
uollJJuuol aql sJZII
-nn
tusrupqJJr.u JJr{louy'asesodsuell
e
Lpo
salnbar
usrueqJJru
Jo
addt
sql
'JJJsueJl
Surrnp
VN(
Jouop 3ur4ue1;
eqt uoJJ uosodsuerl Jql
Jo
eseJIJJ eql
SJAIOAUI
qJII{i\A
';.
lI
:i1r.ji'jli'rrri
UI
UMOqS
usruPqf,Jru Jql JSn
suJ
pue
OIuJ
suosodsuerl
alrsoduor
pue
saruanbas
'uosodsuerl
eq1
1o
fidor
e
aneq
luardLrer
pue
rouop
qloq
os
'pabueqlun
suteual
alts
louop eq1
'e1Ls
luardLrar
p
lp
silosut
qrLqm
'uosodsuetl
aq1
1o
Ador
e
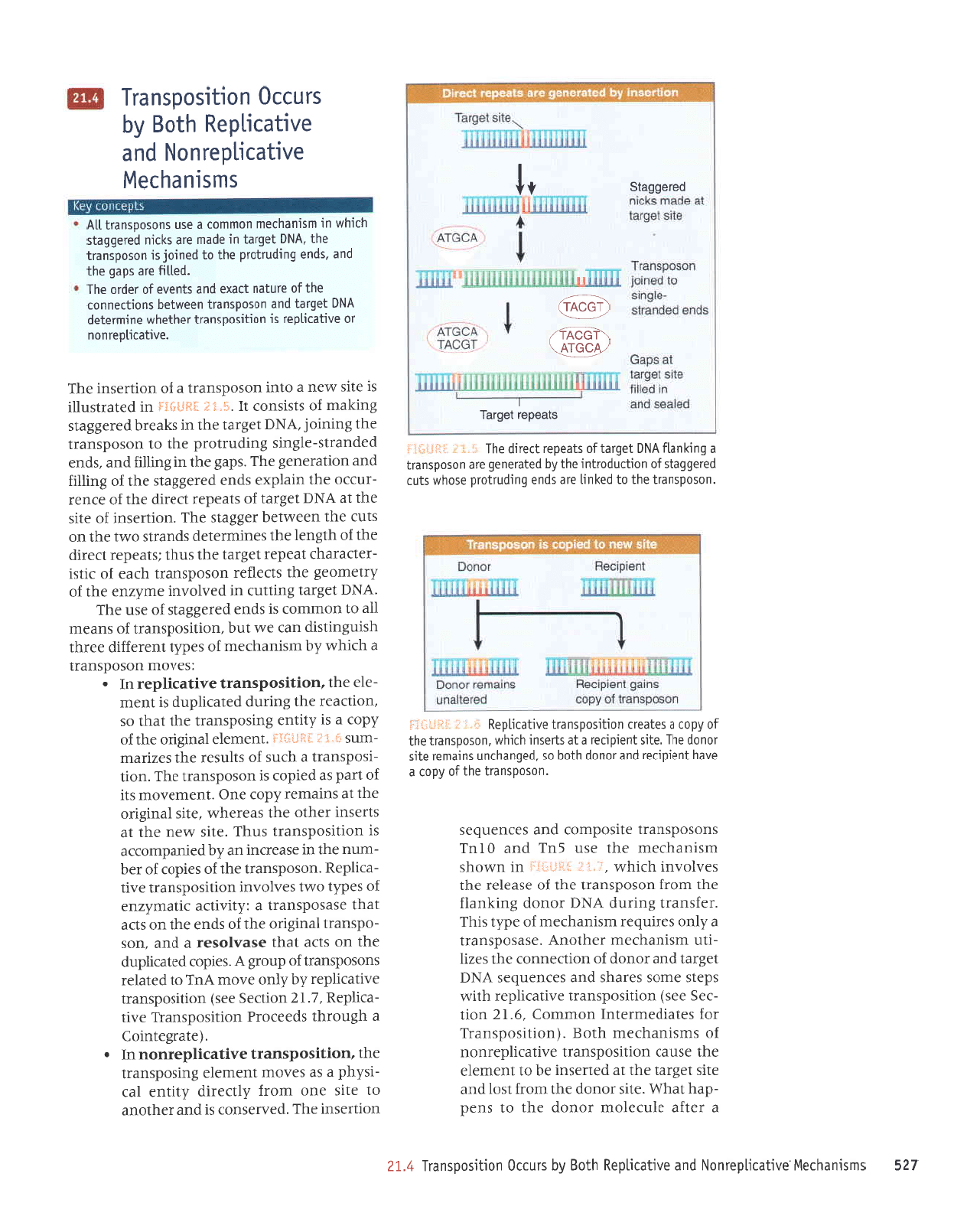
seleerr uotltsodsuerl anLlecLldeX
;1
i,,:i
:irlf ii.]i:i
'uosodsuerl
eql ol
palurl
a.le spua 6urpntlotd
esoqm
slnl
para66e1s
1o
uorllnporlur aql
^q
palpreue6
ale
uosodsuetl
e 6uL1ue11
y116
1a6re11o
sleadar
lrarrp
aLll
,-'1,.r it:.:iilLri.:i
uoruesur
JqI'pe^resuof,
sI
pue
JJqloue
ol Jlrs
euo
rxoJJ
dllrartp
'(lllua
Ier
-rs.{qd
e se salour
luJruJle
Sutsodsuerl
eqt
/uolllsodsue,rl
e^IlEJIIdaJUorr
u[
r
'(arer8atulo)
e
q8norqr spJaJoJd
uolllsodsuer1,
eAIl
-er11dag
'4'I
Z
uollJes
aas) uotltsodsuerl
anrlerlldar
z{.q,{po
elolu
vu1
ot
pelelar
suosodsuerl;o
dnor8
y
'sardor
palerqdnp
Jr-ll
uo sDP
leql
eSPAIOSaJ
P
pue
'uos
-odsue;l
1eu€r.to
aql
Jo
spuJ
er{l uo
sDe
lBql
aSesodsueJl
P
:dltnrlre
lrletu,{zua
;o
sadLl
oM1 sallolut
uorlrsodsueJl
JAI1
-ergdag
'uosodsue:l
aqt;o
sardor;o
raq
-I'unu
Jql uI
JSPJJ)ul
ue
r{q
palueduofJe
sr
uorlrsodsuert
sneJ
'alIS
,lrJU eql
]e
suJSuI
JJqlo aql
seeJJqat
'a1rs
leut8tro
eql
le
sulelual
r(dor euo
'luaIuJAOIrr
SU
yo
ged
se
patdol
st
uosodsuerl
eq1,'uou
-rsodsuerl
E
q)ns
Jo
sllnsJJ
eql
sazlJeru
-uns
';":.;1
:iijii:igj
'tueuala
pur8rro
aqt;o
Ldor
e sr ,{.lltua
Sutsodsuerl
Jql
1eql
os
'uortJeal
aql
Surrnp
parerudnp sI
lueru
-JIe
aql'uolllsodsupJl
aaIlBJIIda'r
u1
.
:sJAOrU UOSOCISUeTI
p
qJrqu Lq rusrueqraru;o
sad[1
luereJJlp
aerql
qsrn8urlslp
ueJ e,lr
1nq
'uorllsodsueJl
Jo
sueeur
ile
01 uouruo)
sI spua
para83e1s
Jo
asn
JqI
'y51q
la8rer
Suptnt
uI
pe^lolul :ru.dzua aql;o
Lrlaruoa8
aql
SDJIJaJ
uosodsuBrl
qJea
Jo
JIIsI
-JJtJeJeqJ
lBadar
la3.rel
aqt snql
lstead:r
1)JJIp
aql;o
qfual
aql sauIIrJJlJp
spueJls
o^{l
aql uo
stnJ
eqt
ueeMraq
raSSBts
aqJ
'uonresul
Jo
JlIs
aqr
le
vN(I
1a3rel
1o
sleada:
Derlp
aql
Jo
JJUaJ
-rnJJo
aql ureldxa
spua
paraSSels aql
1o
3u191
pue
uouerrua8
aql
'sde8
aqr
u1 8utt1g
pue
'spua
papuerts-a18uts
Surpnrlord
aqr
01 uosodsuerl
aql 3u1ulo['YNC
lr8rer
eql ul s>learq
para38e1s
Suqeru
Jo
stsrsuoJ
tI
':;'t
it .?di1*.i
uI
pelerlsnlll
sr elrs
,lreu
e olur
uosodsueJl
P
Jo
uoluJsul
JqJ
'aAqel!l0eluou
ro
enrlerqdar
sL uorlrsodsuell
iaqleqM
0uLulalap
ypq
1ebre1
pue uosodsuell
uoaMleq
suollleuuol
aql
Jo
ornlPu
llPxa
puP
slua^a
J0
raplo
aql
'pallg
o.rP sdeb
eq1
pue'spua
6urpnrlord
aq1 o1
pautolst uosodsuell
aqr'VN6
1a6re1
ur apeul
ate
s4rru
pa.re66e1s
qltq/v\
ul
ulstueqleu
uotllulol
e asn
SuosoosuPll
llv
sustueqla[^l
e^qelrldajuoN
puP
a^qPlrldau
qlo8 Aq
sleedei
labtel
/f,ocE\
'.{PcYY
(@
pere66ers
^l
sJnll0
uoLlLsodsuejl
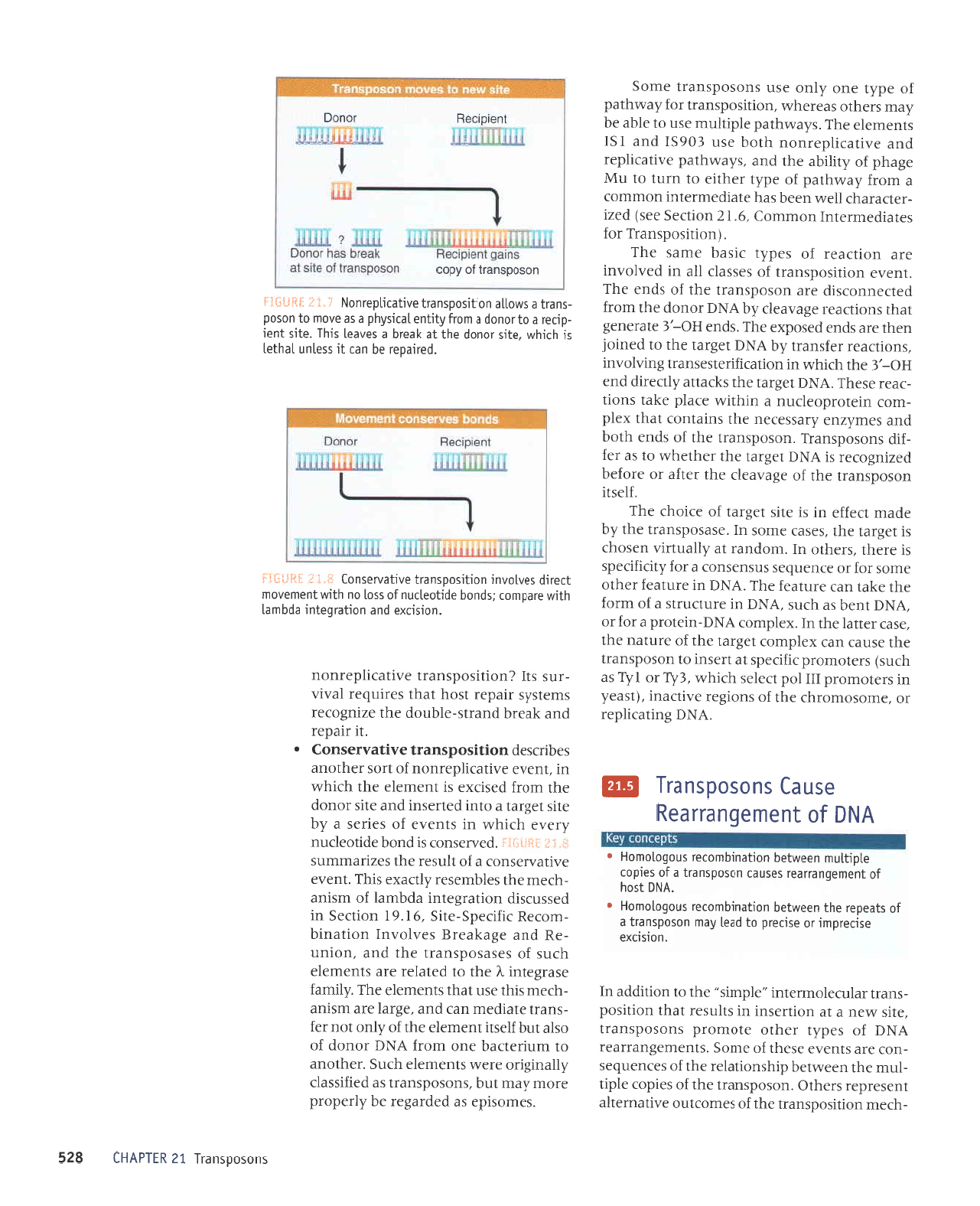
suoso0suerl
IZ
ulldvHl
829
-qreu
uorlrsodsuerl
eql
Jo
sJruoJlno
JArteuJJtlp
luasJJdJr
sJJr{tO
'uosodsueJt
Jql
Jo
serdo)
Jldrl
-lnur
Jql uJaMlJq
drqsuortelar
aqt;o
saruanbas
-uoJ
eJe sluJna
JSJqI
]o
aruos
'sluJrua8uerrear
VN(
Jo
sadLt
raqro
alouord
suosodsuBrl
'Jlrs
,l.rJU
P
le
uoruJsur
ur
sllnsal
teql
uorlrsod
-sueJ1
Jelnf,eloruJalur
,,a1durs,,
eqt ot
uottppp
uI
'uotsDXa
asnardur
to
asrratd
o1
peal
Aeu
uosodsuetl
e
J0
slpadal
aq] uaaMleq
uotJpurqulolol
snoboloruog
o
'vN0
lsoq
1o
luetuabueleet
sasnpl
uosodsuetl
e
1o
sardor
aldrllnul
uaoMlaq
uorleurqulola.l
sno6o1otuo11
o
vNo
Jo
luauabuPrrPeu
asnPl
suosodsuell
@
'y519
Surlelrldar
ro
'aurosoruorqJ
Jql;o
suor8ar
JlrtJeur
,(tseaL
ur
sJetoruoJd
III
Iod
Dalas
q)rq,lr
,t
^I
to
I AI
sp
qrns)
sralourord
rr;oads
le
uasur
ol uosodsuerl
Jqt JSneJ
uer xaldruor
ta8rel
aqr
Jo
arntpu
Jql
'eseJ
Jaltel
aqt u1
'xaldruo)
VNO-uralord
e ro; ro
'vN(
luaq
sP
qJns
',vNo
ul Jrnlf,nJts
e
Jo
ruJo}
eql
J>lel UPJ
JrnleJJ
eql
'vN(I
ur JrnleeJ
lJqlo
Juros roJ
ro aruanbas
snsuJsuoJ
e ro; .,{1nr;r:ads
sr aJJqt
'sJJqto
uI
'tuopueJ
1e
Lllengt.
uesoqJ
sr
la3:e1
eql
'sJSe)
eruos
uI
'asesodsuerl
aql .{q
JppLu
llJIlJ
ut sr
Jlts
ta8ret
;o
J)toq)
JqI
.JIrsll
uosodsuen
aqt;o
aBeneJIJ
Jr{t lJlJe
Jo
JJoJJq
pazruSorar
sl
VNC
ta8ret
aqt
rJqteqM
ot se
rJI
-;rp
suosodsuerl
'uosodsueJl
ar{l
Jo
spuJ qtoq
pue
saru.dzua
,{ressarau
eq}
surpluot
reql
xald
-ruoJ
urJloJdoalrnu
p
urqtrM
areld
a>lel
suorl
-JeJJ
eseqJ'VNC
ta8rel
eqt
s>lJpne f,prarrp pua
HO-,€
Jql
q)rqm
ur uorleJrJrralsasueJl
Surnlonur
'suorlJeJJ
JeJSueJt
Iq
VXC
ta8ret
aqt
ot
pauroI
uJqt
JJe spua
pasodxJ
JqJ'spuJ
HO-,€
ateraua8
teql
suort)eel
a8eneap
[q
VNq
Jouop
Jq1
rro]J
pJlf,Juuo)srp
are
uosodsupJl
Jql
Jo
spuJ
JqJ
'luJle
uorlrsodsuerl
Jo
sJSSelJ
IIp
ur
pJAIoAur
JJe uoll)eer
;o
sadLl
)rseq
Jrues_
er{J
'(uorlrsoctsuPJJ
JoJ
satelpJurrJlul
uolutuoJ
'g'I
z
uorpes
aas)
pazr
-JeppJeqJ
IIJM
uJJq
spq
alerpeuJelul
uouruoJ
e rroJJ ,{e.trqled
;o
ad,{1
rJqtlJ
ol
urnt ot nW
a8eqd
yo
.{.trgqe
eq1 pue
's,{e^,vrqted
a,.r.r1err1dar
pup
r^rlef,rldaruou
q10q
asn
€06sI
pue
ISI
sluJruJle
aq1
'sLemqted
a1dr11nur
esn ol
alqp
Jq
,deur
sraqlo
seJrJqM
'uorlrsodsuerl
JoJ ^emqled
1o
ad,,(1
auo ,{.1uo
asn
suosodsueJl
Jtuos
'sauosrda
se
papre8ar
aq.{lredord
aroru.deru
1nq
'suosodsueJl
se
pagrsselJ
.dleur8rro
aJaM
sluaruJlJ
qtnS
'Jeqloue
ol urnrJJlJpq
Juo ruoJJ
vNQ
Jouop
Jo
osle
lnq
]lJSll
luJruJIJ
Jql
Jo
,{Juo
tou
ra.;
-suerl
Jlerpeur ue)
pue
'a3re1
aJe ursrup
-q)Jlu
srql
Jsn
lpqt
stuJtrlJIa
eq1
'{1ruej
aser8atut
y Jqt
ol
pelelJr
Jrp
stueruJlJ
qJns
Jo
sasesodsuert
aql
pup
'uorun
-JU
pue
a8e>1earg
sJAIoAUI
uorleurq
-ruoJJU
rr;rlad5-atr5
'9I'6I
uortf,es
ur
pJSSn)stp
uorter8atur
epquel
Jo
rusrue
-qJJru
Jql selqruJsJr
f,prexa
srqJ'luele
JAI1PAJSSUOJ
P
}O
1INSEJ
Jql SEZIJEUIUINS
*'i I
:iif
i:::*i
'pa^-rJsuoJ
sr
puoq
JprloJlJnu
d:ana q;rq.rn
ur
sluJla
;o
sauas
e ,{.q
ells
1J3Je1
P olul
peuJsur
pup
alrs Jouop
Jql
ruor}
pesr)x3
sl
lueualJ
Jql
qJrqM
ur
'luJAJ
anrlelldaruou
Jo
tJos
Jaqloup
sJqrJJSJpuoglgsodsueJleAIleAJasuoJ
.
'11
JIeOeJ
pup
>leJrq puprls-elqnop
aql
azruSorar
suals.{s
rredar
lsoq
lpql
sarrnbar
lerun
-Jns
sll
2uorlrsodsuert
a.trlelldeJuou
'uorsoxa
pue
uoLletbelur
epqupl
qltm
ereduor
jspuoq
aptloepnu
Jo
ssol ou
qltm
luauonoul
]larrp
sa^lo^ur
uorlrsodsuell
a^rlp^lasuol
*.;i _lHi]*id
'palrP00l
aq
uet
]r
ssalun
leqlal
st
qlrqM
'alrs
.louop eql
lp
)paiq
e
so^pol srql
.alrs
]uar
-doel
e o1 rouop
e uo4 A1L1ua
lerrsAqd
e se o^oru
o1 uosod
-suPrl
e sMolle
uorlrsodsuell
a^rlplrldelu0N
t'l.i
$lit"t*i-J
uosodsuell
1o
{doc
].
j."
:
,ll -i::
:
luerdrcag
rouo6
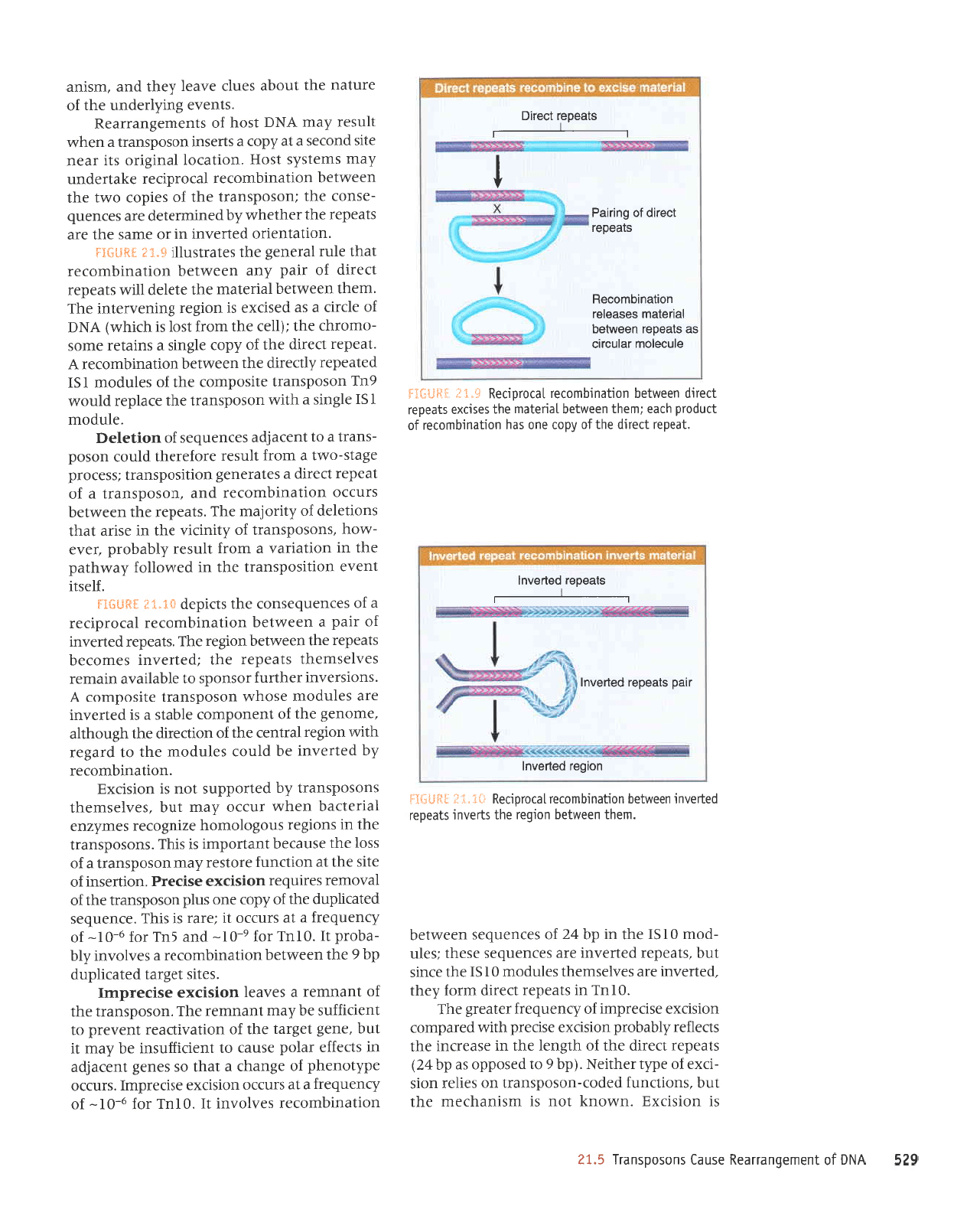
629
vNo
Jo
luauabuElleeu
asnpl suosodsuell
g'Iz
sr
uorsrJxg
'u/!|ou>l
lou
sr usrupqJeru
aql
lnq
'suorlJunJ
pepoJ-uosodsueJl
uo
sJrler
uols
-r)xr
Io
ed&
rrqlreN'(dq
6
or
pasoddo
se
dq77)
steadar
DeJIp
eqt
yo qfual
aql
ut asearJul
eql
soagar,{.1qeqo:d uorsnxa asoard
qtu,r pareduror
uorsrJxJ asrlardrur
;o,{.ruanbar;
ralear8
aq1
'0IuI
q
stPJdJr
trarrp
ruroy
z(aqt
'peue^ur
eJP sJAIJsruJql sslnpou
0I sI
eql JJUIS
1nq
'sleadar
pauelur
are saluanbJs
Jseqt
lsJln
-pou
0ISI
aql ur dq
tZ
Jo
seruenbas
uaa^,tataq
'uloql
uaeMlaq uotbar aql s]le^ut
sleeoal
peile^ur
ueoMlaq uoqeurquolo.l
lerorollau
ilc'5
f ;t!frlFIj
uoroal
peua^ul
:led
sleeder
peuo^ul
L______l_________
sleoooJ
pouo^ul
'leeder
lralrp
aq1;o
fidor
auo
seq uotlpurquolol
]o
pnpold
qlee
lueql uaeMlaq
leualeu
oql
sestlxa
slPodal
llarrp
uaaMleq
uorlPurquola.l
lel0.l0Dau
s'lt
.i{**g*
olncolouJ
relnclrS
se slpaoor uoo^qeq
lEuoleu
seseoler
uolleulquloceH
sleeoeJ
lcarlp
lo
Ouuled
L_________J
sleeder
1calt6
t
uolleulqluoJaI
sJAIoAul
1I
'0IuJ
JoJ
e-0I-
Jo
,buanbary
e
le
sJnJJo
uoISIJXa
asnardrul
'srnJJo
adLlouaqd
;o
a8ueqr
e
leql
os saua8
luarefpe
ur slJJJJe
relod
asner
ol
luJIJIJJnsut
aq
,{eru
1t
1nq
'aua8
1a8re1
aqt
Jo
uollelltf,ea.r
lua,rard
o1
luarJrJJns
aq
deu
lueuruJJ
aql
'uosodsuPr
aql
Jo
lueuluer
p
sJAeJI
uolslJxe
aspa,rdrul
'satrs
la8rel
palerudnp
dq
6
aql uJamlaq
uolleqqrxoJeJ
e
sallorrur
r(1q
-eqord
1I
'0IuJ
ro;
6-0I-
pue
EUJ
roJ
e-0I-
Jo
.druanbar;
e
lp
sJn)Jo
1l
IJJeJ sl slqJ
'aruanbas
parerqdnp
aqr;o
,(dor
auo snld
uosodsuerl
aq1;o
Ienorual
serrnbar
uolsl)xe
asIJaJd'uoluasul
Jo
elrs
Jql
le
uoIlJUnJ
JrOlSJl
,(eur uosodsueJl
e
Jo
ssol aql
Jsnef,eq
luBuodrut
sl slqJ
'suosodsuerl
aql
ur suo€ar
snoSoloruoq
aztuSolar
saur,{.zua
IPrJellEq
ueqM
rnrlo
z{eru
lnq
'se^Iasluaql
suosodsuerl
,{q
palroddns
lou
sI UoISIJXa
'uorlPulquroJJJ
z(q
pagaaur Jq
plnol
salnpou
eqt
01
pre8ar
qtp,l
uor8ar
leJtual
ar{l
Jo
uopterrp
aql
q8noqtp
'aruouaS
eqt
Jo
luauodruor
a1qe1s
P sI
paualul
eJe salnpou
esoqM
uosodsuPJt
atlsodruor
y
'suorsJelul
requnJ
rosuods
01 JIqPIPAe
uIPIueJ
se^lasuaql
sleadar
Jq1
jpJua^ul
seruoJaq
sleadar Jql
ueaMlJq
uor8ar
aq;
'sleadar
peuJAuI
;o
rred
p
uJJMlaq
uolleulquotar
lerordrlar
e
1o
saruanbasuoJ
eql
sprdap
ftr.En SHfillIi
.JIASTI
luelJ
uortrsodsuerl
Jql uI
pamolloJ [emqted
Jql uI
uolrelrel
p
ruoJJ
tlnsar
z(lqeqord
,:aaa
-Moq
'suosodsuerl
1o
Lltunn aql
ut aslJe
leql
suorlJlJp;o
.d1rroferu aq;
'sleadar
aqt
uJaMleq
sJnf,Jo
uorleulquoJeJ
puP
'uosodsuerl
e
;o
leadar
peJIp
e saleraua8
uorltsodsuerl
lssarord
a8els-o^4,t1
p
IuoJJ
llnsal
eJoJJrJql
ppol
uosod
-supJ1 p
ol
luarefpe
sa:u:nbas
jo
uollalec
'elnpour
151
afurs
e
qllrlr uosodsuerl
aqt
areldar
plnoM
6u;
uosodsupJl
allsodluoJ
aq1
Jo
sJlnpou
ISI
pateadar.dp)JJIp aqt
uaamtJq
uolleulqruoJal
Y
'leadar
DeJIp
Jql
;o
,{dot a18uts
e sulelJr
eluos
-ouorqr
rq1
j(llal
aqt
ruorJ
lsol
sr
qrlq1\{)
vNa
Jo
JIIJTJ
p
se
pJSIJXa st uor8ar
Suuan:a1ut
aq1
'ruJql
ueaMleq
PITJIPII
Jql elJIJp
111.lzr
sleada.r
DeJrp
yo
rted
z(ue
uaaulaq
uolleulqruoJar
teql
alnr
leraua8
eqt
selerlsnlll
S'8fr
3Hfi*ii
'uorleluJlJo
paUJAUI uI JO AIUes
aq1
ere
steadar
aqt
Jeqlaqm
Lq
paunuralap are saruanb
-JSuoJ
aql
luosodsueJl
Jq1
;o
satdor
oml
Jql
uJJ,lrlaq
uolleulquorar
lerordnaJ
e>leuepun
,(eru srualsz{s
lsoH
'uolleJol
leut8tro
sll JPJU
alrs
puof,Js e
le
ddol e suJSuI
uosodsuerl
e uaqu
tlnsar
.deru
vNC
rsoq
;o
sluaruaSueJrPag
'slualJ
Surdpapun
aql
;o
eJnleu
eql
lnoqe
senlJ
J^eel
,(eqr
pue
'luslue
puP
'Iu
'z-I'[l
Jql JlnlrlsuoJ
lpq]
sJJuJnbas
dq
ZZ
eql ot
spurq
elrs Surpurq-snsuasuoJ
JqI'suau
ur
l)e
ot asesodsuert
Jql
Jo
suunqns
sladruor uoltf,p
Jo
Jpour rrJql pue
'ygq
3ur1e1n
-drueur
ro; satrs
ol^t spq
VnW
'VNC
nW
Jo
spuJ
qloq
uo uollf,eJI peleulpJooJ
P sJJnsuJ
leql
trel. e
ur suorlJunJ
,lrou
lJrupJlJt
aqJ
.uosod
-sueJt
eqt
Jo
spue oMl
Jqt
;o
srsdeu,{s
sJ^Jtqf,e
sIqJ'JaruerlJl
e uroJ
sJus
esJqt
ol
punoq
slrun
-qns
VnW
'uosodsuert
Jqt
Jo
pua
LIJeJ
ol
lsJsoll
sJlls
oMl aql ,{1uo
sJAIoAur
slqJ'it:':
:j
==;-:=l_r
ul
pJleJlsnllr
sa8ets
aarqt
aqt
q8norqr
sassed
atrs
ta8ret
e 01
VNCI
uosodsuerl
nW
Jqt
Sururol
'sdats
tuanbas
-qns
pue
aSeneap pupJls
roJ
tou lnq
'xaldruor
Jql
Jo
uollPluroJ
JoJ ,{ressarau
aq ot sreadde
1r
1reJlf,
tou
sr
Jtrs
IpurJtur
Jql
Jo
JIor
JLIJ'xalduor
e suJoJ
elrs
IeuJetur
Jql
tp
pue
spua
tq8rr
pue
Url
Jrlt
qloq
te
VnW
Jo
Surpurg
'aruoua8
a8eqd
Jql
ur alrs
IeuJJlur
up ol
spurq
osle
vnw
.Jlrs
qJpe
ot
purq
ueJ
VnW
Jo
Jaruouour
y
'pua
lq8rr
aI{]
lP
Jre
€U
pue',zu',Iu
lpue
]JJI
Jql
]p
Jre
€f
pue
'e-I'II
'vNC
nw
Io
pue
q]ea
le
pale)ol
JJe snsuJsuot
dqZe
e
qtrM
satrs
Surpurq-VnW
JarqJ'esesodsuerl
\/nw
aqt Lq
pauro;rad
are
ygq
a8eqd
eq]
Jo
suortplndrueru
Ieprur
rql
'luJrJJJrp
eJe
suorlf, eJ-r
luanbasqns
Jqt
tnq
'tJBrel
str
pue
uosodsuerl
Jql uJJMleq
uorlJeJJ
yo
adLl
Jtups
Jqt Jnlolur
uortrsodsue'tt
yo
sad^,(1 qlog
'uoltlsodsuerl
arrrlerrldar ,{q
palyrldrue
sr sardol
Jo
Jequnu
Jql
'ap{r
rr1d1
Surnsua
aql Surrnp luortrsodsuert
aa.rl
-errldaruou
Lq
auoua8
aqt
otur saler8alur
n141
'llJJ
tsoq
e SurlraJur
uodn
'sLe.ra
o,l,at
ul uortrsod
-sueJl
Jo
ssarord
eql
sesn
qJIr{M
,nry
a8eqd qlrna
pelpJAeJ
ISJIJ
seM
u(e,ltqted
Srql
Jo
qJnW
'.r{.1snor.tard
srnrro
.,(11e
-n1re
lnq'a8eleap
ratJe
uuoqs
sr
a8els srsdeu.ds
aql
'saSenealr
aql
SurzvrolloJ
ul ,{llrlldurls
roJ
lssarord
srqt
ur raqlaSol
tq8noJq
are uosodsuerl
eql
Jo
spua oazrl
a{J
'la8ret
eql
pue
uosodsuerl
aql
ueemlJq
uorlf,euuof,
luelelof,
e
alerauaS
ol
JSIMSSOJ]
pauroI
are spuJ pJ>l]ru
JqJ
.spueJls
qtoq
uo
pJ>lJru
sr
atrs
ta8rel
Jqt
pup
spuJ qloq
te
pJ>lJru
sr uosodsuerl
Jqt
tpqt
smoqs
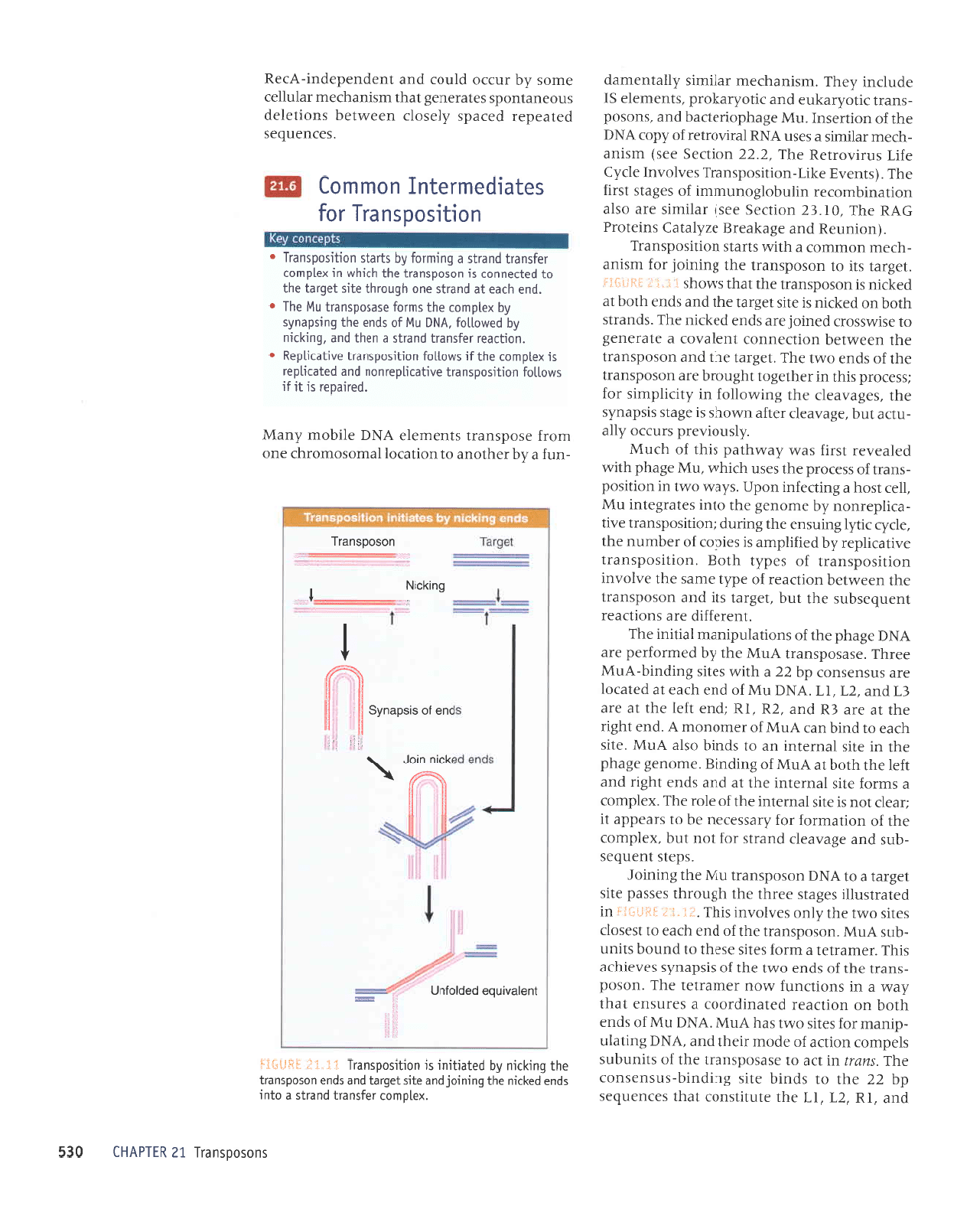
i
i
.i
jj
i$ll:jij
'la8ret
stl
01 uosodsuerl
Jql
SururoI
JoJ tusrup
-qlJu
uoruuroJ
p
r{lr.,t.t.
supls
uorlrsodsuerl
'(uorunau
pue
a8e>1earg
azLlele)
surJloJd
9VU
JqJ'0I'{,2
uoItJeS
aas)
relturs
ale
osle
uorlpurqruof,ar
urlnqolSounurlur
Jo
sa8els
ls,rrJ
aql'
(sluanE
J>lrl-uortrsodsuerl
sa,r1onu1
ap.d3
aJI'I snJrlorlJu
JqJ
'Z'Ze
uolq)eg
aas)
rusrue
-q)eru
Jelrrurs
e sasn
VNU
Iplrloltar;o
,{dor
y1qq
Jql
Jo
uoIUJsuI
'nW
a8eqdouapeq
pue
'suosod
-suerl
rrlo.{,re>lnJ pue
Juo^Je>loJd
,slueualJ
SI
apnlJur z{aq1'rusrueqJeu
relrurs .{lletuaruep
suosodsuerl
IZ
UlIdVHl
'xal0ulol
teJsuPll
puPlls
e olut
spue
pa)rru
aq1 6ururol
pue
elLs
1a6re1
pue
spua
uosodsuerl
aq1 6uLlrLu fq paletlrur
sr
uoLlLsodsuetl
li':.i:
=Si]*;i
0€9
iiir
'1 il
iiii
aj
lualp^rnoo
paplolun
ftft
V\.
)lcru
uror
',,
r!
rua
ro
qsdeu^s
ll ll
KJ
1
I
6ur>1c;11
uosodsuerl
-unJ
e Aq lJqloue
ol uorlef,ol
IeuosoruoJqJ
Juo
uror; asodsueJl
sluJtuelJ
17NC
alrqou
z(uery
'paJrPoar
sr
1r
Jr
sMolloJ
uorlr.sodsuetl
anrlerqdel uou
pup peletqder
s[ xelduo]
oql
Jl
sMolloJ
uorlrsodsuerl
anqerLldag
r
'uorJleal
loJsupll
puptls
e uaql
pue
'6uL1rru
r\q
pemollot
'VN6
n14
Jo
spua aq1 6ursdeur\s
fq xalduor
eql
sut.loJ esesodsuetl
n[4] ar.l]
r
'pue
qlpa
lp
puerls
auo
q6norql
alrs
1abre1
eq1
ol
pallauuol
sr uosoosuel]
oql
qrrqM
u[
xalduo]
la1suerl puells
e 6urturo; r\q
syels uorltsodsuetl
r
salPrpaujelul
uoru ruol
'saruanbas
pateadar
pareds
z{1asop
uJemleq
suortelJp
snoaueluods
saleraua8
leql
usrueqJeru
relnllaJ
auros ,(q
Jnlf,o
plnoJ
pue
luapuadapur-ytag
uor.lr.sodsuejl
loJ
