Lewin Benjamin (ed.) Genes IX
Подождите немного. Документ загружается.

10
20
30
40
50
60 70
^
80 90 100
.t1o
GGACCTGGAATATGGCGAGAAAACTGAAAATCACC'GAAAATGAGAAATACACACTTT
AGGACGTGAAATATGGCGAGIAAACTGAAAAAGGTGGAAAATTTGAAATGTccAcTGTA.
GGACGTGGAATATGGCAAGAAAAOTGAAAATCATGGAAAATGAGAAACATCCACTTG
ACGACTTGAAAAATGACGAAATCACTAAAAAA@TGAAAAATGAGAAATGCACACTGAA
200 210 220 230
lI+:.j$il
ti":?+
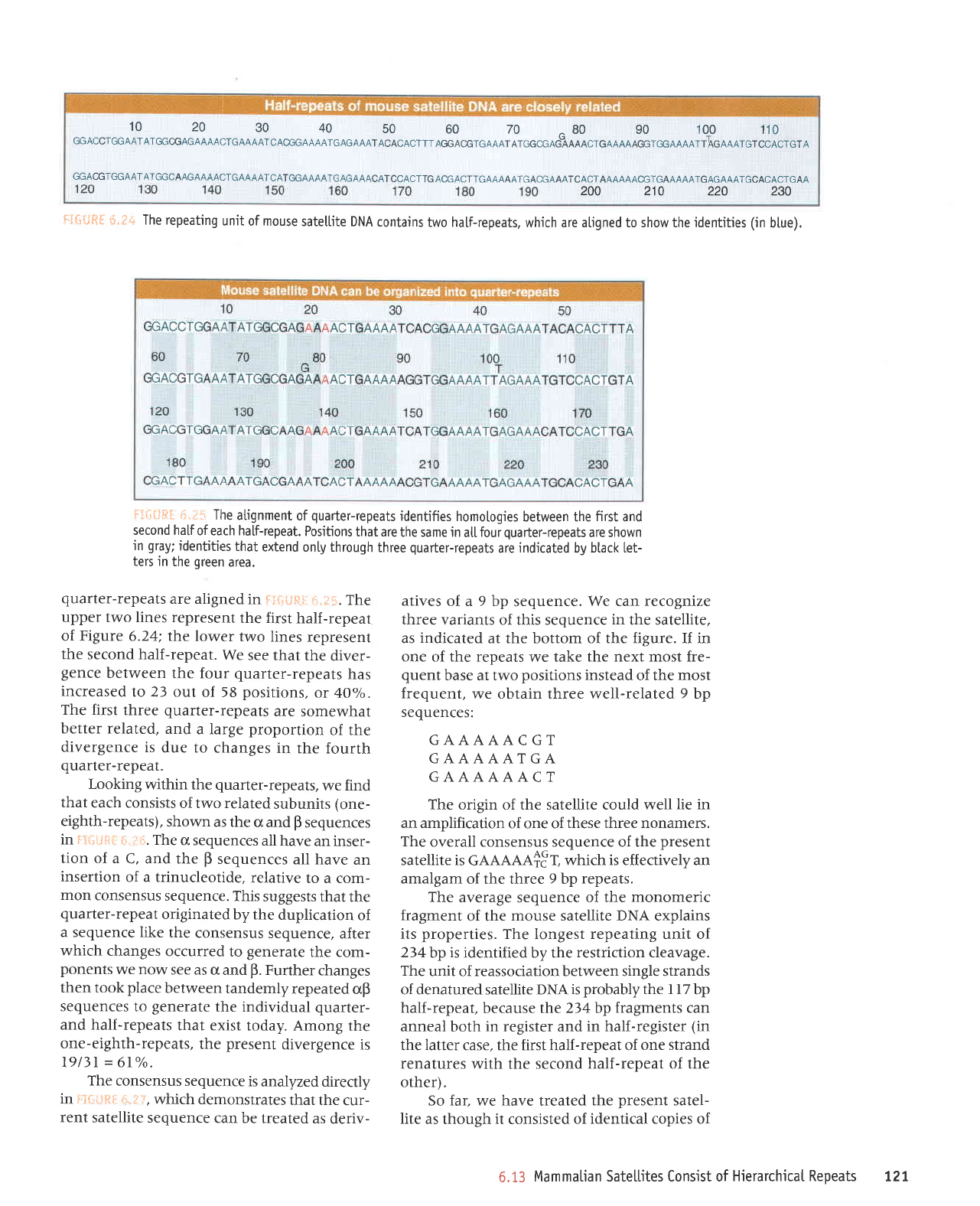
The repeating
unit
of mouse
satellite DNA
contains
two half-repeats,
which are atigned to show the
jdentities
(in
btue).
l:{-irin*i:
ii.;::..
The
atignment
of
quarter-repeats
identifies
homologies
between the
first
and
second half
of each half-repeat.
Positions
that are
the same in
atl
four
quarter-repeats
are shown
in
gray;
identities
that
extend
onty through
three
quarter-repeats
are
indicated
by
btack tet-
ters in
the
green
area.
quarter-repeats
are aligrred
in
$i.*#*l
ri=itl,. The
upper
two lines represent
the first
half-repeat
of Figure
6.24; the lower
two
lines
represent
the
second half-repeat.
We see
that the
diver-
gence
between
the four
quarter-repeats
has
increased
to 23 out
of 58
positions,
or 407o.
The
first three
quarter-repeats
are
somewhat
better related,
and
a large
proportion
of the
divergence
is
due to
changes in
the fourth
quarter-repeat.
Looking
within
the
quarter-repeats,
we find
that
each consists
of two related
subunits
(one-
eighth-repeats),
shown
as the
cr and
B
sequences
in
Fli-=ijli:
{r.;-:r-:. The
O( sequences
all
have an inser-
tion of a
C, and the
p
sequences
all have
an
insertion
of a trinucleotide,
relative
to a com-
mon
consensus
sequenc(:.
This
suggests that
the
quarter-repeat
originated
by the
duplication
of
a sequence
like the
consensus
sequence,
after
which
changes
occurred
to
generate
the com-
ponents
we now
see as
cx, and
B.
Further
changes
then took
place
between
tandemly
repeated
ap
sequences
to
generate
t.he individual quarter-
and
half-repeats
that exist
today.
Among the
one-eighth-repeats,
the
present
divergence
is
19 l3I
=
610/o.
The
consensus sequence
is
analyzed directly
in l3:ii.rtiii *-"1,i,
which denlonstrates
that the
cur-
rent
satellite sequence
can be
treated as
deriv-
atives
of a 9 bp
sequence.
We can recognize
three
variants of this sequence
in
the satellite,
as
indicated
at the bottom of the figure. If in
one of the repeats
we take
the next most fre-
quent
base at two
positions
instead
of the
most
frequent,
we obtain three
well-related 9
bp
sequences:
GAAAAAC GT
GAAAAAT
GA
GAAAAAAC T
The origin of the satellite could
well lie in
an amplification
of one of
these three nonamers.
The
overall consensus sequence of the
present
satellite is
GAAAAAf."t,
*hi.h is effectively an
amalgam
of the three
9
bp
repeats.
The average
sequence
of the
monomeric
fragment
of the
mouse satellite DNA explains
its
properties.
The longest repeating unit of
234bp is
identified by the
restriction cleavage.
The
unit of reassociation between single strands
of denatured satellite DNA is
probably
the I I 7
bp
half-repeat,
because
tl:'e 234 bp
fragments
can
anneal
both
in register
and
in half-register
(in
the latter case, the first half-repeat of
one strand
renatures with
the second
half-repeat of the
other).
So
far,
we have treated
the
present
satel-
lite
as though it consisted of
identical copies of
6.13
Mammatian Satettites Consist of
Hierarchical Repeats t2t

g1
GGACCTGGAATATGGCGAGAA
AACTGAA
pl
AATCACGGAAAATGA
GAAATACACACTTTA
CI2
GGACGTGAAATATGGCGAGAGA
AACTGAA
92
AAAGGTGGAAAATTTA
GAAATGTCCACTGTA
O3 GGACGTGGAATATGGCAAGAA
AACTGAA
O3
AATCATGGAAAATGA
GAAACATCCACTTGA
A4 CGACTTGAAAAATGACGAAAT
CACTAAA
F4
AAACGTGAAAAATGA
GAAATGCACACTGAA
cont"n.ty.tt,#ffi$ffiffi
ffiw.'..
:
ta.
. . . .
Ancestral?
AAACcTGAAAAATGA GAAATGCACACTGAA
a:i:iiii+
ii.iil The atignment of eighth-repeats
shows
that each
quarter-repeat
consists of an
s
ha[f and a
B
hatf. The consensus
sequence
gives
the
most common
base at each
position.
The
"ancestra["
sequence shows a
sequence
very closety
retated
to
the consensus sequence,
which coutd
have
been the
predecessor
to the cx
and
B
units.
(The
satettite sequence
is con-
tinuous, so that
for
the
purposes
of deducing
the consensus
sequence
we
can treat
it as a cjr-
cutar
permutation,
as
indicated by
joining
the
last GAA triptet
to the
first
6 bp.)
the 234 bp
repeating unit. Although this unit
accounts for the majority of the
satellite, vari-
ants of it also are
present.
Some of
them are
scattered at
random throughout the satellite;
others are clustered.
The existence of variants
is implied by our
description of the starting material
for the
sequence analysis as the
"monomeric"
frag-
ment. When the satellite is digested by
an
enzyme that
has
one
cleavage site in the
234
bp sequence, it also
generates
dimers,
trimers,
and
tetramers relative to the 2)4bp length.
fhey arise when a repeating unit
has lost the
enzyme cleavage site as the result of
mutation.
The monomeric
234
bp unit
is
generated
when two adjacent repeats each
have
the
recog-
nition
site.
A
dimer occurs
when one unit has
Iost the site, a trimer is
generated
when
two
adiacent units have lost the site, and so on.
With
some restriction enzymes, most of the satellite
is cleaved into a member of this repeating series,
as
shown
in the example
of
*ii,t::+*,
*.::ij;.
The
declining number of dimers, trimers, and so
forth shows that there is a random distribution
of
the repeats in which
the enzyme's
recogni-
tion
site
has
been eliminated by
mutation.
Other
restriction
enzymes show a different
type of behavior with the satellite DNA.
They
continue
to
generate
the
same series of bands.
Ihey
cleave,
however,
only a small
proportion
of the DNA, say 5oh-looh. This implies that a
certain reqion of the satellite contains a con-
CHAPTER
6
Ctusters and Repeats
centration
of the
repeating units with this
par-
ticular
restriction site. Presumably the
series of
repeats in this domain
all are derived
from an
ancestral
variant that
possessed
this recogni-
tion site
(although
in the usual way, some
mem-
bers since have
lost it by mutation).
A satellite DNA
suffers unequal recombi-
nation. This has additional
consequences when
there
is internal repetition
in
the
repeating unit.
Let us
return to our cluster consisting of
"ab"
repeats. Suppose that
the
"a"
and
"b"
compo-
nents of the
repeating unit are themselves suf-
ficiently well related to
pair.
Then
the
two
clusters can
align in half-register, with the
"
a"
sequence
of one aligned with
the
"b"
sequence
of the other.
How frequently this occurs will
depend
on the closeness of
the relationship
between the two
halves of the repeating unit.
In mouse satellite
DNA, reassociation between
the denatured satellite
DNA strands invitro com-
monly occurs
in
the
half-register.
When a recombination event occurs out of
register, it changes the
length
of the
repeating
units
that are involved in the reaction:
ffisl*.ffiffi,$i*ffiffii*hg.i$aba
ba babababababy
'{'
xa ba ba ba ba bababiili*,F.i{,Hffitrtftii$iilHi*tifi$i#iii
J
xababababababababaababababababababv
t'
xababababa ba ba ba bbabababababababv
722
GGACCT
GGAATATGGC
GAGAAAACT
GAAAATCAC
GGAAAATGA
GAAATCACT
TTAGGACGT
GAAATATGGC
G
A G AGA
A A
C T
GAAAAAGGT
G G
A A
A A
TTT
A
G
A A
A T-C
A C T
GTAGGACGT
GGAATATGGC
AAGAAAACT
GAAAATCAT
GGAAAATGA
G A A
A C-C
A
C T
TGACGACTT
GAAAAATGAC
GAAATCACT
AAAAAACGT
GAAAAATGA
G A
A A T-C
A C T
GAA
GzoAro&r A2oA12A17T8
G11 As
Tz%&QTrs
c?
.
indicates
inserted
triplet in
B
sequence
C
in
position
10
is extra
base in
cr sequence
li...iiSi:: r:.":.:,
The
existence
of an
overatI consensus
sequence is
shown
by
writing
the sateltite
sequence in
terms
of a 9
bp
repeat.
In
the upper
recombinant
cluster,
an
"
ab"
unit has been
replaced
try an
"aab"
unit. In the
lower
cluster,
lhe
"ab"
unit has
been replaced
by a
"b"
unit.
This
type
of event
explains
a feature
of the
restriction
digest
of rnouse
satellite
DNA.
Figure
6.28
shows
a fainter
series
of bands at
lengths
of Y', l'/r, 2Yr,
an<I 3/z repeating
units, in
addition
to
the stronger
integral
length repeats.
Suppose that
in the
preceding
example,
"ab"
represents
the 234
bp repeat
of
mouse satellite
DNA,
generated
by cleavage
at a site in
the
"b"
segment. The
"a"
and
"b"
segments
correspond
to the I I7
bp half
-repeats.
Then,
in the
upper recombinant
cluster, the
"aab"
unit
generates
a
fragment
of I
% times the
usual
repeating length.
In
the lower
recombi-
iriirr.iii;l
ir,,1
:l
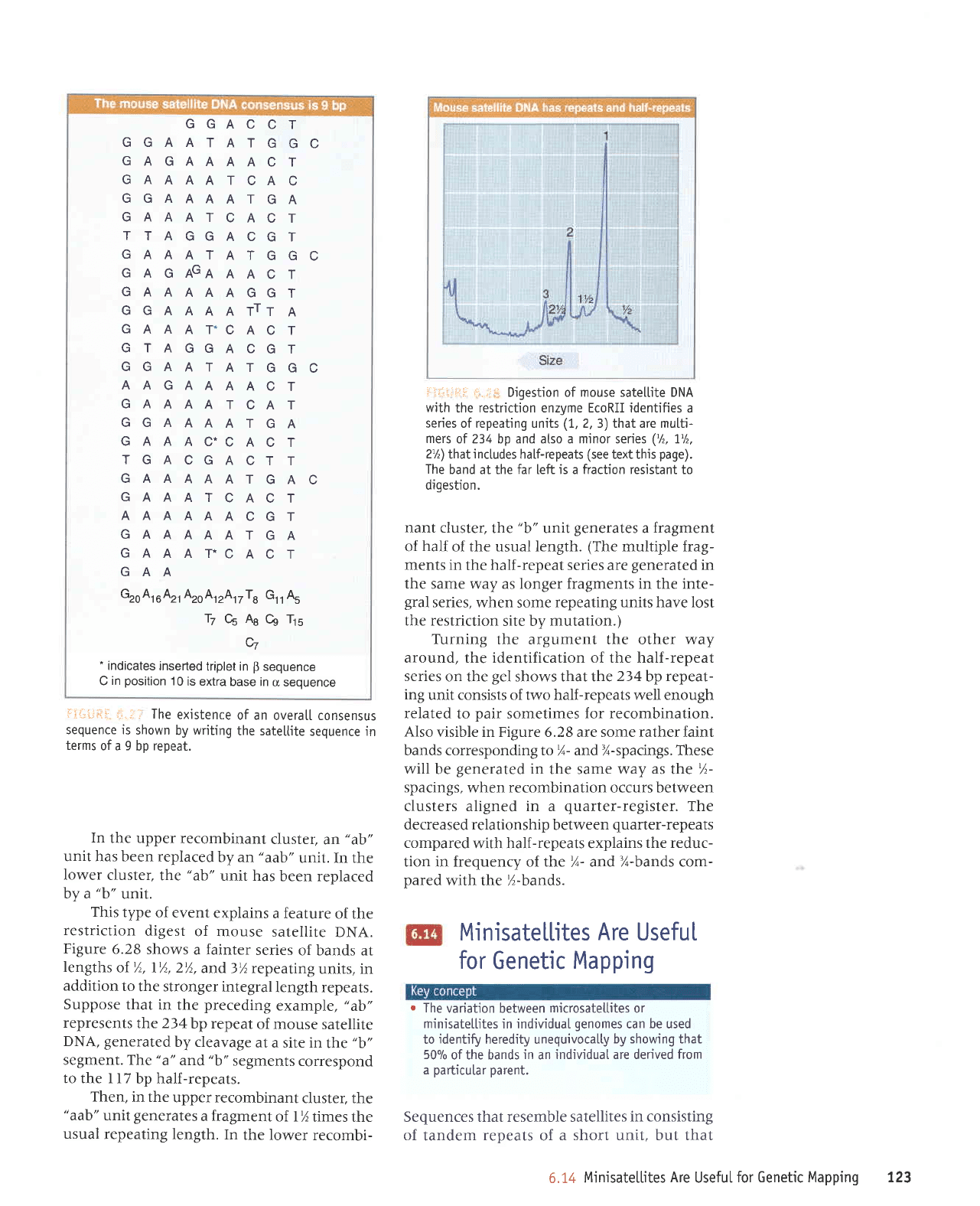
Djgestion of mouse satetlite DNA
with
the restriction enzyme EcoRII identifies a
series of repeating units
(1,
2, 3) that are mutti-
mers
of 234 bp and atso a minor series
('/,,
1'/,,
2%)
that
inctudes
half-repeats
(see
text this
page).
The
band at the far left is a fraction resistant to
digestion.
nant cluster,
the
"b"
unit
generates
a
fragment
of
half
of the usual length.
(The
multiple frag-
ments in
the half-repeat series are
generated
in
the
same way as longer fragments in the inte-
gral
series,
when some
repeating units have lost
the restriction
site by
mutation.)
Turning
the argument the other way
around, the identification of the half-repeat
series
on the
gel
shows that the
234bp repeat-
ing
unit consists
of
two half-repeats well enough
related
to
pair
sometimes
for recombination.
Also
visible in Figure 6.28 are some rather faint
bands corresponding to',4- and
Z-spacings.
These
will
be
generated
in
the same way
as
the z-
spacings,
when recombination occurs between
clusters aligned in a
quarter-register.
The
decreased relationship
between
quarter-repeats
compared
with half-repeats explains
the reduc-
tion in frequency of r}":'e
/^-
and %-bands com-
pared
with the
Z-bands.
Minisatellites
Are
UsefuL
for
Genetic
Mapping
.
The
variation between
microsate[tites
or
minisatellites
in individuaI
genomes
can be used
to
identifo
heredity unequivocalty
by showing that
50%
of the bands
in
an
individuaI are derived
from
a
particu[ar parent.
Sequences that resemble satellites
in consisting
of
tandem
repeats
of
a short unit, but that
6.14
Minisatettites
Are
Useful
for Genetic Mapping 723
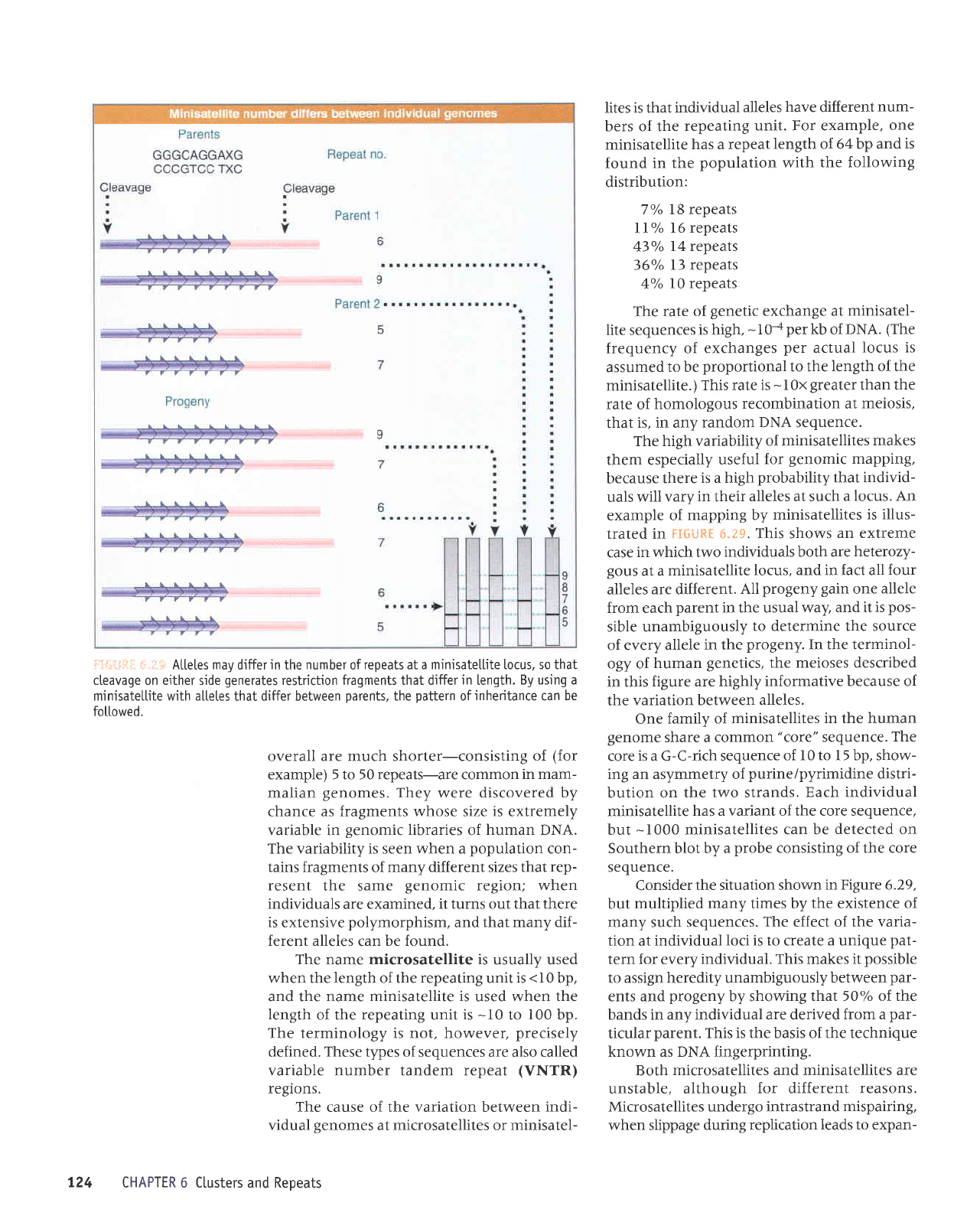
ii.i--l,riii:::.,;.:r Attetesmaydifferinthenumberofrepeatsataminisatellitetocus,sothat
cteavage on either side
generates
restriction fragments that differ
in
length.
By using a
minisatettite with altetes
that differ between
parents,
the
pattern
of
inheritance
can
be
fottowed.
overall are much shorter-consisting
of
(for
example) 5 to 50 repeats-are common
in mam-
malian
genomes. They
were discovered
by
chance as fragments whose size is extremely
variable in
genomic
libraries
of
human DNA.
The variability is seen when a
population
con-
tains fragments
of
many different sizes that
rep-
resent
the same
genomic
region;
when
individuals
are examined,
it turns out that there
is
extensive
polymorphism,
and
that many dif-
ferent alleles
can
be found.
The
name microsatellite
is
usually used
when the length of the
repeating
unit
is
<10
bp,
and the name
minisatellite is
used when
the
Iength
of the repeating unit is
-10
to
100
bp.
The terminology is not, however,
precisely
defined. These tlpes of sequences are also called
variable number tandem repeat
(VNTR)
reglons.
The cause of the
variation
between indi-
vidual
genomes
at microsatellites or
minisatel-
124
CHAPTER 6 Ctusters and Repeats
lites
is
that
individual
alleles
have
different
num-
bers of the
repeating
unit. For example,
one
minisatellite
has a repeat
length of 64 bp and
is
found
in the
population
with
the following
distribution:
7% 18 repeats
II%
16 repeats
43%
14 repeats
360/o
13 repeats
4% I0
repeats
The
rate of
genetic
exchange at
minisatel-
lite sequences
is high,
-I0aperkb
of
DNA.
(The
frequency of
exchanges
per
actual
locus is
assumed
to be
proportional
to the
length of the
minisatellite.)
This rate
is
-l0x
greater
than the
rate of
homologous
recombination at
meiosis,
that
is, in any
random DNA sequence.
The high
variability of minisatellites
makes
them especially
useful
for
genomic
mapping,
because
there
is a high
probability
that individ-
uals will
vary in their
alleles at such a
locus. An
example of
mapping by
minisatellites is
illus-
trated in
FISUR{ $"t9. This shows
an extreme
case
in
which
two
individuals both are
heterozy-
gous
at a
minisatellite
locus, and in fact all
four
alleles
are different.
AII
progeny gain
one
allele
from each
parent
in the usual way,
and it is
pos-
sible unambiguously
to determine the source
of every
allele in the
progeny.
In the terminol-
ogy of
human
genetics,
the meioses described
in this
figure are highly
informative because of
the variation between
alleles.
One
family of minisatellites
in the human
genome
share a common
"core"
sequence.
The
core
is a G-C-rich sequence
of l0 to l5 bp, show-
ing an asymmetry of
purine/pyrimidine
distri-
bution on
the two strands.
Each individual
minisatellite
has a
variant
of the core sequence,
but
-1000
minisatellites can be
detected on
Southern blot by a
probe
consisting of the core
sequence.
Consider
the situation shoq'n
in Figure 6.29,
but
multiplied many times by
the existence of
many such sequences.
The effect of the varia-
tion at
individual loci
is
to create a unique
pat-
tern for every
individual. This makes it
possible
to assign heredity unambiguously
between
par-
ents and
progeny
by showing
that 50% of the
bands in any individual are derived
from a
par-
ticular
parent.
This is the basis of the technique
known as DNA fingerprinting.
Both
microsatellites and minisatellites are
unstable, although
for different reasons.
Microsatellites undergo intrastrand
mispairing,
when
slippage
during replication
leads to expan-
sion
of the repeat,
as
shown in i:jr,rljFri:
ri .ii,r.
Sys-
tems
that repair
dameLge
to DNA-in particu-
lar
those
that recognize
mismatched
base
pairs-are
important
in reversing
such
changes,
as shown
by a large increase
in
frequency
when
repair genes
are inactivated.
Mutations
in repair
systems
are an important
contributory
factor
in
the development
of cancer,
thus
tumor cells
often display variations
in
microsatellite
sequences.
Minisatellites
undergo
the
same
sort of unequal
crossing-over
between repeats
that we have
discusse
d for
satellites
(see
Fig-
ure
6. 3
)
. One telling
carse
is that increased
vari-
ation is associated
wit.h
a meiotic
hotspot.
The
recombination
event is not
usually
associated
with recombination
b,etween
flanking
mark-
ers,
but has a
complex form
in
which the new
mutant
allele
gains
inlormation
from
both the
sister
chromatid
and the
other
(homologous)
chromosome.
It is
not clear
at w.hat repeating
length
the
cause
of the variationL
shifts from
replication
slippage to recombinartion.
@
Summary
Almost
all
genes
belorrg to
families,
which are
defined
by the
possession
of related
sequences
in
the exons
of indivirlual
members.
Families
evolve
by the duplicat.ion
of a
gene (or genes),
followed
by divergence
between
the
copies.
Some
copies suffer
inactivating
mutations
and
become
pseudogenes
that
no longer
have any
function.
Pseudogenes
also may
be
generated
as DNA
copies
of the mRNA
sequences.
An evolving
set
of
genes
may remain
together
in a cluster
or may
be dispersed
to new
locations
by chromosomal
rearrangement.
The
organization
of existing
clusters
can
sometimes
be used to infer
the series
of events
that has
occurred.
These events
act
with regard
to
sequence rather
than function,
and
therefore
include
pseudogenes
as well as
active
genes.
Mutations accumu.late
more
rapidly in
silent
sites than in replacement
sites
(which
affect the
amino acid
sequence). The
rate of
divergence at
replacement
sites
carL be used
to establish
a
clock, which can
be calibrated
in
percent
diver-
gence
per
million
years.
The clock
can then
be
used to calculate the
time of
divergence between
any two members
of t)re family.
A tandem
cluster consists
of many
copies
of a repeating
unit that includes
the transcribed
sequence(s)
and a nontranscribed
spacer(s).
rRNA
gene
clusters
cocle only
for a single rRNA
irri.il+lr:
L;
.
.,
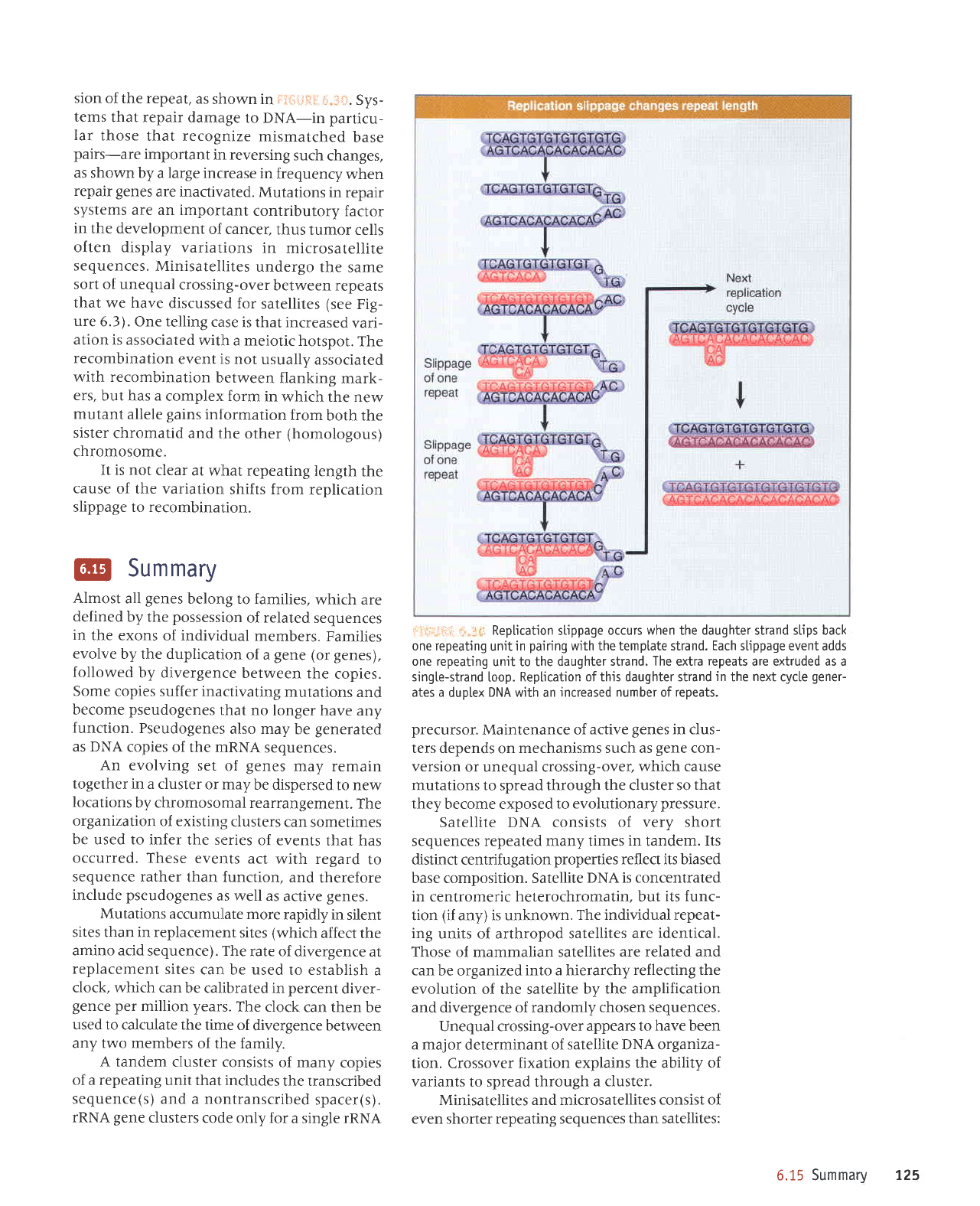
Rep[ication stippage
occurs when the daughter strand stips back
one repeating
unit
in
pairing
wjth the template strand. Each slippage event adds
one repeating
unit to the
daughter strand. The extra
repeats
are extruded as a
singte-strand
[oop.
Reptication of thjs daughter strand
in
the
next
cycte
gener-
ates a
duplex
DNA
with an
increased number of repeats.
precursor.
Maintenance of
active
genes
in clus-
ters depends
on
mechanisms such as
gene
con-
version
or unequal crossing-over,
which cause
mutations to
spread
through the cluster so
that
they
become exposed
to evolutionary
pressure.
Satellite DNA consists
of
very
short
sequences repeated many times
in
tandem.
Its
distinct centrifugation
properties
reflect its biased
base composition. Satellite
DNA is concentrated
in centromeric heterochromatin, but
its func-
tion
(if
any) is unknown.
The individual repeat-
ing
units of arthropod satellites
are identical.
Those of mammalian satellites
are related and
can
be organized
into a hierarchy
reflecting the
evolution of the satellite
by the amplification
and divergence of randomly
chosen sequences.
Unequal crossing-over
appears to
have been
a major
determinant of
satellite DNA organiza-
tion. Crossover fixation explains
the ability of
variants to spread through
a cluster.
Minisatellites and microsatellites
consist of
even shorter repeating sequences
than satellites:
6.15 Summary
725

<10
bp for microsatellites and
I0
to
50 bp for
minisatellites. The number of repeating units
is
usually 5 to 50.
There is high variation in the
repeat number between individual
genomes.
A
microsatellite
repeat number
varies
as the result
of slippage during
replication; the frequency
is
affected by systems that
recognize and repair
damage
in DNA. Minisatellite repeat
number
varies
as the result of
recombination-like events.
Variations in repeat number can be used
to
determine hereditary
relationships
by
the tech-
nique known as DNA fingerprinting.
References
Gene
Duptication Is a Major Force
in Evolution
Resea rch
Bailey, J. A., Gu,
2.,
Clark,
R. A., Reinert, I(.,
Samonte, R. V., Schwartz, S.,
Adams, M. D.,
Myers, E. W., Li, P. W, and Eichler, E. E.
(20021.
Recent segmental duplications
in
the
human
genome.
Science 297, 1003-1007.
][
Gtobjn Ctusters
Are Formed
by
Dup[ication and Divergence
Review
Hardison, R.
(1998)
Hemoglobins from
bacteria
to
man:
evolution of different
patterns
of
gene
expression. J. Exp.
Biol.2Ol, l}99-ll17.
@
The Rate of Neutral Substitution Can
Be
Measured from Divergence
of Repeated
Seouences
Resea rc h
Waterston, R. H.
et al.
(2002\.Initial
sequencing
and comparative analysis of the mouse
genome.
Nature
420,
520-562.
Pseudogenes
Are Dead Ends of Evotution
Resea
rch
Hirotsune, S., Yoshida, N., Chen,
A., Garrett, L.,
Sugiyama,
F., Takahashi, S.,
Yagami, I(.,
Wynshaw-Boris,
A., and
Yoshiki, A.
(2003).
An
expressed
pseudogene
regulates the mes-
senger-RNA
stability of
its homologous coding
gene.
Nature
42), 9 l-96.
Crossover
Fixation Coutd
Maintain
IdenticaI Repeats
Review
Charlesworth,
B.,
Sniegowski,
P., and Stephan, W.
(19941
. The evolutionary
dynamics of repeti-
tive
DNA in eukaryotes.
Nature 771,215-220.
Minisatettites
Are Usefu[ for Genetic
Mapping
Research
Jeffreys,
A. J.,
Murray,
J.,
and
Neumann,
R.
(
1998). High-resolution
mapping of
crossovers
in human sperm defines a
minisatellite-associated
recombination
hotspot.
Mol.
Cell
2, 267-273.
Jeffreys,
A. J., Royle, N. J., Wilson,
V.,
and Wong, Z.
(1988).
Spontaneous
mutation rates
to
new
length alleles at tandem-repetitive hypervari-
able
loci in human DNA. Nature 332,278-281.
Jeffreys, A. J.,
Tamaki, I(., Macleod, A., Monck-
ton, D. G., Neil, D.
L.,
and
Armour,
J.
A.
(L994).
Complex
gene
conversion events in
germline mutation at human minisatellites.
Nat. GeneL6, l)6-145.
Jeffreys,
A.
J.,
Wilson,
V.,
and Thein, S. L.
(1985).
Hypervariable minisatellite regions in human
DNA. Nature il4, 67-73.
Strand, M., Prolla,
T. A., Liskay, R. M., and Petes,
T. D.
(1991).
Destabilization of
tracts
of
simple
repetitive DNA in
yeast
by
mutations
affecting
DNA mismatch
repair.
Nature )65
,
27 4-27
6.
726
CHAPTER 6 Clusters
and Reoeats
LZI
a6od
Wu
uo
panuquoJ
'der
,E
aq1 o1 filparLp
purq
Aeur
qlat
leulup
Jo
aselfiuepeep
eql
o
'uorpalrp
,E€-,€
aql ur
ll0/v1
lpql
sase
-allnuoxa
lp.lalas
Jo
xelduor
e
sesn fervrqled
lseefi
teqlouy
r
'
,€.<-,g
ruor; uorleperEap
rrlfloalrnuoxa
sonlonur fiemqled
lsear\
eug
r
'(y)r{1od
,€
oqt
pup
dpl
,g
aql
jo
le^oual
satrnber
ypXtu
1seefi
1o
uoqeperba6
r
saqnqlv
aldqlnhj sa^lo^uI
uoqeper6a6
ypgur
'(y)[1od
1o
ssol Aq
pera66u1
aq feu uoqezrlrQp]se(
o
'sllaJJa
Durzrtrqels0p
lo 6urzrlrqpls eneq feu
VNUru
up urqlLrvr
seruanbas
tJJoodS
.
'sasPallnuoxa
r{q
uoqeper6ap
lsuLeEe lL
lralord
VNUtU
Jo
spua
qloq
le
suotlplqr.pour
arlf
r
aruanba5
pup
alnllnlls
sll uo
spuada6 f1g11qe15
y11pLn
'/E<-/€
ulolJ
luaubelJ
eql
J0
uorleper6ap rqAloalrnuoxa
fq parvrollol
sabe^eall lrl^l
-oallnuopua
Jo
uotlpurquol
aql ulol; sllnser
uorlepet0e6
r
',€-,9
sl
VNUU
leuallpq
1o
uoqeperbap
10
uoqlelrp
llpla^o
aql
.
saur{zu3
aldr}ln1l1 sa^lo^ul
uoqeper6a6
VNUu
leuappg
'uoLleper6ap
lsurebe
VNUU
aql sozltrqels (y)[1od
aq1
o
'(agVa)
uralord
r4oads e r{q
punoq
sr
(y)A1od
aq1
r
'
uoLldursuetl
ta11e
ldursuetl
leollnu
e ol
peppe
sr 6uo1 sapLloaltnu
0OZ-
(y)ri1od;o q16ue1 y
r
palelfuapeflod
sI snuru.uaf,t
aql
'aursouenb
lpurulal
Mau
aql
Jo
osoqu
lo aspq aql ol
poppp
ere sdnorb
lriqlau
ea:q1
ol aug
'luu
,g-,9
e er
ldursuerl
aql
Jo
aspq
leurutal
aql ol
g
e burppe Aq
patuo;
sr der
,9
y
r
paddel
sI
VNUUr
l]]o&plnl
Jo
pul
,E
aql
'palelduol
uaaq e^eq
suorlelqrpout
11e
rale Aluo
Luseldoflr
aql o] snellnu
aql uol1
palrodxe
sr
[t{!ur
o{]
o
'pua
,€
aql
1e
(y)f1od
;o
eruanbas
e
pue pua
,9
aql
le
der
pa1e1r{q1au
p
}o
uorltppp
aql
apnlrur suogelglpoul
a{l
o
'uorldursuerl
rege f11oqs
ro 6ur
-rnp
snallnu aql ur
palJrporu
sL
ldursuerl
y;1gu
rqo&e1na
y
r
u0ll0ulsuPll
sll rauP ro 6uun0
pa$lpoli|
sI
vNurx
lqo&plnl
'sauaE
luare;;rp
luasardal leql
suor6el
6urpor
lpro^as
6uLlreq
ur lruollso^lod eq r\eu
ypgtu
lpualreQ
!
o
's0lnurur
rvral e ,{luolo ajll-JlPrl P
seq
puP
elqPlsun sl
vNUr.u
leua|les
r
'palalduor
ueeq sPq srsoql
-uAs
s1r aloJaq
VNUrU
ue 6ur1e1suer1 ur6eq
sauosoqu sp'pu
-apeq
ur {lsnoauellnuLs rnllo uollelsuell
pue
uoqdursuetl
r
vNU
raEuessew
leuapeg ,o
allfl aJll aql
'vNUu
aql
6uo1e uorssarbord
1o
abels
lualoJJrp
p
lp
sr auosoqu
qlel
'seuosoqu
lpra^as
Aq
pelplsupll
flsnoaueqnurs sr
!trl!ur
u!
r
vNUu
auo ol
pu!8
sauosoqtu
AUPy\|
',E
ol
,g
urorj
VNUrx
ue
6uolp soAou aulosoQp
!
r
'suopol
1a1du1
6urpuodsoliol aql ol asuodser
ur urpql
apqdadrtlod 6uLtrtotb
aql ol sptlP ouil.uP
ppP
SVNUI
lfireourLue
qlrqm
ur
luauuorlnua
aq1 saptnotd aulosoQu a{l
o
'(SOl
ro
5gg)
lrunqns
llpurs
e
pue (salortelna pue
eual
-rpq
roJ
S09
ro
S0g)
trunqns
e6rel e;o stslsuor tt::::lj
.
(
saui0s
-oqu
rLlortelna
loJ
SO8
pue
saurosoqu
leualleq
ro;
S0/)
uorleluaurrpes
ro
olpl .lroql
fq
pazuelrereql
arp saulosoQt!
r
sauosoqru
fq
palelsuerl
sI
vNU
la6uassaw
'DUe
raqlo
eql
lP
ru.le uopolrlue
eql
puP pua
auo
le
ule
loloellP
aql
ql$
ornpnrls
Arerlal
pedeqs-1
up suuoJ
JpapaAolr
ell
o
arnpn.lls
rteqtal
aqt
Jo
spul
]e
arv uopolltuv
pue
ulats loldallv aql
y11911[reouruP
aql
Jo
Altrgnads
eql .loJ alqrsuodsar
rtlalos sr uopolrlup aq1
lo
eruenbes alf
r
'p!lP
ourup aqJ
Jo
dnor6
H00l
aqr ol ulP roldarle
eql
]o
puo
oql
lp
prrp
rqfuape
eq1
1o
dnor6
H0
,€
lo
,z
aql
uolJ
lutl
lalsa up 6urLuro;
fiq
yp1llAreoutue
ulo; o1
pebteqr
s!
VNUI
o
'(sVt1Ul
ra6uol aq1 ur ulp
leuorlrppp
ue
pue)
sulp
luplsuor
rnoJ
qltm
alnllnlls
f.repuoras
Jpapa^oll
P olur sploJ
leql
saseq
96
olrl
J0
aluanDas
e se!
![l] !
r
JPaua^oll
P suuol
vNU
ralsuPll
'VNU
olul
poqulsupll
sl
VN6
]o
spue.rls oul aq1
1o
euo [1ug
r
patelsuPll
sI
puP
uoqdulsuell
r{q parnpol6 sI
vNulll
u0!pnpollul
sNrlrn0 usrdvHS
01 ^lrlrqp eqt seq
tl
pue
'lueuodrur
sr aJntf,nrts
IeuorsuJrurp-aeJqt
sll
:VNU
Jo
asn eqt Sururano8
sJruJql uoururo)
Jql
Jo
oMI JJS J^^'vNul uI
'll
uorJ
pJJnp
-ord
sr
teqt
uratord
Jo
lunorue
Jqt eroJJreql
pue
,{ttnttre
s1r 3ur11or1uoJ
ur alor
luelrodurr
ue Leld uer-uorSar
Surpor aqt
Jo
aprs JJqtra
uo saruanbas
aqt relnrrlred ur-vN6ru
aql
Jo
eJnpnJls Jql
'JJAJMo11 'uratord
Surpuodsar
-JoJ
Jql ur
prf,p
ourure ue stuasardal
VNUru
Jql
;o
uor8ar Supor
Jqt urqlrM
laldrrt
eprtoelJnu
qJeJ :JJnlea;
luelrodrur
aql sr J)uJnbJS
sll
'VNU
ra8uassaru
JoC
'llurlsrp
sr sJseJ eseql
Jo
q)pa
ur sLeld
yNU
reqr
rlor
Io
ad.{r ar{l
'ureqJ
aprtdad.{lod e
otur sprJp ourure
SurzrrauLlod dllenpe
roy snlereddp Jqt
sJprnord
qrrqlr pue
'sluauodluoJ
VNU
slr sp
IIJM
se suralord
.dueru
sureluoJ
leql
xalduor
uratord
-oepnuoqu
a8rel e
'Juosoqu
aqtJo stuau
-oduor
are
(y111gr)
sVNU
IpruosoQlU
.
'vNuru
ur uopoJ .relnrrlred
rIJeJ ol Surpuodsar
-roJ
spr)p ourure apnord
01
pJsn
JJe
lpql
svNU
llerus
are
(ypg1)
svNU relsue{ .
'uratord
sluasardar
teqt
aruanbJS
VNO
e;o Ldor
aqt sarJJeJ
tpqt
Jlerpeurelur
ue saprnord
(VNUur) yp6
ra8uasgW
.
:sureloJo
Jo
uorlJnp
-o:d
aql ur
panlolur,{prarrp
are
VNU
Jo
sJsselJ
Joleru eeJl{f
i
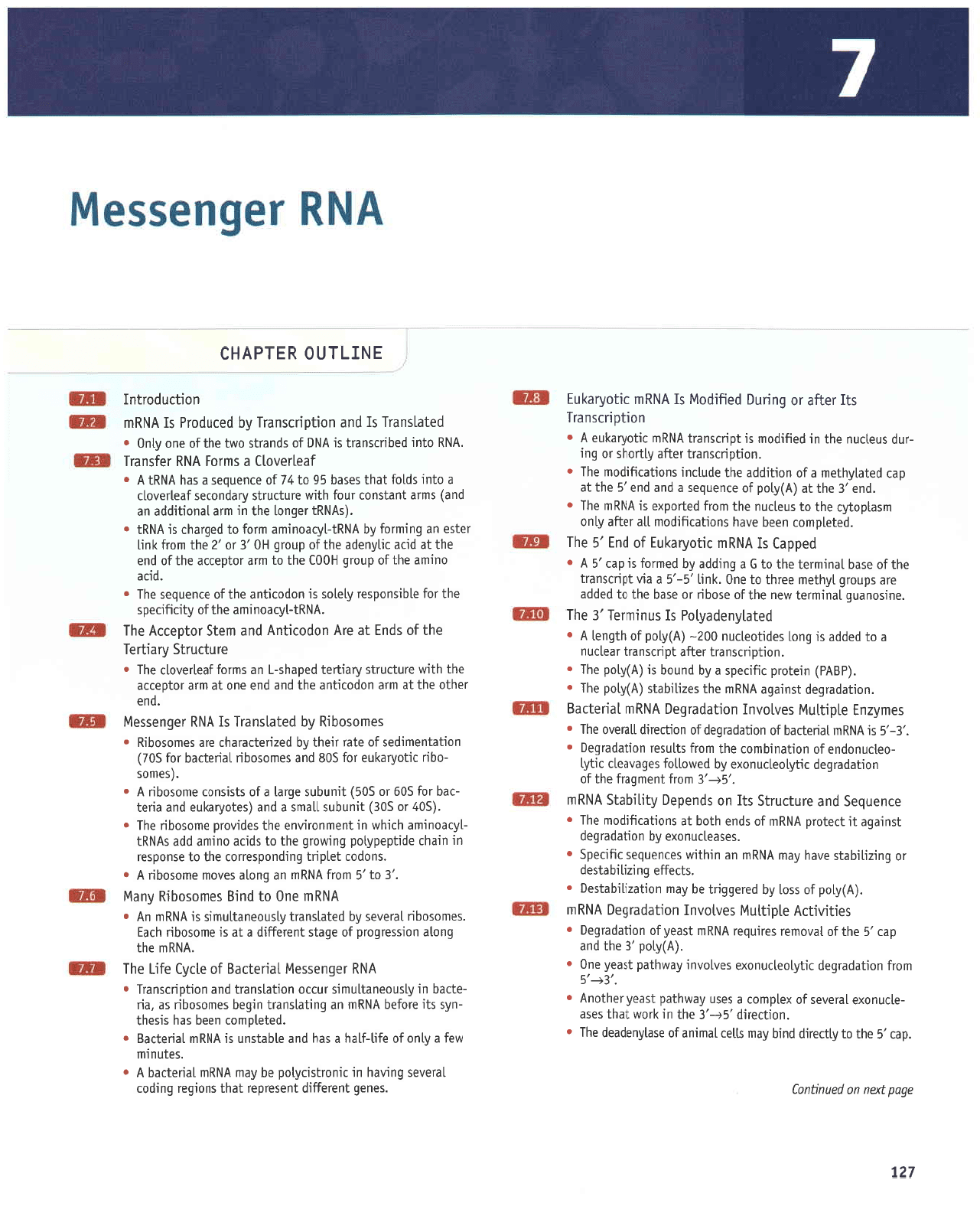
i
:i..]irij:::1
uI
pJzlrplutuns
sv_
'AruJrJrJla
AIqe
-qord
pue
f,tllllesran
uI JSeJJf,UI
luanbasuor
e
qllm'sulJtoJd.{.q
rano
ue>lel ro
patsrsse
.{lruanb
-esqns
arem suorl)unJ
JSJqI
;o
.{.ueyg
'uor1
-eruJoJur
)rlaue8 Surssardxa
pue
Sururelureru
ur
tuauodruoJ
enrtJe
aql
.dlleur8rro
spM
VNU
qJIqM
ur
,,plJoM
vNu.,
ue ur
pJAIoAe
a,req.deru
ssJ)oJd erllue Jql
leql
MerA
leraua8
Jql suod
-dns
uorssardxa
auaS
qll,lr pJuJJJuoJ
suorl)unJ
dNU
robuassau{
/
ulldvHl
'(srseqlufis
uralold
rol
luauluorrnua
eq1 saprnord
lpql
auosoqu eqt;o
lueuodruol
roleLu e)
VUUI
pue
'(uopor
qlea
ol 6urpuodsauor
pop
oururp aq1 sepnord)
ypgl'(aruanbas
6uLpor aq1
seurer)
VNUUr
orp uotsserdxa euab
ro1
patrnbar
Alesrantun
y15
1o
sed^l aajql aql
i
'.r-
ri,,:iili]!j
Lueur
ur
VNU
Jo
tuJruJ^lolur
aq1
'uorssardxa
auaS;o sa8els raqlo
le
seloJ
leuoll)unJ
Jo
IpJnl
-rnr1s
.,{.e1d
teql
perJ^oJsrp
ueJq J^eq svNU
:aqlo
.{ueru
uaql eJurs
lnq
'srsaqluLs
uralord
ur elerpeurJlur up sp
pezrJelf,eJeqJ
lsJrJ
seM
U
'uorssardxa
aua8 ur
radeld
IeJtuJJ
e sI
vNU
uoqlnporlul
8Z\
seseq
96-tl
aouer azrs
:aJnlcnlls
fuepuoces oArsuelxa
qlrm
vNU ll€rus
e
sl
vNUl
saseq
OOO'O!-969
e6ue: ezrg
uralord
lueserdoJ leql
soseq
lo
ocuonbas e spq
VNHu.r
AlPutuns
'pnq
aq1 ur Aluo
punoj
sr
uralotd aq1
1eq1
os
'pnq
aql ur
palplsuptl
pup pa.loqlue
sr
lI
o
'pnq
ralqbnep
oql olur
sluaupl!J urlre
6uo1e
1r
syodsuerl
toJoru
!
o
'.ro1oru
ursofru
p
o] spurq
leql
ural
-ordoelrnuoqu
e
su.loJ
VNUu
lqsv
lspa1
.
pazrlelol
r{11ergrrad5
ag uel
vNUru
'sllal
luPlo
uaa/v\]aq saluPlsrp
6uo1
1aner1
upt svNUrll
.
'eupuoqlolrru
olur
po]Joour
a.lP asPalJnuoqu
P
Jo
]uauooulor
VNU
aql
puP
s![rll]
o
'snatlnu
eql
urolJ
palodxa
oq
lsnu
useldofr
aql
ur uorllunJ
1pq1
sVNU rrloAre>1na
11y
r
'e1rr1ed
uLelordoalrnuoqu e
sp ausrqueu e
q6norql payodsuerl
s!
VNU
o
palodsuerl
arv sVNU l!]o&Plnl
'suuoM
pue
lseefi
ur
puno1
ueeq eleq
LualsAs uoLleper6ap
aq1
rol 6uLpor seuo!
e
'p0perDap
aq
0l
VNUl.ll
asnel suotlPlnU oSUaSUo[rl
o
uals[5 a]upllra^tnS
e ra66u1 suoqeln111 asuasuoN

6Zl
palelsuell
sI
pue
uoqdulsuelf
Aq
pe:npor6
sI
VNUrU
Z./
'spr.lP
ourulp
ual toJ apol
qlrqM
'dq
0€
ureluol
xtloq
alqnop
vNo
aqlJ0 sulnl
aalql'p|'rp
0urue auo 01u!
seseq
J0
laldul
qlea
sppal
uotlplsupll
'puetls
6urpor
VN0
eql se
aluonbos
aups
aql spq
pue puetls
alelduel
VN6
aql ol
fielueur
-alduol
s!
leql
VNU
ue seleraue6
uoLldursuerl
r ,r.
:i.f
riitij
'pazrsaqluAs
sr
1r
reqe
ro
uJq,lt.
IIal
eqt ur uorle)o1
radord
sll spurJ
uralord
p
Moq;o
uorlsanb
Jqt ol
uJnt eM
'uorlpzlleJoT
uralord
'g
1
raldeq3
uI
'VNUru
yo
aruanbas e
;o
SutuBaru
aq1
lardralur
ol
pesn
sr apoJ JrlJueB
aqt.dem
Jqt ssnrsrp
J,lr
'Jpo)
)rlJueg aql
Sursn
'6
raldeq3
uI'pJzrsJqlu.,{s
sr uratord e
qtrq.,vr,{q
ssaro:d
aql ssnlsrp
a,ra
'srsaqluLs
ulJtord
'g
JJI
-deq3
u1
'stsaqlu.ds
uratord ro; a1e1drua1 e se esn
str
pue
VNUrU
ssnJsrp am ratdeqt
slrlt
ul
'puerls
asues ro
pueJls
8ulpot
eql
pJIIpJ
sr
pue
(n
Jo
peelsur
1
Surssas
-sod
ro;
ldarxa)
VNUIU
aqt
se JJuJnbJS
au4ss
arql sJeeq
puPJls
VNq
lJqlo JI{J .
(.vNuru
ot
LreluarualdruoJ
sr
leqt
VNU
ro
VNC
Jo
aruanbas
p
eqrJJSJp
ol turJl
leraua8
e
se
pJSn
s asuas!ruV)
'pueJls
asuasllup
Jo
puPJls
aleldruat
Jql
pellet
sr SurJred
aseq
fueluaualdruor
erl
VNUtu
Jqt
Jo
srs
-aqlu,{s
stf,JJrp
teql
VN(
Jo
pueJts
Jr{I .
:r,
j
rrilitiih
i
ur
palrrdap
sp
vNC
Jo
spuells
o^{t Jql
qstn8utlsrp
e^A
'vNu
ra8uassaru
e otur
pJqIrJS
-ueJl
sr xaldnp
VN(
e
Jo
puelts
auo z(1u6
'prf,e
ourrue
auo sluasardar
uo6ar Surpot
rrll
Jo
(uopor)
1a1drr1
eprloJlrnu
qree
:epoJ
JrleueS aqt .dq
e)uenbJs uralord
p
ol
pateleJ
sr
teql
uor8ar
Surpor auo
lseJl
lP
sureluof
vNuur
q)pJ
lnq
'pJlpl
-suprl
lou
sl
vNuru
ue
Jo
r{l3uel eJrluJ
aql'uralord e Sursrrdruor sptre oulup
Jo
eJuenbJs Jq1 olur
yNgru
;o
aruanbas
JplloJllnu Jql SUa^uoJ
uollP{suPJf,
o
'ygq
xaldnp rqt
lo
spuens aql
Jo
Juo
qllm
aruanbes
uI
Ierlturpl
VNU
papuens-a13urs
e sJleJJuaB uorldursue4l .
'ssarord
a8els-ozlrl
e dq s;nlo uorssardxa auag
'vNU
olu!
poqulsuerl
sl
VNg
Jo
spuerls oml aq1;o euo fi1ug
.
palelsuPrl
sI
puP
uorldrrlsuerl
nq
palnpord
sI
vNULU
'uorssardxa
aua8 ur suorpunJ;o
.d1aue.t
p
qtrm
sygg 8ur
-po)uou
Jo
sselJ Jepeorq
qJnlu
E
;o
saldruexa
are
VNur
pup
VNUI
1nq
'anbrun
sr
VNuru
Jo
uorpunJ
Surpor eqJ
'selnJaloru
VNU
Jeqlo rprM
Llerr;nads
DprJlul
ol
pue
aJnlrnrls Lrepuoras
str urJoJ 01
qloq
'Suured
aseq
uodn dllerrlrrr
puadap
suortJunJ slr
leql
sr
'eJaqMesle
pup
srs
-aqluzls
uralord
qloq
ut
'yNu
Jo
saIlIAItJp Jql
;o 11e
q8norqt
sunr
ler{l
eueqt aqJ
'snteredde
leur8rro
aqt JoJ srseq
eqt uJJq JAeq [eru sygg
eql
leql
rJqruauJJ
plnoqs
rl!\
puP
'svNu
rrqlo
dq ro surelord raqlra
.dq
uorleln8ar
ro;
1a8re1
e
Jq upJ
leqt
pue
JIoJ a^rlJe ue sz(e1d
leql
lueu
-odruoJ p
se
lr
,lrerl
ol JAeq
aa,r
'srsaqluLs
ural
-oJd
ur
VNU
Jo
JIor eql JeprsuoJ a,lr sp
'leql
sl
punorS>peq
slql
lnoqe
Surqt
tueuodrur
aq1
'svNuJ
Jql
Jo
Juo ur
seprsar,{lrnrlre
srql
'uralord
olut
palerodroJul
sI
prJe
ourue ue
qJrqm.{q puoq
aprldad
p
Jo
uorl
-pruJoJ
aql az[1e1er 01
^UIqe
aqt
sr JruosoqrJ
Jql
Jo
serlr^rpe
IerJnrJ
Jql
Jo
Juo
'auosoqrr
Jql
Jo
sJrlr^rlre
eql uI ,(lDarrp seledrJrued osIP
1I
'qJene
suralord
pruosoqu
qJrqM
ot
>lro,lrerueJJ
e
Suprnord ur
'lernlrnrts
sl
vNU
Jo
oloJ aug
'.d1r
-^rtJe
Jo
ad,{.1 raqloue eJS
JM
'yNUJ
r{llM
'pr)P
oulluP su dupues
-a.rdar
1a1drr1
epltoelrnu
Jql
qllM
srted
(uopor
-uup
Jql) JJuenbas
1a1dr4
goqs
e uJqM'3utrrcd
aseq
.{q
pJIIoJtuoJ
sr
pesn
st
yggl-lLreoulrue
up
q)rqm qtlm
^trJrJnads aq1
'stsaqluLs
utal
-ord
ro;
pesn
sr
tpql
JrntJnJls eql se
paztuSora.r
sr
qJrqM
'VNdl-1,{reoururp
up seleeJJ e8e
-Iull
JqJ
'prJe
ourue rgnads e o1 a8elurl ro; a1e
-rrdordde
sr
teqt ta8rcl
e Sutpurord
se atu.{zua
ue ,{.q
lsrq
pazruSorar
sI
eJnlJnJls
leuolsuJrulp
-aarqt
rqJ'(VNUru)
VNU
taqtoue
qltu
rrcd aseq
vNU
rabuassay{
/
ulldvHl
o€I
'pr)e
ourue ue ol
pa>lurl
eq uP)
dnor8
gg-,9
Jo
-,2
aaJJ
JSoqM aruanbas
pa;redun
uP ur spuJ
leql
uals
parred
-JSeq
e
Jo
stsrsuo)
urre roldal)p Jr{l o
:uorlf,unJ
ro eJnlJnJls rraql
roJ
perueu
Jre sruJe rofetu
JnoJ aI{J
'}"'
j
:ilt+r,jj
j
ul
IIelJp
aJoul uI
palEJl
-snllr
sr
Jealre^olJ
Jql
Jo
uopJnJlsuoJ aqJ
'urpr{f,
apltoapnu.{lod Jql
Jo
srs
-aqluu(s
Jaue sJseq
prepuels
JnoJ Jql
Jo
uorle)
-lJlporu
,{q
paleraua8
are
leqt
saspq
,,lensnun,,
apnl)ur sJ)uJnbJs rlJql
'VNUI
Jo
sruJe
eqt
pJIIeJ
ere srlntJnJts
dool-ruets aq1
'sdoo1
papuerls-a18urs
JoJ sruals
suroJ Surrrpd aseq
[reluarualdruoJ
q)rqm
ur
'r'r.
]$i;].i::
ur
pJlerl
-snllr
?palreaolJ
e
Jo
uJoJ eql ur uJllrJAr Jq ueJ
eJnl)nrls
Lrepuoras
VNUI
aqJ
'sarnlrnrls
Lrerl
-Jal
pue
fuepuoras uoruuroJ
a^pq syNUl
IIv
'Suured
aseq Lreluaualdurol
erl uopoJ Jql
JZIU
-3orar
or
VNU1
aql salqeue uopo)rlup
aql'pna lutwo stl 6u4uasatdat
ulpu aLfi
o1 fualuau,taplun sr
qJrrlrM'uopoJllue
aqt'aruanbas eprloelJnulJt
p
surpluof,
1I
o
'pa4u!t
,filua1anot
sr
1r
qJrqM
ol
'prJp
oururp
a13urs e sluasardar
lI
.
:sarlradord
IprJnJ)
oltl se{
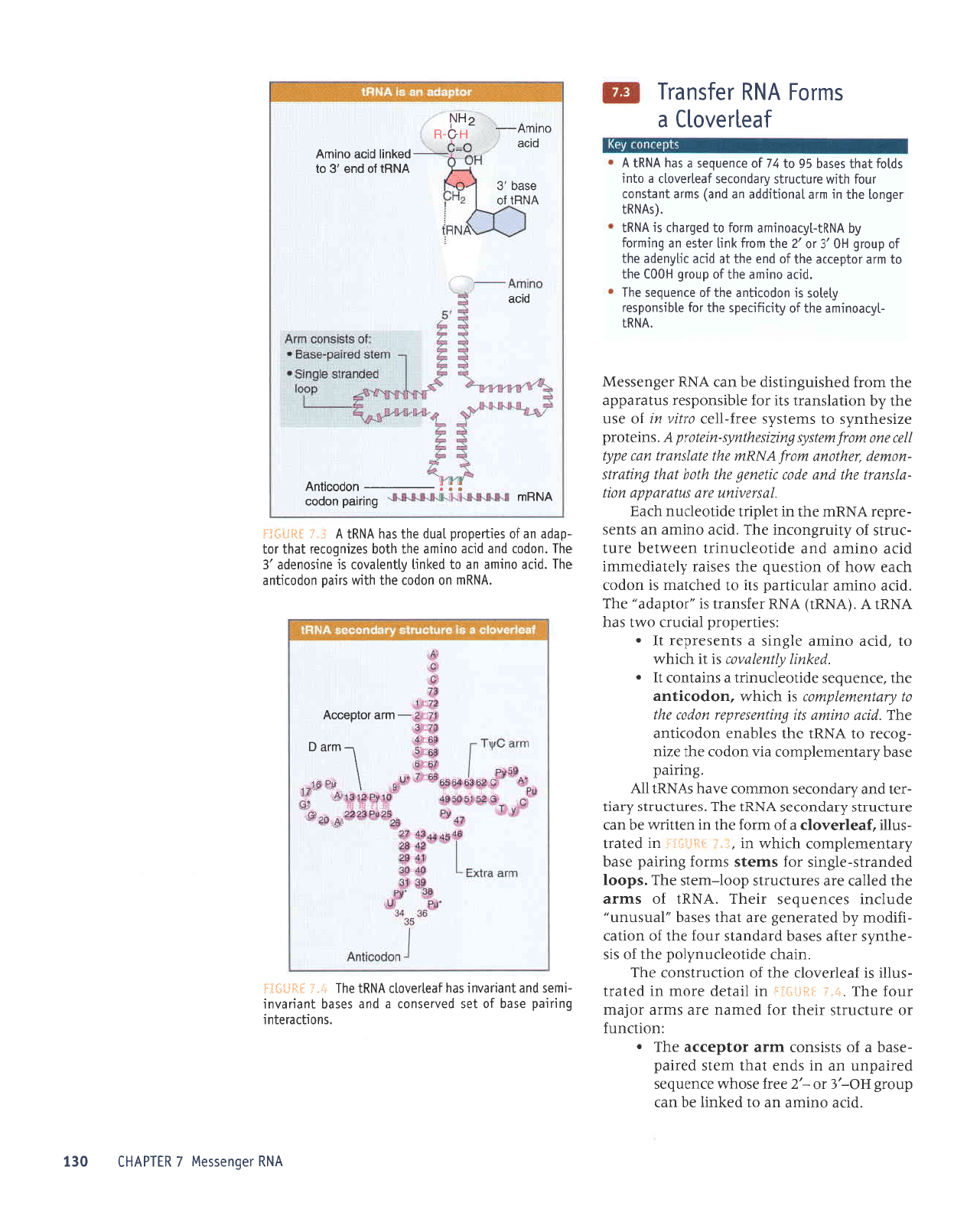
VNUI Y'(VNUI) VNU
raJsuerl
sl
,,roldepe.,
aqJ
'prJp
ounup relnllred
slr 01
pJqJleru
sr uopoJ
q)pa
,lroq
;o
uorlsanb Jqt
sJsrer
,{.lalerparurur
pIJE
OUIIIIe
puP
JprlOJIJnUrJl
UeeMleq aJnl
-JnJls
Jo
,{.lrnr8uorur
JqI
'prJp
ourrue ue sluJs
-ardar
VNUIU
aql ur
taldr:l
eprloJllnu
qJeA
'lastautun
ata sruatadda uoq
-olsuoli
at4t
pua
apot c4aua6 aLU Llllq
p41
6ut1a4s
-ulu.tap
'Ja4puo
ruo{
y1tryw
a4l apFuDJt uat ad[l
iln
aul u,tott utalsts
Cut2tsa4luts-utalotd
y'suratotd
azrsaqlur(s
o1 srualsz(s aJJJ-llJJ
1Jit^ ut
Jo
esn
Jq1 ^q uorlplsueJl
slr JoJ elqrsuodsar
snle.redde
aql ruorJ
paqsrn8unsrp
aq
up)
ygg
ra8uassary
.VNUl
lfreouLue
aq1
1o
rtlLrrlLrads
aq1 rol alqrsuodser
r\1e1os
sr uopolque
aq1
1o
eruenbes aqg
'poP
ourule
aq1
1o
dnotE
H00l
aql
o1 ure roldarre
aql
Jo
pua
aql
1e
pLre
rqr\uape aq1
;o
dnor6
H0,€.lo,Z
aql
ulotJ
Iurl
jalsa
ue 6uruuol
r\q
y1g11r{reourue
ulo} o1
pabreqr
sl
VNUI
.(SVNUl
ra6uol aq1 ur ulp
lpuorlrppe
ue
pue)
suue
]uplsuol
rno1
qllm
arnllntls A.repuores jpapa^oll
e olur
sploj
lPr.]l
sasPq
96
ol
,L
lo
aluanDas e
spq
vNUl v
JPaua^oll
P
'su0tllelelut
6uuLed aspq
Jo
las
pelrasuol
p
pup
sospq
lupuenut
-tulos
puP
lueue^ut
seq
Jee|e^olr
VNUI
aq]
1':i
-tjii*i:i
'VNUru
uo uopol aql
qlrm
srred uopoltlue
aqj
'pop
ouuilp ue ol
palurl Allualenor
sr aursouapp,€
aql
'uopol
pup prre
oururp aql
qloq
saztuboral
lpql
rol
-depe
ue
1o
sarladord
lpnp
aql spq
VNUI V
i'd
S*ll*:j
VNUur
Ouuted uoPoc
3!3-uopocrluv
VNU} 10
oseq
,e
p!ce
oull..llv-
vNUt lo
pua
/e
ol
palurl prce
oururv
HC_U
-l
6HN
p!ce
gdeS..
-,
;5
fi8
{il*'
uire
o
ure roldeccy
t
srurol
vNU
leJsuerl
