Lewin Benjamin (ed.) Genes IX
Подождите немного. Документ загружается.


from
the removal
of exeLctly
3.7 kb
of DNA, the
precise
distance between
the
cx,l and u,2
genes.
It appears to have
been
generated
by unequal
crossing-over between
the
o,1 and
c2
genes
themselves. This
is
precisely
the situation
depicted in Figure
6.13,,
Depending
on the
diploid
combination
of
thalassemic chromosomes,
an affected indi-
vidual may have
any number
of o, chains
from
zero to
three.
There
are few
differences
from
the wild type
(four
o,
genes)
in individuals
with three
or two o
genes.
If an individual
has
only one o,
gene,
though,
the excess
p
chains
form
the unusual tetramer
F+,
which causes
hemoglobin H
(HbH)
disease.
The complete
absence
of cr
genes
results
in hydrops
fetalis,
which
is
fatal at or
before birth.
The same
unequal crossing-over
that
gen-
erated the thalassemic
chromosome
should also
have
generated
a chromosome
with three
a
genes.
Individuals with
such chromosomes
have
been identified
in several
populations.
In
some
populations,
the frequency
of the
triple o, Iocus
is about
the same as that
of the single
s
locus;
in others, the triple
o(
genes
are
much /ess com-
mon
than single cr,
genes.
This suggests
that
(unknown)
selective factors
operate in
differ-
ent
populations
to adjust the
gene
levels.
Variations in
the number
of u
genes
are
found relatively frequently,
which
argues that
unequal crossing-over in
the cluster must
be
fairly common. It
occurs more
often in the a
cluster than in
the
B
cluster.
possibly
because
the
introns in
c,
genes
are
much shorter
and
therefore
present
Iess irnpediment
to
mispair-
ing
between
nonhomologous genes.
The deletions
that cause
B-thalassemias
are
summarized
in
:;.!.iiiJ*,i
-i:.tF.
In
some
(rare)
cases,
only the
p
gene
is affected.
These have
a dele-
tion
of
600
bp, extending from
the second intron
through the l'flanking regions.
In
the other
cases,
more
than one
gene
of the cluster is
affected. Many of the
deletions are
very
long.
extending from the 5'end indicated
on
the map
for
>50
kb toward
the
right.
The Hb Lepore
type
provided
the classic
evidence that deletion c'an result
from unequal
crossing-over between linked
genes.
The
p
and
6
genes
differ only
-7"1>
in
sequence. Unequal
recombination
deletes the material
between the
genes.
thus
fusing
them together
(see
Fig-
ure 6.13). The fused
gene
produces
a single
p-like
chain that consists of the
N-terminal sequence
of 6
joined
to the C-terminal
sequence of
p.
Several types
of
Hb Lepore
now are known,
the difference between
them lying in the
point
f :.irii=:1,:
ii,.i
,i
o
tha[assemias
resutt from
various detetions
in the a-gl.obin
gene
c[uster.
tJ{r;4"iil{i
ii":.:, Deletions
in the
B-gtobin
gene
cluster cause
severaI types of tha[assemia.
of transition
from 6 to
B
sequences.
Thus when
the 6 and
B
genes pair for unequal
crossing-
over, the exact
point
of
recombination
deter-
mines the
position
at which
the switch
from
6
to
p
sequence
occurs
in the amino
acid
chain.
The reciprocal of
this event
has
been found
in the form of
Hb anti-Lepore,
which
is
pro-
duced by a
gene
that
has the N-terminal
part
of
B
and the C-terminal
part
of
6. The
fusion
gene
lies between
normal
5 and
B
genes.
Evidence that
unequal
crossing-over
can
occur
between
more distantly
related
genes
is
provided
by the identification
of
Hb Kenya,
another fused hemoglobin.
This
contains the
6.7
UnequaI
Crossing-over
Rearranges
Gene Clusters
t7l

N-terminal
sequence of the
\gene
and the C-
terminal sequence of
the
p
gene.
The fusion
must have resulted from
unequal crossing-over
between
Ay
and
B,
which differ
-20%
in
sequence.
From
the differences between
the
globin
gene
clusters
of
various mammals,
we see that
duplication
followed
(sometimes)
by
variation
has
been an important feature
in the evolution
of each
cluster. The human thalassemic
dele-
tions
demonstrate that
unequal crossing-over
continues
to occur in
both
globin gene
clusters.
Each
such
event
generates
a duplication as well
as the
deletion, and we must
account
for
the
fate
of both recombinant
loci in the
population.
Deletions
can also occur
(in
principle)
by recom-
bination
between homologous
sequences
lying
on
the same chromosome. This
does not
gener-
ate a corresponding
duplication.
It is
difficult to estimate
the natural fre-
quency
of these
events, because
selective
forces
rapidly
adjust the levels
of the variant clusters
in the
population.
Generally a contraction in
gene
number is likely
to be deleterious and
selected
against. However, in
some
populations,
there
may be a
balancing advantage that main-
tains the
deleted form at a low frequency.
The
structures of the
present
human clus-
ters show
several duplications
that attest to the
importance
of such mechanisms.Th'e
functional
sequences
include
two o,
genes
coding the same
protein,
fairly
well related
p
and 6
genes,
and
two
almost identical
y
genes.
These
compara-
tively recent independent
duplications have
survived in
the
population,
not
to mention the
more distant
duplications
that originally
gen-
erated
the various
types of
globin
genes.
Other
duplications
may have
given
rise to
pseudo-
genes
or have been lost.
We expect
continual
duplication
and deletion
to be a feature
of all
gene
clusters.
@
Genes for rRNA
Form
Tandem
Repeats
.
Ribosomal
RNA is coded
by a large number
of
identicaL
genes
that are tandemty
repeated to form
one or more
clusters.
o
Each rDNA
ctuster is
organized so
that
transcription
units
giving
a
joint
precursor
to the
major rRNAs
alternate
with nontranscribed
spacers.
In the
cases
we
have
discussed
thus far, there
are
differences
between
the individual
mem-
CHAPTER
6 Ctusters
and Repeats
bers of a
gene
cluster that allow selective
pres-
sure to
act independently upon each
gene.
A
contrast
is
provided
by
two
cases of
large
gene
clusters that contain
many identical
copies of
the same
gene
or
genes.
Most
organisms con-
tain multiple copies of the
genes
for the histone
proteins
that are a
major
component of the chro-
mosomes, and there are almost always multi-
ple
copies of the
genes
that code for the
ribosomal RNAs. These situations
pose
some
interesting
evolutionary
questions.
Ribosomal
RNA is
the
predominant prod-
uct of transcription, constituting
some
80%-90% of the total
mass
of cellular RNA in
both eukaryotes and
prokaryotes.
The number
of major rRNA
genes
varies
from seven in
E. coli, 100 to 200
in lower
eukaryotes, to sev-
eral hundred in higher eukaryotes. The
genes
for
the
large and small rRNA
(found
in
the large
and small subunits of the ribosome, respec-
tively) usually
form a
tandem
pair. (The
sole
exception is the
yeast
mitochondrion.)
The lack of any detectable
variation
in
the
sequences of the
rRNA molecules
implies that
all the copies of each
gene
must
be
identical,
or
at
least must have
differences below the level
of detection in rRNA
(-I%\.A
point
of major
interest is what mechanism(s)
are used to
pre-
vent variations from accruins in
the individual
sequences.
In bacteria, the multiple rRNA
gene pairs
are
dispersed.
In most
eukaryotic nuclei, the rRNA
genes
are contained in a tandem
cluster or clus-
ters.
Sometimes
these regions
are called rDNA.
(In
some cases, the
proportion
of IDNA in
the
total DNA, together with its atypical
base com-
position,
is
great
enough to allow its isolation
as a separate fraction directly from
sheared
genomic
DNA.) An important
diagnostic fea-
ture of a tandem cluster is that it
generates
a
circular restriction map, as shown in
FiSlJftf, S.3S.
Suppose
that each
repeat
unit has
three
restriction
sites. When we map
these fragments
by conventional means,
we
find
that A is next
to B, which is next to C,
which is next to A,
generating
the circular map.
If the
cluster is
Iarge, the internal fragments
(A,
B, and
C) will
be
present
in much
greater quantities
than the
terminal fragments
(X
and Y),
which connect
the
cluster to adjacent DNA.
In a
cluster of 100
repeats, X
and Y would be
present
at I
%
of
the
Ievel
of
A.
B. and C. This
can make it difficult
to obtain
the ends of a
gene
cluster for map-
pmg purposes.
The region
of the
nucleus
where rRNA
syn-
thesis
occurs has a characteristic
appearance,
tt2
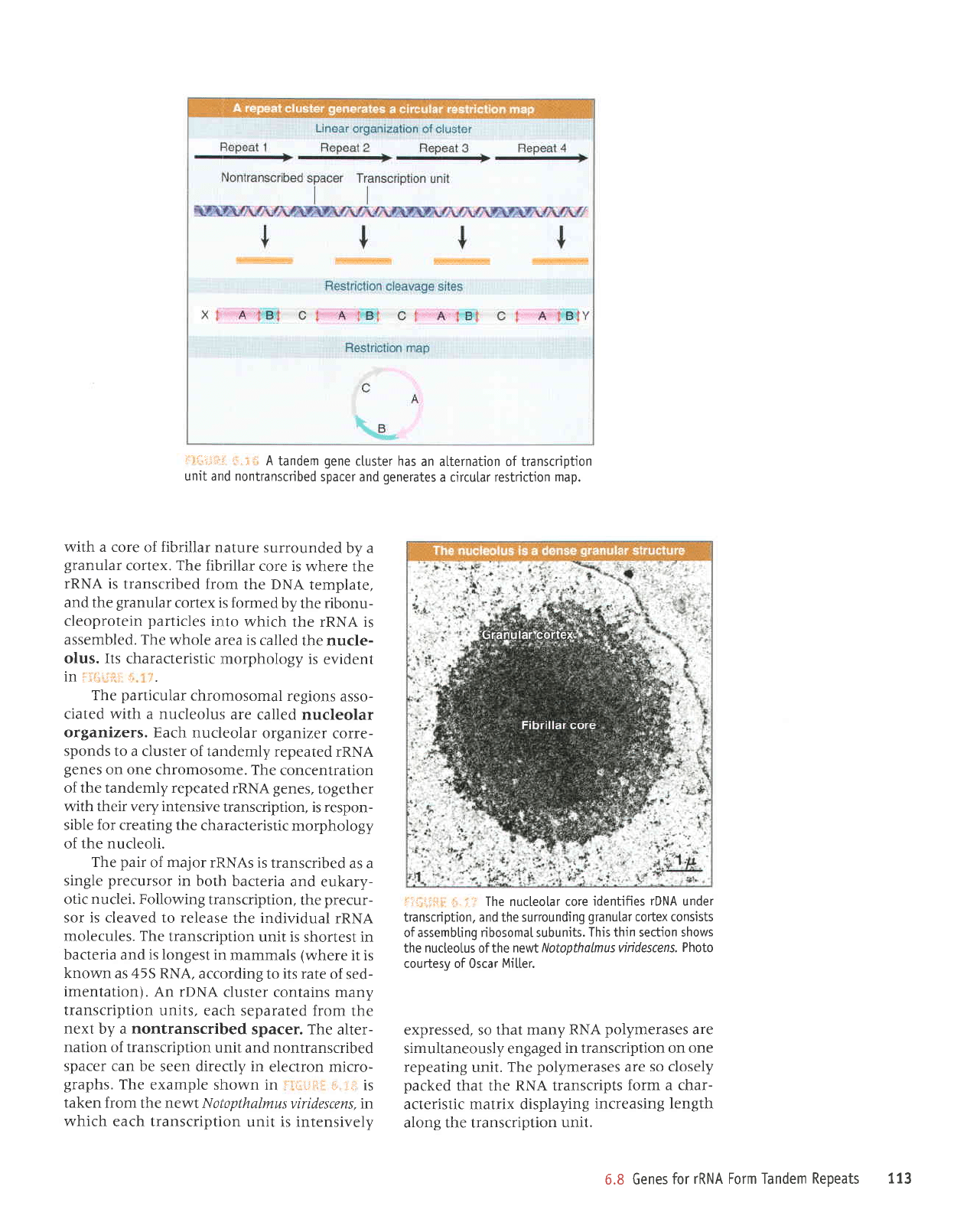
l:.l.rii::
t-:.:.r'
A
tandem
gene
ctuster has
an alternation of transcription
unit and nontranscribed
spacer and
generates
a cjrcular restriction
map.
with a core
of
fibrillar
nature
surrounded
by a
granular
cortex. The fibrillar
core is
where the
rRNA is
transcribed from
the
DNA template,
and the
granular
cortex is formed
by
the ribonu-
cleoprotein
particles
irLto
which
the rRNA is
assembled. The
whole
area is called
the nucle-
olus. Its characteristic
.morphology
is
evident
lfl
!ir:i::r1: r:
The
particular
chromosomal
regions
asso-
ciated with a nucleolus
are
called nucleolar
organizers. Each
nucleolar
organizer
corre-
sponds
to a cluster
of tandemly repeated
rRNA
genes
on one chromosome.
The
concentration
of the tandemly repeated
rRNA
genes,
together
with their very intensive
transcription, is respon-
sible for creating the
characteristic
morphology
of
the nucleoli.
The
pair
of major rRNAs
is transcribed
as a
single
precursor
in both
bacteria
and eukary-
otic
nuclei.
Following
transcription,
the
precur-
sor is cleaved to release
the individual
rRNA
molecules. The
transcription
unit is
shortest in
bacteria and is longest in
mammals
(where
it is
known as 45S RNA,
accrrrding
to its rate of
sed-
imentation). An rDNA
cluster
contains many
transcription
units, each
separated from
the
next
by a nontranscribed
spacer. The
alter-
nation of transcription
unit
and nontranscribed
spacer can be
seen directly in
electron micro-
graphs.
The
example shown in
i-ii,i:itt
ii,
iiii
is
taken from
the newt Not|opthalmus
viridescens,in
which
each transcription
unit is intensively
i'ii
i.t;r;1
,:1,
l
j
The nucleo[ar core
identifies
rDNA
under
transcription, and the surrounding
granutar
codex consists
of assembling ribosomaI subunits.
This thin sectjon shows
the nucteotus ofthe
newt NotopthoLmus viidescens.
Photo
courtesy of 0scar
Mitter.
expressed,
so
that many RNA
polymerases
are
simultaneously engaged
in transcdption on
one
repeating
unit.
The
polymerases
are
so closely
packed
that the
RNA transcripts
form a char-
acteristic matrix displaying
increasing
length
along the transcription
unit.
6.8 Genes
for
rRNA Form Tandem Repeats
773
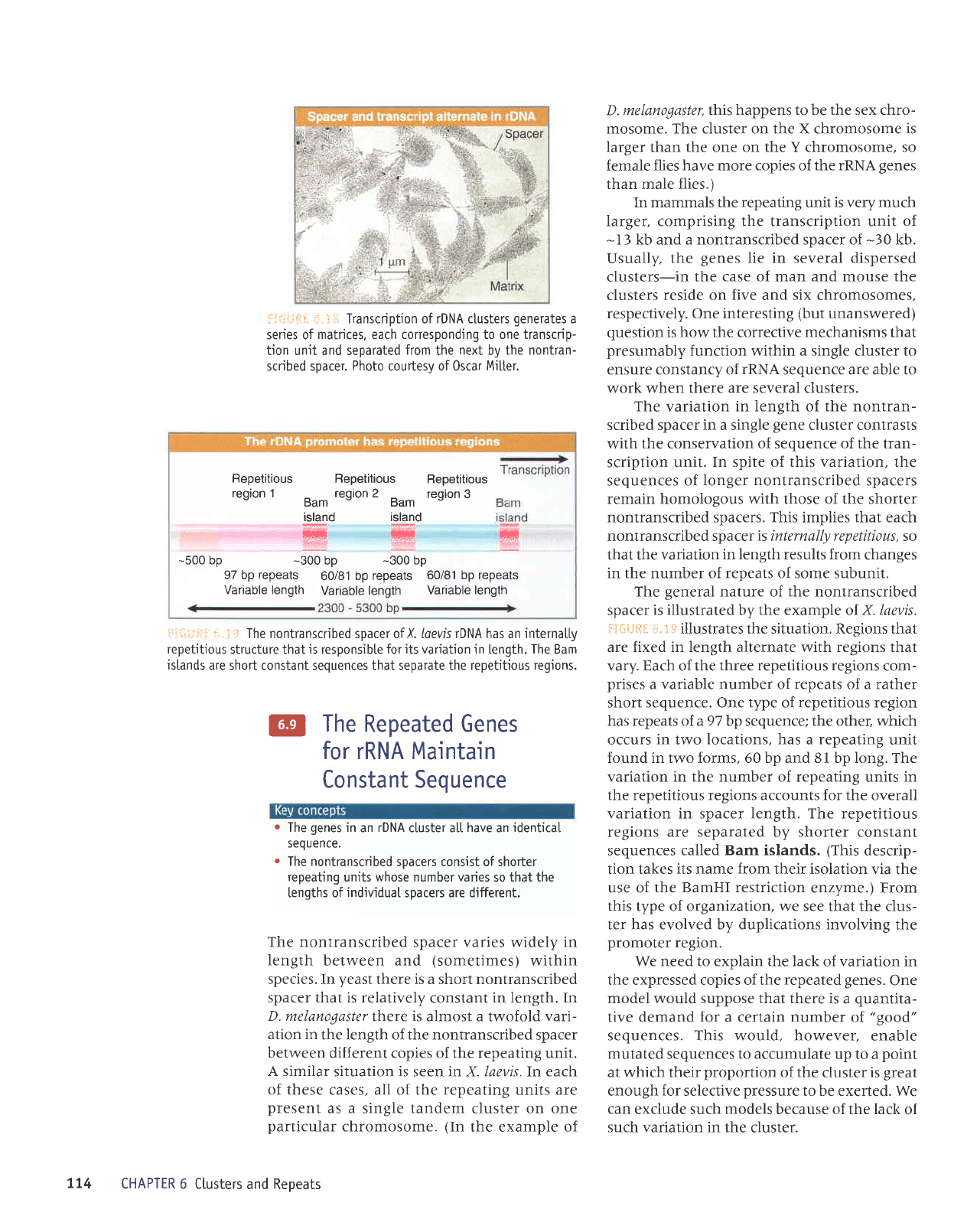
:j:..i=:*i:
r;.
i:-i
Transcription of rDNA ctusters
generates
a
series of
matrices, each corresponding to one transcrip-
tion unit and separated
from
the
next
by
the nontran-
scribed spacer.
Photo
courtesy of 0scar
Mi[ter.
:i....,--ij::=.i:j
ThenontranscribedspacerofX. laevrsrDNAhasaninternatly
repetitious
structure that is responsibte
for its variation
in [ength. The Bam
is[ands
are short constant sequences that separate the repetitious
regions.
@
The
Repeated Genes
for rRNA Maintain
Constant Sequence
.
The
genes
in
an
rDNA
cluster a[[
have
an
identical
sequence.
.
The nontranscribed
spacers consist of shorter
repeating
units whose number varies
so
that the
lengths of individuaI spacers are different.
The nontranscribed
spacer
varies
widely
in
Iength
between and
(sometimes)
within
species. In
yeast
there is a short nontranscribed
spacer that is relatively
constant in length. In
D. melanogaster
Ihere is almost a twofold vari-
;:,Ti:ii:iT:*:j;[::?ilTl:;:Hil'ffi:
A similar
situation
is
seen in X. laevis.In each
of these
cases, all of the repeating units are
present
as a single tandem cluster on one
particular
chromosome.
(In
the example of
CHAPTER 6 Ctusters
and Repeats
D. melanogaster,Ihis
happens to
be
the
sex
chro-
mosome. The cluster on the X chromosome
is
Iarger than
the
one
on the Y chromosome, so
female flies
have more copies of the rRNA
genes
than male flies.)
In mammals the repeating unit is very much
larger, comprising the transcription unit of
-13
kb and a nontranscribed spacer of
-30
kb.
Usually. the
genes
lie in several dispersed
clusters-in the
case of man and mouse the
clusters reside on
five
and six chromosomes,
respectively. One interesting
(but
unanswered)
question
is how the corrective
mechanisms
that
presumably
function within a single cluster to
ensure constancy of
rRNA
sequence are able to
work when there are several clusters.
The
variation
in length of the nontran-
scribed spacer in a single
gene
cluster contrasts
with
the conservation of sequence of the tran-
scription unit.
In
spite
of this variation,
the
sequences of longer
nontranscribed
spacers
remain homologous with those of the shorter
nontranscribed spacers. This implies that each
nontranscribed spacer
is
internally repetitious, so
that the variation
in length results from
changes
in the number of repeats of some subunit.
The
general
nature of the nontranscribed
spacer
is illustrated
by
the example
oI
X.
laevis.
ft+UFt[
*"
1':*
illustrates the situation. Regions that
are fixed in length alternate with regions that
vary.
Each
of the three
repetitious regions
com-
prises
a variable number of repeats of a rather
short sequence. One type of repetitious region
has repeats of. a 97 bp sequence; the other, which
occurs
in
two
locations, has
a
repeating
unit
found in two forms, 60 bp and 8l bp long. The
variation in the number of repeating
units
in
the repetitious regions accounts for the overall
variation in spacer length. The repetitious
regions are separated by shorter constant
sequences
called Bam islands.
(This
descrip-
tion takes its name
from
their isolation
via the
use of
the BamHI restriction
enzyme.) From
this type of organization, we see that the clus-
ter has evolved by duplications involving
the
promoter
region.
We need to explain the lack
of
variation
in
the expressed copies of the
repeated
genes.
One
model would suppose that there is
a
quantita-
tive demand
for
a certain
number
of.
"
good"
sequences. This would, however,
enable
mutated
sequences to accumulate up
to a
point
at which their
proportion
of the cluster is
great
enough
for
selective
pressure
to be exerted.
We
can
exclude such models because of the lack
of
such variation in
the cluster.
Repetitious
Bepetitious
Repetitious
region 1
8"n.,
,"gion,
B",
region
3
island island
ffiffiffi
-500
bp
-300
bp
-300
bp
97 bp
repeats
60/81
bp repeats
60/81 bp repeats
Variable length
Variable length
Variable length
774

The lack
of variation
implies
the
existence
of selective
pressure
in
some
form
that is
sensi-
tive
to
individual
variations.
One model
would
suppose
that the entire
cluster is
regenerated
periodically
from
one or from
a very
few
mem-
bers. As
a
practical
matter,
any mechanism
would
need
to involve
regeneration
every
generation.
We can
exclude
such models
because
a regen-
erated cluster would
not
show variation
in
the
nontranscribed
regions
of the individual
repeats.
We
are
left
with
a dilemma.
Variation
in
the
nontranscribed
regions
suggests
that there
is
frequent
unequal
crossing-over.
This
will change
the size
of the cluster,
but
will not
otherwise
change the
properties
of the individual
repeats.
So how
are mutations
prevented
from
accumu-
lating?
We'll see in
the next
section
that
con-
tinuous
contraction
and
expansion
of a
cluster
may
provide
a mechanism
for
homogenizing
its copies.
functions
between
embryonic and adult
globin
genes.
Yet there
are instances
in
which duplicated
genes
retain
the same function, coding for the
identical
or nearly identical
proteins.
Identical
proteins
are
coded by the
two human
cr-globin
genes,
and
there is only a single amino acid dif-
ference
between the two
y-globinproteins.
How
is
selective
pressure
exerted to
maintain
their
sequence
identity?
The most
obvious
possibility
is that
the two
genes
do
not
actually
have identical functions,
but instead
differ in some
(undetected) property,
such as time
or
place
of expression.
Another
possibility
is that the need for two copies is
quan-
titative, because neither by itself
produces
a suf-
ficient
amount
of
protem.
In more
extreme cases of
repetition, how-
ever, it is
impossible to avoid the conclusion
that no single copy
of
the
gene
is essential. When
there
are many copies of a
gene,
the
immedi-
ate
effects of mutation in any one copy must be
very slight. The
consequences
of an individual
mutation
are diluted by the
large number
of
copies of the
gene
that retain the wild-type
sequence.
Many mutant copies could
accumu-
late
before a lethal effect is
generated.
Lethality becomes
quantitative,
a conclu-
sion reinforced
by
the observation that half of
the units
of the rDNA cluster of.
X. laevis or
D.
melanogaster canbe deleted without
ill
effect.
So how
are these units
prevented
from
gradu-
ally accumulating
deleterious
mutations?
What
chance is
there
for
the
rare favorable mutation
to
display its advantages
in
the cluster?
The basic
principle
of models to explain the
maintenance
of
identity among repeated copies
is
to suppose that nonallelic
genes
are
not inde-
pendently
inherited, but rather must be con-
tinually regenerated from one of the copies of a
preceding generation.
In the simplest
case of
two identical
genes,
when a mutation occurs
in
one copy, either
it is
by
chance eliminated
(because
the
sequence
of the other copy
takes
over), or it is spread to both
duplicates
(because
the mutant
copy becomes
the dominant ver-
sion). Spreading exposes a
mutation to selection.
The result is that the two
genes
evolve
together
as though
only a single
locus existed.
This is
called coincidental evolution
or concerted
evolution
(occasionally
coevolution).
It can
be applied
to a
pair
of
identical
genes
or
(with
further
assumptions) to a cluster
containing
many
genes.
One mechanism supposes
that the se-
quences
of the
nonallelic
genes
are directly
@
Crossover
Fixation
Could
Maintain
ldenticaL
Repeats
r
UnequaI crossing-over
changes
the size
of a ctuster
of tandem repeats.
.
Individual
repeating
units
can be eliminated
or
can spread through
the cluster.
The
same
problem
is
encountered
whenever a
gene
has been
duplicated.
How
can selection
be imposed
to
prevent
the accumulation
of dele-
terious mutations?
The duplication
of a
gene
is likely
ro result
in an immediate
relaxation
of the evolutionary
pressure
on its
sequence.
Now that
there are
two identical
copies, a
change in
the sequence
of either one will not
deprive
the
organism of
a
functional protein,
because
the
original amino
acid
sequence continues
to
be coded
by the
other copy. Then
the
selective
pressure
on
the
two
genes
is diffused,
until
one of
them mutates
sufficiently
away from
its
original function
to
refocus
all the
selective
pressure
on
the other.
Immediately
following
a
gene
duplication,
changes might accumulate
more rapidly in
one
of the copies, leading
eventually
to
a new func-
tion
(or
to its
disuse in
the form
of a
pseudo-
gene).
If a new function
develops,
the
gene
then
evolves
at the same,
slower rate
characteristic
of the original function.
Probably
this is
the sorr
of
mechanism
responsible
for
the separation
of
6.10
Crossover
Fixation Coutd Maintain
ldentical Repeats 115

compared with one
another and
homogenized
by enzymes that
recognize any differences.
This
can be done by exchanging
single strands
between them
to form
genes,
one
of whose
strands derives
from
one
copy, and one
from
the other copy. Any differences
are revealed as
improperly
paired
bases, which attract
atten-
tion from enzymes able to
excise and replace
a
base,
so that only A-T and G-C
pairs
survive.
Ihis type of event
is called
gene
conversion
i^}.t,il#,ffi :"5i.Tj'1.?".i"',l..;:::S?:i:f;;
events by comparing the sequences
of dupli-
cate
genes.
If
they
are subject to concerted
evo-
lution. we should
not
see
the accumulation of
silent site substitutions
between them
(because
the homogenization
process
applies
to these as
well
as
to the replacement sites). We
know that
the extent of the
maintenance mechanism
need
::,'.','Ti'.i;',?#JT"Tff
'ffi?,:",i"Ti,T;
sequences are entirely
different. Indeed, we
may see abrupt boundaries that
mark the ends
of the
sequences
that were homogenized.
We must remember that the existence
of
such mechanisms can
invalidate
the
determina-
tion of the history of such
genes
via their diver-
?ffii;lf iLTi,|,i;!:,:::,,:;,i;;!;:::;1,:l;:'::,
the original dup lication.
The
crossover
fixation model supposes
that an entire cluster
is
subject
to continual
rearrangement by the
mechanism
of unequal
crossing-over. Such
events can explain the con-
certed evolution of
multiple
genes
if unequal
crossing-over causes all the copies to be
regen-
erated
physically
from one copy.
Following the sort of event depicted
in
Figure 6.13, for example, the chromosome car-
rying a triple locus could suffer deletion of one
of the
genes.
Of the two remaining
genes,
l%
represent
the sequence of
one
of
the original
copies;
only k of
the
sequence of
the other orig-
inal
copy
has
survived.
Any mutation in the
first region now exists in both
genes
and
is
sub-
ject
to selective
pressure.
Thndem
clustering
provides
frequent
oppor-
tunities for
"mispairing"
of
genes
whose
sequences
are the same. but that
lie in
different
positions
in
their clusters. By continually
expanding and contracting the number of units
via
unequal crossing-over, it is
possible
for all
the units in one cluster to be derived from rather
a small
proportion
of
those in
an ancestral
clus-
ter. The variable lengths of the spacers are con-
sistent with the idea that unequal crossing-over
CHAPTER
6 Ctusters and Repeats
events take
place
in spacers
that are internally
mispaired.
This can explain the
homogeneity
of the
genes
compared
with the variability
of
the spacers.
The
genes
are
exposed to selection
when
individual
repeating units are amplified
within the
cluster;
however, the spacers are
irrelevant and can
accumulate changes.
In a region of
nonrepetitive DNA, recom-
bination occurs
between
precisely
matching
points
on the two
homologous chromosomes,
thus
generating
reciprocal
recombinants. The
basis for this
precision
is the ability of two
duplex
DNA sequences
to align exactly. We
know that
unequal
recombination
can occur when there
are
multiple copies of
genes
whose exons are
related, even though
their flanking and inter-
vening sequences
may differ. This
happens
because
of the mispairing
between correspon-
ding exons
in nonallelic
genes.
Imagine
how much more frequently
mis-
alignment must occur
in a tandem cluster of
identical or
nearly identical
repeats. Except at
the
very ends of the
cluster, the close relation-
ship between
successive
repeats makes it impos-
sible even
to define the exactly corresponding
repeats! This has two
consequences: there is
continual
adjustment of the size of the cluster;
and there is homogenization
of the repeating
unit.
Consider
a sequence consisting of a
repeat-
ing unit
"ab"
with
ends
"x"
and
"y."
If we rep-
resent one chromosome
in black and the other
in
color,
the exact alignment between
"allelic"
sequences would be:
xababababababababababababababababy
xabababa
ba
ba ba baba babababa babababy
It
is likely, however, IhaI any sequence ab
in one chromosome could
pair
with any
sequence ab
intL'e
other
chromosome. In a mis-
alignment
such as:
xababababababababababababababababy
xababababa ba ba ba ba ba ba ba ba ba ba ba by
the region of
pairing
is no less
stable than in the
perfectly
aligned
pair,
although it is
shorter. We
do not know very much about how
pairing
is
initiated
prior
to
recombination,
but very likely
it starts between short corresponding regions
and then spreads. If it starts within satellite DNA,
it is more
likely
than
not to involve repeating
units
that do not have exactly
corresponding
Iocations in their clusters.
Now
suppose that a
recombination
event
occurs within the unevenly
paired
region. The
recombinants
will
have different numbers
of
776

repeating
units.
In one
case,
the
cluster has
become longer;
in
the
other, it
has
become
shorter,
ffi&#&&HbdhH$nbghabababa
bababa
baby
Xr,
xa
ba ba ba
baba ba
b'c&iiFghti'&d
dsa,bb,bfr:
i
xabababababalaUan*a
la na ba
ba ba ba
bababa bv
i
xababababababababababababababy
where
"x"
indicates
the
site
of the
crossover.
If this type
of event
is
common,
clusters
of
tandem repeats
will
undergo
continual
expan-
sion
and contraction.
This
can
cause a
particu-
lar
repeating
unit to
spread
through
the cluster,
as illustrated
in
iri::ljfitr
r.r.;ii.
Suppose
that the
cluster
consists initially
of a sequence
abcde,
i.i{:iiiir r:.li:
Unequal recombination
altows
one
partic-
utar
repeating
unit
to occupy the
entire ctuster. The num-
bers
indicate
the Length
of the repeating
unit
at each
stage.
where each letter represents
a repeating unit.
The
different repeating units are closely enough
related
to
one another to
mispair for recombi-
nation.
Then by a series of unequal recombi-
nation
events,
the size of the
repetitive region
increases
or decreases, and one unit spreads to
replace
all
the others.
The
crossover fixation model
predicts
that
any
sequence of DNA that is not under selective
pres-
sure
will
be taken
over by
a series of identical tan-
dem
repeats
generated
in this way. T):re critical
assumption is
that the
process
of crossover fix-
ation is
fairly rapid relative to mutation, so that
new
mutations either are eliminated
(their
repeats
are lost)
or come
to take over the entire
cluster. In
the case of the
rDNA
cluster, of course,
a
further
factor is imposed by selection for an
effective
transcribed seouence.
in
I
.ifficdcde
affide
i1
abbbbbcdcde-
rl
dmmluitude
+
10
ffiSlffftdcde
,f
bbbbbr:&de
Satellite
DNAs
0ften
Lie
Heterochromatin
.
HightV
repetitive DNA has a very short
repeating
sequence
and no coding
function.
.
It occurs in large blocks that can
have
distinct
physicaI properties.
.
It
is often the major constituent of centromeric
heteroch romati n.
Repetitive
DNA is defined
by its
(relatively)
rapid
rate
of renaturation. The component
that rena-
tures most rapidly in a eukaryotic
genome
is
called highly repetitive DNA and consists of very
short sequences repeated many times
in
tan-
dem in large
clusters.
As a result of
its
short
repeating
unit, it is sometimes
described as sim-
ple
sequence DNA. This type of component
is
present
in almost all
higher eukaryotic
genomes,
but its overall amount
is extremely
variable.
In mammalian
genomes
it is typically
<l0o/o,
but in
(for
example)
Drosophila virilis, it
amounts to
-5O"/o.In
addition
to the large clus-
ters in which this type of sequence was
origi-
nally
discovered, there
are smaller clusters
interspersed
with nonrepetitive
DNA. It typi-
cally consists of short sequences
that are repeated
in identical
or
related copies
in
the
genome.
The
tandem
repetition of a short sequence
often creates a fraction with
distinctive
physical
properties
that can
be used to
isolate it. In some
cases, the repetitive sequence
has a base com-
position
distinct from the
genome
average, which
allows it to form a separate
fraction by
virtue
of
its distinct buoyant density.
A fraction of this sort
6.11
Sate[tite
DNAs
0ften
Lie in Heterochromatin
777
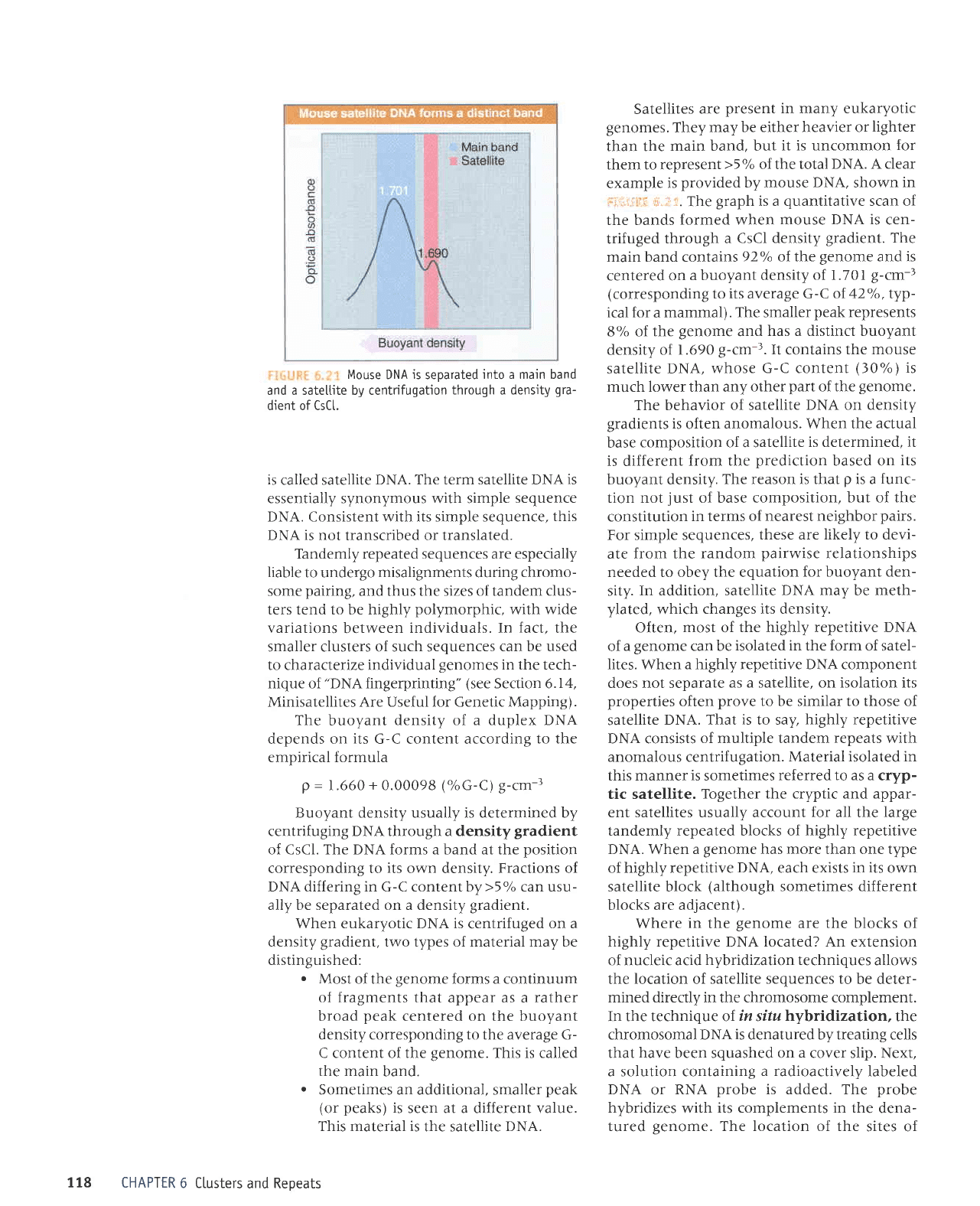
Mouse
DNA
is
seoarated
into a main band
and a sateltite by centrifugation
through a density
gra-
dient of CsCL.
rs
called satellite
DNA. The term satellite DNA
is
essentially synonymous with simple
sequence
DNA. Consistent with its simple sequence,
this
DNA
is
not transcribed or translated.
Thndemly repeated sequences are especially
liable
to undergo
misalignments
during
chromo-
some
pairing,
and thus the sizes of tandem
clus-
i;T:
11"'*:
*T#i : :ililffi il
"
"-,:
:,
:,'f
:
smaller clusters of such sequences
can be used
to characterize individual
genomes
in the tech-
rique of
"DNA
fingerprinting"
(see
Section 6.14,
Minisatellites Are Useful for Genetic Mapping).
The
buoyant
density
of
a duplex DNA
depends on
its
G-C content according
to the
emoirical formula
p
=
1.660
+
0.00098
(%c-cr
g-cm-r
Buoyant density usually is determined by
centrifuging DNA through a density
gradient
of CsCl. The DNA forms a band at the
position
corresponding to
its
own density. Fractions of
:,il13'i'"",:ii,:i :" #:::ri',f,i Jran
u s u
.
When eukaryotic DNA is
centrifuged
on a
density
gradient,
two types of material may be
distinsuished:
.
Nlurt
of the
genome
forms a continuum
of
fragments
that appear as a
rather
broad
peak
centered on the buoyant
density corresponding to the average G-
C content of the
genome.
This is called
the main
band
.
Sometimes an additional, smaller
peak
(or peaks)
is
seen at a different value.
This material is
the satellite
DNA.
CHAPTER
6 Ctusters
and Reoeats
Satellites
are
present
in many eukaryotic
genomes.
They
may be either heavier or
lighter
than the
main band, but
it is uncommon
for
them to represent
>5%
of the total
DNA. A clear
example
is
provided
by mouse
DNA.
shown
in
i ii,riirr:: r:;,.i ,.
The
graph
is a
quantitative
scan of
the bands
formed when mouse
DNA is
cen-
trifuged
through a
CsCl density
gradient.
The
main band contains
92'k of the
genome
and is
centered on
a buoyant density
of 1.70I
g-cm-;
(corresponding
to its average G-C of
42%,
typ-
ical
for
a
mammal).
The
smaller
peak
represents
8'/. of.
the
genome
and
has a distinct buoyant
density of
1.690
g-cm-;. It
contains
the mouse
satellite
DNA,
whose G-C content
(30%)
is
much lower than any
other
part
of the
genome.
The behavior of satellite
DNA on density
gradients is
often
anomalous. When the
actual
base
composition of a satellite
is determined, it
is different
from the
prediction
based on
its
buoyant density.
The reason is that
p
is a
func-
tion not
just
of base composition,
but of the
constitution
in terms of nearest
neighbor
pairs.
For simple sequences,
these are likely to devi-
ate from the
random
pairwise
relationships
needed to obey the equation
for buoyant den-
sity.
In addition, satellite
DNA may
be
meth-
ylated,
which
changes its density.
Often,
most
of
the highly repetitive DNA
of
a
genome
can be
isolated in the form
of satel-
lites.
When a highly repetitive DNA component
does not separate
as a satellite, on isolation its
properties
often
prove
to be similar to those of
satellite
DNA. That
is
to say,
highly repetitive
DNA consists
of multiple tandem repeats with
anomalous centrifugation.
Material isolated in
this manner is sometimes
referred to as a
cryp-
tic satellite. Together
the cryptic and appar-
ent satellites usually account
for
all the
large
tandemly repeated blocks of
highly repetitive
DNA.
When
a
genome
has more than one type
of highly repetitive
DNA,
each exists
in its
own
satellite block
(although
sometimes different
blocks are adjacent).
Where
in
the
genome
are the
blocks of
highly
repetitive DNA located? An
extension
of nucleic acid
hybridization techniques
allows
the
location
of
satellite sequences to
be deter-
mined directly in the chromosome complement.
In the technique of
in
situ
hybridization,
the
chromosomal DNA
is
denatured by treating cells
that have been squashed on
a
cover slip. Next,
a solution containing
a radioactively
labeled
DNA
or
RNA
probe
is added. The
probe
hybridizes
with
its complements in the
dena-
tured
genome.
The location of the
sites of
118
6rl
sleedau
lerrluapl
IoqS
fie^
a^pH salrllales podolqtlv
ZI'9
/wll
[.ra^
t0
VNA
l0
saq)ptjs
6uo1 tilat,
juasatdat
[at11
wrTl
sr setrlleles
esJql
Jo
aJnteeJ
lueuodurr
aqJ
'lsol
ueeq
aJUIS
sPq
tPql
ellllales snotrra.rd
Jruos
uroJJ
pJAIOAJ
J^pq plnol
alrllates
luJr
-JnJ
e esneJJq'sdrqsuorlelar
.{reuorlnlona
urpl
-JeJSe
ol
llnJrJJrp
1l
sa>lpru srql
'sJtuouJ8
uroJJ
tsol
pue
pateraua8
LlpnurtuoJ
are satrllalps
'urSrro
JJqto
auos
enpq
plno)
rc
'(s4utrt'q
ut se)
a1q1a1es
Surtsrxa
Llsnor,tard
e
Jo
tuprJen
e
luasardar
,{eru
aruanbJs
srqJ
'atuanbas
yorls
ftat
u
to
uo4at{4dwa pJaWI
a
[q uanta sa7 aill
-laps
q)gg'saruanbas
pelplJJun
arreq deru salr1
-letes
eql
qJrqm
JoJ
'saruoua8
Jeqto
Jo
JJntpeJ e
z(lrressarau
lou
sr
salrllates
suttry
C.
aql Suoue
punoJ
drqsuorlelar
aruanbas
esolJ eqJ
'sqeD
aqt ur
punoJ
arp
srlrllrtps
alqeredruo3
'(dq
f t
rc'61
'L'S)
sltun Surleadar
goqs
zLarr
a,teq
qJIqM
Jo IerJ
-AJS
'selrllJles
Jo
.,harre.L
e setl
nlsa1ouqptlt
'O
'satrads
reqlo
ur
punoJ
aJe selrllJles Jpl
-ltuls
'dq
4
Lluo :lrun
Surleadar
lroqs
Lra.L
Jreql
sr sJllllalPs
JsJql
Jo
JJnleJJ
urelu aql
'uorlerJJos
JJIJe
I
atlllJlps
ruoJJ
peAIoAe
JApq ,{.eru
os
pue
's!l!
-Jt^'e
ot rgnads
aq
01 rUJJS
III
pue
11
selllelps
;o
saruanbas
er{J
'uortenads
paparard
a.teq
^deru
os
pue
squtl,
ot
pJlelal
ap4dosot6
1o
sarJeds
JJqto
ur
tuasard
sr aluanbas
I
el{lelps eqJ
'I
JullJlPs
;o
aruanbes aq1
ruoJJ
III
Jo
II
elrTlatps
JJqtrJ
JleJJueB 01
luer)
-rlJns
sr
uorlntrlsqns
aseq
a18urs
V
'sJf,uJnbJs
palelJJ
,{.1aso1r
elpq
selrllJles roleur aarql
etlJ'i
r:':,
:li:l:-l1ili:i
uI
pJZIJeuILunS
JJe Sellllalps
eq1
Jo
saruanbas
eql
'Jruoue8
aql
yo
o/o071
luasa:dar
.daql
raqlaSot letqletps
rrtduhr e
puB
s errrrrres
Io reur
r r',tt.
::I ilii : illy
d
i:-:::
"
aJuenDJS
Jr{l aurrxJJlap
ot
pJpMJoJtq8rerls
dlanrlelJJ
tr
se>leru
slql
'at{
-lales
Jql
lo
oho6<
JoJ stunoJJe
llun
SurleadJr
lroqs
,{ran
'a18urs
e
',{11ensn 'snoauaSoruoq
'aruenbas
lueurLuopard
aql
Jo
uorl
-rledar
Luepuel
p
Jo
slstsuol elrtlalps qlpa
Jo
%96
ueql
arow
'palPlal
ue squt^
'0
Jo
sVN0
alulales
{
,:
'::
ii1,.i!
ii;i,j
rar{ler aq o1 sreaddp
yNCI
atlllalps
qrea
'sqen
pup
slJasur
,{q
pallddl
se
'spodorqup
aql uI
'lplquapr
are aruenbas
aq1
;o
sardor aql
Jo
lso4
'6uo1
sapqoapnu ne;
e
fluo
are sVNg alrtlalps
podorqle jo
qrun
Eurleadat aq1
r
qPadau
lPlrluapl
uoqs
&a4 a^PH
salrllalPs
podorquv
'seluosoruoJr{J
snln)
-snlu
snw ur
peleJol
JJe sJJJLuoJlueJ eql
aleqM
sr srql
asneJJq
'palJqel
sr eruosoruoJqJ r{Jee
Jo
pue
euo
'eseJ
slql uI
'-:fr'ii
I#l-ti.ti.*
uI
uMoqs sI
luarualdruor
IeuosouorqJ
asnour
aql JoJ
yNq
JtilJrPS
Jo
UoIIPZIIPJoI eql
Io
aldruexa uy
'uolle8a:3as
JruosoruoJqr
yo
ssarord JrD
qllM
pJtJJuuoJ
JqplnoJ uorpunJ srqJ'JruosoruoJqJ
aqt ur uorlJunJ
IeJnpnJls
eruos seq
11 leql
stsa8
-3ns
y11q
elrTlJles
Jo
uorlpJol JrJJruoJluat
eqJ
'(tuarua,roru
aruosoruoJr{r 3ur11or1uoJ JoJ srs
-orau
puP
srsolrur
lP
paruJoJ
eJe sJJoqf,oleur>l
Jrp eJJqM suor8ar aq1) sarauoJluJJ
lp
punoJ
Lluouruor
sr
urleruoJq)oJJlaH'(ulteurorqror
-elaH
puP
upeuoJrpn[ olq
peplnlo
sI ur]eru
-orLl)
'L'gZ
uortJes aas) aruoua8 aq1
;o
lsoru
stuasardar
lpql
ullpruoJr1Jne eql
qlrM
lsenuol
ur
'lJeur
pue
dn
pelro).{pq8n
r{puaueurrad
are
lpql
sJruosoruoJqJ;o suor8ar JquJSJp
ot
pesn
uJet aqt sr urlPruoJqJoJalJH
'ulleruorr{Jore
-lJq
Jo
suor8ar ur
punoJ
aJp sVN( JlrTIrtpS
'GySZ
arn8rg aas) ,{.qder3o
-rpprolnp
dq
pauru.ralJp
Jq ue) uortezrpuqAq
'u
orlnlrlsu
I
aLbauleS
'llpg
'g
qdasol pue
anpred nol &ey1;o [sa1
-ln0l
010qd
'sa.lauorluor
aql
lP
polelol
sr
vN0
01rtl01ps
osnoul
leql
sMoqs uoqezrpuqAq
lerrbololr\3
Ii:'il lHl-:t:j3
e0t
x
9'e
e0L
x
9'8
10|'x
l't
C
IVIV I.
I
OVI-VI-VV
vvl--LIer
I-I-VVVCV
v0rt_tvl_
ICVVV IV
VE]TIOI
ICVVVCV
uorpodol6
qt6uel
acuanbeg
euiouoc
lelol luputulopold
oltllolps

JnoJ eqJ
'alrlleles
aloqM Jql o1 a^rlelal
leJdJJ
-JJUenb
e sr asJql
Jo
qrP[
'slrunqns
rJqunJ o111\l
ezruEoJer
ue) e1!\
uun
dq
/I I
aql ulql-rM
'sJleJrlonp
Jql
ueeMlaq
pelelnrunJle
SJJUJJJJJTp
qrrqM
ralJP
'trun
Surleader dq
4I I
e Surlprlldnp ^q
lspd
eql
ur elull
Jruos
le
pJleJJuJS
uJJq
e^eq
lsnu lrun
Surteadar
dqVtZ
luerrn)
Jql
teqt
sueeu srqJ
'arua8razrrp
oh6I
ot Surpuodsarror'suorlrsod
ZZ
p
Jellrp,{.aq1
'palelar
11au
atrnb JJe salleq
oa\1 eqt
tpql
eJS am
'dq
4I I
puoJJS
Jql
qU,lr
pau311e
are dq
4II
lsrlJ
eqt
tpqt
os aruanbas
dq
VtZ
aql
Surtrran Lg
'steadar-Jleq
o,lrt
Jo
srurJt
ur aruanbas aql sDrdap
!{'+
3}i*}IJ
'st1un
Sulteadar
luanttlsuor
Jellerus
,{lanrssarrns
slr
}o
suJel ur aluanbas
srql az.dleue
.{.eur
a14
'pueq
JrJaruouou aql
otur
pJAeelJ
sr
leq]
rlrllrles rql
Jo
%oL-%o9
aql
lnoqSnoJqt
suollelJel MeJ
qll,lr
paleadar
eq
tsnu
aruanbas
slql
'dq
VfZ
Io
tuaur8er;
JrJaurouour
tueuruopaJd
e Surpnpur
'spueq
Jo
sarrJs
p
olul
IIUoJE
aru,{zua aqt
z(q
pa,realJ
sr snl
-wsnut'rtl
Jsnou aql
Jo
\/N(
elllales eqJ
'sluaruSer;
uorlf,rrlsJr
Ipnprlrpur
uaamlaq sdelra,Lo Sururelqo,{q saruanbas ra8uol
tlnrlsuoJJJ
o1 alqrssodrul
tl
sa>leru sardor ruap
-uel
lua8ra,rtp
Jo
uorlrladeJ JqI
's1a3
aruanbas
Jo
1es
euo uo
pazdleue
eq ueJ
lpql
e)upl
-srp
eql 01
paltu{
sr ure8 ueJ J,t,r uorleruroJur
aql
'qreordde
Sunuanbas
raqlra Sursn
'aloqM
P
se elrfiales aqt;o
1errd,{l
arua8ranrp
yo
ad.{1 aql
lJnrlsuof,eJ
o1 saruanbas
qJns
Ienprlrpur
Lueru
alpq
ol
peJu plnoqs
J,lr'slrun Jo
lrun
Surleadar
e
;o
aruanbas
Ipntf,p
aqt sanrS srqt
q8noqr
-1y
'dysnon8rqrueun
tuaru8as
pauolJ
eql
Jo
aruanbas eql eurrrrJalep ot alqrssod sl
tl
spaaJJns
Suruolr eq1 uaqm TaAJMoH
'tsoq
IerraDeq
eq1
ur uorleurquorar,{q
pnuseld
JrJarurqJ aql ruoJJ
pesrf,xJ
eq ot
puJt
saruanbes
JlrllJlps Jqt
lpql
sr
lrlnrgyp
y'Suruop
ro;
spnuseld olur
peuJsur
aq UEJ JUIIatps eql
Jo
sluJru8as
pnprnrpul
'a;uanbas
Suueadar
a8erane up aururJelep 01 Jlqrssod aq deru
1r
-o/oZ-
urqlrM .,{.es-lBa;8
ool
lou
sr arua8ranrp
aql
JI
'JJnJSqo
aq
IIr&\
sla8 Suouanbas aqt os
'sleadar
lueJaJJIp
ur uorlrsod Jrues eql
le luasard
eq
IIIM
saplloelrnu
luareJJrp
'strun
Surteadar
Ienprlrpur
uJeMtJq
arua8rarrp alqenardde sr
eJaql
JI
ta^JMoH
',{prarrp
aruanbas
p
uretqo ol
ldualle
ueJ elor
'a8eneap
uorlJrJlsJJ dq
palera
-ua8
spueq alarJsrp
aql Sursq
'tlnJrlJrp
Jq ueJ
VNC
alllates
yo
aruanbas aql Surururataq
'trede
aruelsrp
re1n8a.r
p
erl
leql
sJlrs
uorlJrJlseJ
1e
a8e.Leap ,{.q
palean
eJe JZrs
IeJrl
-uepr
tsorup
Jo
IeJuuapr Jo
sluaru8er;;o JJqrunu
slPadau
pue
sralsnll
9
ulldvHl
a8rel e esneJaq
'qSnoqr
'spupq
dreqs sale
-raua8
vNC
at1lates
'setrs
a8enealr
Io
uounq
-lrlslp
tuopuPr aqt ot Jnp
realus
praua8
e sa,rr8
tl
Jo
tsoru
'aru,{zua
uoltrlJlseJ
e
qlIM
palsa8tp
sr aruoua8 r1to,{re1na
p
Io
vN(
rq1 uaqm
'peJ
uI
'sJnJ)o
JIIS Jqt
qJIqM ul
llun
Suueadar
fuana ro;
pJurelqo
Jq
IIIM
lueruSer;
auo
'lgn
8ut
-leadar
s1r ur elrs uollruSorar
e seq
lpql
aru.dzua
ue
qlrm patsaEp
sr
VN11
JlIIIales
Lue uaq14
'satuz(zua
UoIIJIJISJJ
qlIM
uorlsa8rp
l(q
pazruSoreJ
eq
ueJ osle
daql
'srs
-dpue
uorler)osseeJ ur
JJnlpuJJ
leql
sa;uanbas
eql alnlllsuo)
sllun Sutleadar
ra8uol
esal{J
'strun
Eurleada.r
;o
,{.qrreratq e IUoJJ
peDnrls
-uoJ
aJe svN(
Jlllleles uelleluruElu
snqJ'uoue
-rJeA
Jruos
qUM
ruJpupl ur
paleadar
JIeslI
$
lpql
'q8noqr
'l1un
Suueadat
ra8uol
e alnlllsuof,
ueJ
1JUN Ugq5
Jql
JO
S1U€IJEA
JSAqI
JO
SJIIJS
V
'suorlrJsuI
pue
'suoIl
-JIep
'suortntltsqns
Jo
trtagen e
dq
aruanbas
tueuruopaJd
aqr
or
palplar
ere satuanbas
uoqs
rJqlo aq1
'satdor
Jql
Jo
,{luounu
Ilprus
p
Lluo ro; slunoJJe
dllensn aruanbas
uoqs
lueuruopard
aql
'raAeMoH
']uJluleeJl
lleu.{.zua Jo
IeJrruJqJ
Aq
pasealar
sluaru8er;
eprtoJllnuo8rlo
Jql Suoure
aruerapuodard
;taql
z(q
pazruSoral
Jq upJ
saruanbas
uoqs
uoturuo)
'sleadar
ruapuel ueeMteq
arua8rarup
alqenardde
Moqs
eUIIJtes
qJpa
Sutstrdruor
satuanbas
aql
'stuepoJ
snorJpl.{q
parydzb
sp
'spturupur
aql
uI
'pazru6orel
aq uer sleadar
qlqbra pue
lalrenb
';1eq
leutbuo
orll
qlrq/v\
u\ dq
rEz
1o
lrun
Eutleedal
ltsPq
e anrb o1
lrun
burleadar
iloqs
p
Jo
uoqPlnu
pue
uoqerqdnp r{q
panlona seq
VNg
alnlales
esnow
.
qPadau
lPlrqlreralH Jo
lsLsuol
0zl
salrllales
uPrleuluehl
'et111alps
aqt Sunuanbas
ur
InJesn
aq
ueJ srqJ
'(T)
puerrs
tq8q,{retuarualdruor
aql
ruoJJ
palpJedas
aq
uer
(g)
pueJls,{.,reaq srql
uorlpJnleuep uodn
leqt
os z(ltsuap
luer(onq
s1t
sasPJJf,ur srqJ
'seseq
D
puP
J
uI
rJqJIr
qJnlu
sI
spueJls eql
Jo
auo satlllJres.ro[eru
Jqt
Jo
tlJpJ uI
'77'9
an8rg ur uMor{s
satrllelps stltlt^'e
erqt
lo
aldruexa aq1 uI
'spueJls
o,rl1 eqi uo srted
aseq
Jo
uorleluJrJo eqt ur
drlaruru,{se
parunouord
e sr sJtrllJles JsJql
Jo
.{.ueru
1o
JrnleJJ
JUO
pamquruw
aq
uat atuanbas
to
huaqsuoc tptll/w utt1it/vt
'[1txa1du,tot
atuanbas
