Lewin Benjamin (ed.) Genes IX
Подождите немного. Документ загружается.


"?\r\ff\}\F\nl.}\t\if\?\?\}\
Silencing
of one copy takes
-4
million
years
Active Silent
Duplication occurs at
1%/gene/million
years
Divergence accumulates at 0 1"/o/million
years
II*i-3RE {i"4 After a
gene
has been dupticated, differences
may accumulate between the copies.
The
genes
may acquire
different
functions or one of the copies
may become
i nactive.
develop
between the duplicated
genes,
one
of
two types
of event is likely to occur:
.
Both of
the
genes
become necessary.
This can
happen
either because
the dif
-
ferences
between them
generate pro-
teins with different
functions,
or
because
they are expressed specifically
in differ-
ent times
or
places.
.
If this does
not Jrappen,
one of
the
genes
is likely to be eliminated because
it will
by chance
gain
a deleterious
mutation,
and
there will be no adverse selection to
eliminate this copy.
\pically
this takes
-
4 million
years.
In
such a situation,
it
is
purely
a matter of chance in terms of
which of the two copies becomes
inac-
tive.
(This
can contribute to incompat-
ibility between different
individuals, and
ultimately
to
speciation,
if different
copies become
inactive in different
populations.)
Analysis of the
human
genome
sequence
shows IhaI
-5"/o
comprises duplications of
iden-
tifiable
segments
ranging in length from l0 to
100
kb. These duplications
have arisen rela-
tively
recently, that is, there
has not
been
suf-
ficient time
for divergence between them
to
eliminate
their
relationship. They include a
pro-
portional
share
(-6%)
of the expressed exons,
which shows
that the duplications are
occur-
ring more or
less irrespective of
genetic
con-
tent.
The
genes in these duplications
may
be
especially
interesting
because
of
the
implica-
tion that they
have evolved
recently
and there-
fore could be
important
for recent
evolutionary
developments
(such
as the
separation
of
man
from monkev).
GLobin
Clusters
Are
Formed
by
Duplication
and
Divergence
.
Att
gtobin
genes
are
descended
by duptication
and
mutation
from an ancestral
gene
that
had three
exons.
r
The ancestral
gene
gave
rise
to myoglobin,
leghemogtobin,
and cr
and
B
gtobins.
o
The
q,-
and
p-gtobin
genes
separated
in the
period
of earty
vertebrate
evotution,
after
which
dupl.ications
generated
the
individual
clusters
of
separate o-
and
B-[ike
genes.
.
0nce
a
gene
has
been
inactivated
by
mutation,
it
may
accumutate
further
mutations
and become
a
pseudogene,
which'is
homotogous
to the
active
gene(s)
but
has
no functional
rote.
The most common
tlpe
of
duplication
generates
a second
copy of
the
gene
close
to the
first
copy'
In some cases,
the
copies
remain
associated
and
further duplication
may
generate a cluster
of
related
genes.
The
best
characterized
example
of a
gene
cluster
is
presented by
the
globin
genes,
which
constitute
an
ancient
gene
family
con-
cerned with
a
function
that
is central
to the
ani-
mal kingdom:
the
transport
of
oxygen
through
the bloodstream.
RoshanKetab
O2L-66950639
The major
constituent
of the
red
blood
cell
is
the
globin
tetramer,
which
is associated
with
its heme
(iron-binding)
group in the
form of
hemoglobin.
Functional
globin
genes in all
species
have the same
general structure:
they
are
divided
into three
exons,
as shown
previously
in Figure 3.7
.We conclude
that
all
globin
genes
are derived
from a
single
ancestral
gene, and
by tracing
the development
of
individual
globin
genes
within and
between
species
we
may learn
about the
mechanisms
involved
in the evolu-
tion of
gene families.
In adult
cells,
the
globin tetramer
consists
of
two identical
cr
chains
and
two
identical
p
chains.
Embryonic
blood
cells
contain
hemo-
globin
tetramers
that
are different
from
the adult
form.
Each tetramer
contains
two
identical
cr-
like chains
and
two
identical
p-like
chains,
each
of which
is related
to
the
adult
polypeptide and
is
later replaced
by
it.
This
is an example
of
6.3
Gtobin
Ctusters
Are
Formed
by
Duplication
and
Divergence
101
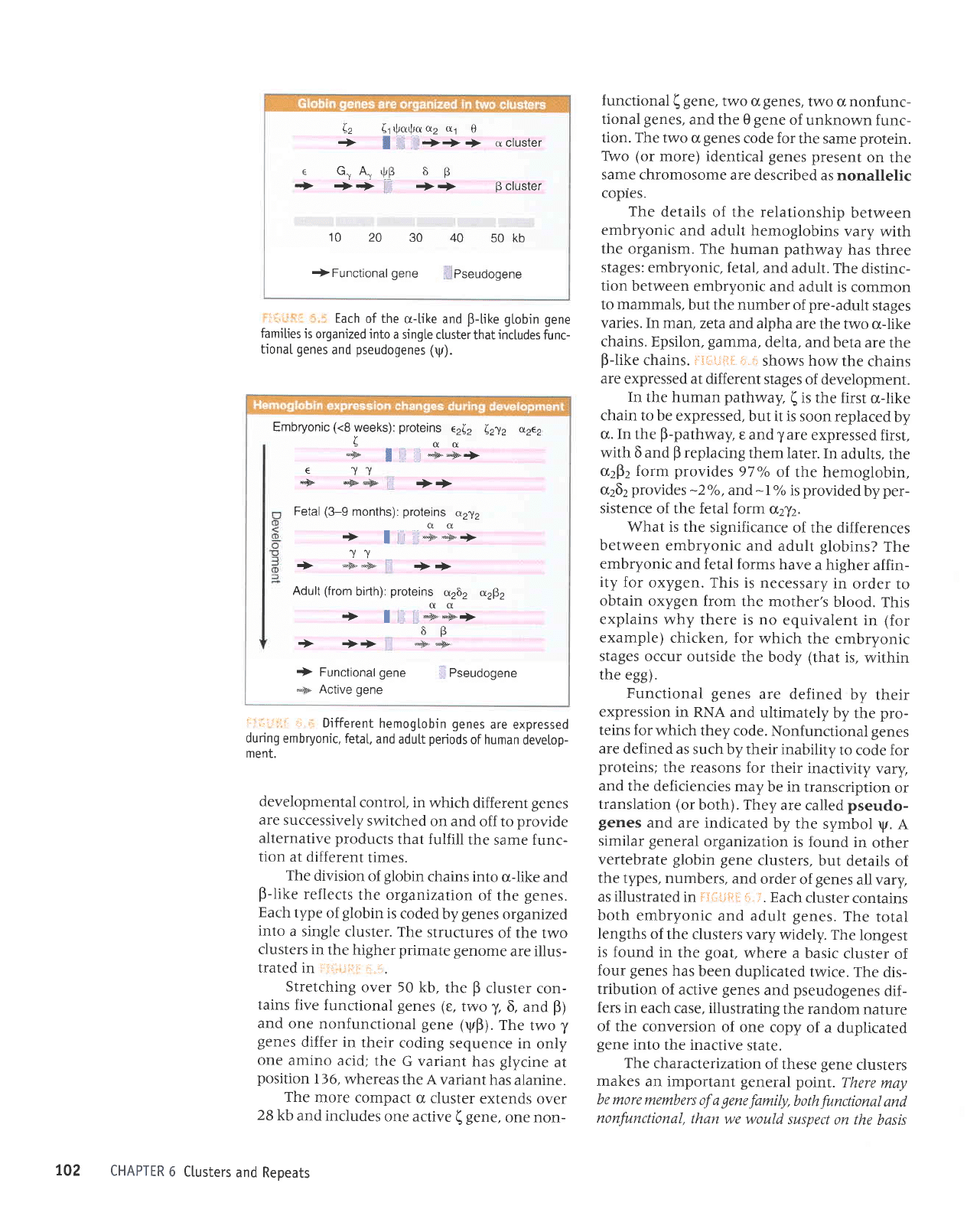
b
(1
rfct$tr ct2 o1
0
+
1i,,,.+++
trcluster
e GrA't_F
6
B
+ ++
I
++
Bcluster
10
20
30 40
50 kb
+Functional gene
;
Pseudogene
;::':.,:
,
,
Each
of the
s-tike and
B-tike
gtobin gene
families
is organized
into
a singte cluster
that
includes
func-
tionaI
genes
and
pseudogenes
(y).
irr.r:.:::
i:,;.
Different
hemoglobin genes
are expressed
during
embryonic, feta[,
and
adutt
periods
of human
devetop-
ment.
developmental
control, in
which
different
genes
are
successively
switched
on and
off to
provide
alternative products
that fulfill
the
same func-
tion
at different
times.
The
division
of
globin
chains
into
a-like and
p-like
reflects
the organization
of the
genes.
Each
type
of
globin
is
coded
by
genes
organized
into
a single
cluster.
The structures
of the
two
clusters
in the
higher
primate
genome
are illus-
trated
in
i:ii;:-:r'i.i
ii,:1.
Stretching
over 50
kb,
the
B
cluster con-
tains five
functional genes
(e,
two
y,
5,
and
p)
and
one nonfunctional
gene (yp).
The
two
y
3:::
i i :['#
J,^,";:
: i':,1
#;
:
ffi ,'l
J""
11
position
I16,
whereas
the
A variant
has
alanine.
The
more
compact
cr cluster
extends
over
28
kb
and includes
one active
(
gene,
one non-
CHAPTER
6 Ctusters
and Repeats
functional ( gene,
two d
genes,
two
cr, nonfunc-
tional
genes,
and the 0
gene
of
unknown func-
tion. The two
clt,
genes
code for
the same
protein.
TWo
(or
more) identical genes present
on
the
same chromosome
are described
as nonallelic
copres.
The details
of the relationship
between
embryonic
and adult
hemoglobins
vary
with
the organism.
The human
pathway
has
three
stages:
embryonic, fetal,
and adult.
The
distinc-
tion
between
embryonic and
adult is
common
to mammals,
but the number
of
pre-adult
stages
varies. In
man, zeta
and alpha are
the
two s-like
chains. Epsilon,
gamma,
delta, and
beta are
the
B-like
chains.
l'g{iLiFii:
il:.i,
shows how
the
chains
are
expressed at
different stages
of development.
In
the human
pathway, (
is the first
o-like
chain to be
expressed,
but it is soon
replaced
by
cr. In
the
B-pathway,
e and
y
are
expressed
first.
with 6 and
B
replacing
them
larer.
In adults,
the
cr2p2 form
provides
97'h
of the hemoglobin,
a262
provide
s
-2o/"
,
and
-1"/o
is
provided
by
per-
sistence
of the fetal lorm
a2y2.
What is the
significance
of the
differences
between
embryonic
and adult
globins?
The
embryonic
and fetal forms
have
a higher
affin-
ity
for oxygen.
This is
necessary
in
order to
obtain oxygen
from
the mother's
blood.
This
explains
why there is no
equivalent
in
(for
example)
chicken, for
which
the
embryonic
stages occur
outside the
body
(that
is,
within
the egg).
Functional
genes
are defined
by
their
expression
in RNA
and ultimately
by the
pro-
teins for
which they
code.
Nonfunctional genes
are
defined as
such by their inability
ro
code for
proteins;
the reasons
for their
inactivity
vary,
and
the deficiencies
may
be in transcription
or
translation (or
both). They
are
called
pseudo-
genes
and
are indicated
by the
symbol
y.
A
similar
general
organization
is found
in
other
vertebrate
globin
gene
clusters,
but
details
of
the
types, numbers,
and
order
of
genes
all vary,
as illustrated
in
il.{i,:*l{i
ii..t.
Each
cluster
contains
both
embryonic
and adult genes.
The
total
lengths
of
the clusters
vary widely.
The
longest
is found
in
the
goat,
where
a basic
cluster
of
four
genes
has
been
duplicated
twice.
The
dis-
tribution
of
active
genes
and
pseudogenes
dif-
fers
in
each case, illustrating
the random
nature
of the
conversion
of one
copy
of a duplicated
gene
into
the inactive
state.
The
characterization
of these
gene
clusters
makes
an important general
point.
There
may
be
more members
of
a
gene
family,
both
functional
and
nonfunctional,
thqn
we would
suspect
on the
basis
Embryonic
(<8
weeks):
proteins
€2{2
L2"12
o2e2
(o0
*&
|
.,
f'
-e*"-*,.>
€
^y^y
-4'
*18$
@.
++
Fetal (3-9
months):
proteins
o2"y2
ct0
+
|
r-*-
-**+
f1
+
*&"&"
.>
+
Adult
(from
birth):
proteins
a262 azgz
u0
.>
I
_+,*w+
6B
.>
->+
@.@
+
Functional gene
;
Pseudogene
*&.
Active gene
702

€0r
aluabra^r0
pue
uoqelrtdno Aq
paurol
alv slalsnll ulqolg
€'9
'euab
lPllsaluP
al6uts
e uolJ suorlPlnu
puP
'suorlrsodsuPll 'suoqeltldnp
Jo
seuos
p
r\q
panlona
a,req seuab urqo16
11y
i:-i'i;
-jtdllti.j
pJrJnJ)o JAeq 01
s;eadde
slqJ
'sluelJen
$
pue
n
eq1
01 asrr arrtS
ol
palerudnp sellt aue8 utqo13
IpJlse)ue
eq1
JJoJaq
uollnlo^e
Jo
auII
aql IUOJJ
paS.ranrp
aneq
tsnur
z(aql os
'uleqJ
utqop;o adrh
a18urs
e [1uo aneq
,,qsIJ
J^ltltuud,,
aLuo5
'JOlSaJUp
uourruoJ
rraq]
luasardJr
ol JrnDnJls
uoxJ-eeJql
aql alet
r(eru a,rzl
os
'saua8
urqo18 se
uolle4ue8ro
eures
eqt seq
aua8 utqolSoz(ru
aq1
'o8e
sreaz( uog1ru
008-
lua)sap
Jo
auq
utqo18
eqt
urorJ
pa8ra,rrp
qJIqM
'utqo18oz(ut
uplleru
-ureru
Jo
ureqJ
a18urs
aql
;o
atuanbas
aqt
,{q
papraord sI IrrJoJ
uJepou
ur aua8
utqop e aJeJl
ueJ
JM
l€ql
>lJeq
lsaqunJ
aqJ
'urJoJ
IPrlseJue
aq1
tuasardar
deru
'saua8
utqo16
aq1 ot
patelJr
sr
qJrqu
's1ue1d;o
aua8 urqoforuaq8al
aql
'E'*
SH**iS
uI
parnlrrd
sr
tuJ)sep
LreuorlnlonJ
Jqt
Jo
,lrall
lua
-sard
rng
'aua8
urqo13
IEJlsaJue
a13uts
e ruor;
sJalsnlf,
aua8
urqop
luasard
Jo
uollnlolJ eql
eJpJt
01 alqe
Jq
plnoqs
aaa
'sanads
;o
;(latrea
e ur sauaE
urqo18
Jo
uoltezlue8ro
aql uror4
'lJol
lenpt,l.lpul
se
punol
are
saua8
lsoru
luoururoJ
tou
JJe ad.{1
sq1
;o
sJJlsnp
JJAaMoH
'saua3
pnplllpu aql SutraptsuoJ
uele IUoJJ
Jo
surJloJd
urqo18-$
umou>l
aq1
Sulztu4nJJs
IrroJJ
lsnf
padxa
plnom
e,t,r ueql
ralear8
qJnlu
sI sIqJ
'sleluurplu
luaJaJJrp
ul
q>l
0zI
ol
0z
Jo
aSuer
e
sarrnbar
surqop
a4t-$
aql
ro1 Supor
leq1
eas
rM
'uoulunJ
Jelnrrued
e roJ apoJ
01
pepJau
sl
VNq
qJnru ,ltoq
yo
uotlsanb
aql ot
pre8ar
qll14
'(slunoue
^^ol ur
ro z(1;arrq
Lpo
passardxa
Ilqeurnsard
pue)
suralord
umoul-ruoJJ
luJreJJIp
lnq
-ot
petplar
aq
deur zlaqt
ro
'saprrdadu(1od
1er
-lluapr
roJ apoJ
reql
salerlldnp
luasardar
,{eru
saua8
leuorlf,unJ
pJtxJ
aq1
'stsr{1aua
utalotdto
'seuab
tnol
eAPq
qlPa
)ltqr
puP
ltqqeX
'sauaEopnesd
orvrl
pue
'seua6
llnpp
oMl'aueb
rtuor\rqure
elpl auo'saua6
rtuofuqure
fllea oMl apnllut
saue6
asnou
ua^os'selelqoila^
ut
punoj
ate sauaEopnesd
pue
seuab
uLqo16-$;o
slalsnll
l'S 3HE1'S€.i
urqol6or.ueqbel
(urqo16o,{ru)
urqol6
;erlsecuy
(qs;peq
p
rtardurel)
eueb
urqoq6 e16u;g
(qs;1
'sue;q;qdue)
seueb
d
,lc
polur-.|
/.::t
* ffifr
acueOrentp
X
uouectldn6
UD
f
*.fi t *{
/
/
saue6
1o
uotletedeg
,
*:
'ffi
ilD
/
: atd
t
I
zn
,
In
slelsnlciouolsueoxf
*
,#,*
s,,#,1
t {t* * #f
zd
rd
(sprlq
B
sleurureu)
esrelsnlc
alej€oos
00r
06
08
no
09
0v
08
0z
0t-
ij;r
i, i:
uslcrt]c
td
nd
d
d
:souoo
llnpv
;
I
i-l
i'-:
i'j
ilqqeg
ddA/")
L'r
:seueb cruofuqu3
f
j
i-r
lj lj
i-l
[1]''i
asno61
ur,Ed
r"'jd
eqh zqh tqd
oqdA
l: I L-:
r-
j
' ,
:-l
i.l
i-l
ij
i-ileoe
e€l
drir
,nr
n, od
dhnr,
iltr
cd
dA
ilt
t,

'luaruJf,eloal
prJe
ouIIuP
eql
Jo
llnsJJ
aql
uo spuedJp
(snoa8etuBnpe
ro
'leJlneu 'snouelalap)
uortplnru
Jqt
Jo
paJJa
aqJ
'papoJ
sr
leqt
prJe
oururp aql
sJallp uorlplnur
e
'satrs
luaurareldar
lV
.
:salrs
luaus
pue
salls
luaurareldar
Iertualod
olur
uor8ar Eurpor
e
;o
:tuanb:s
JpuoJlJnu
Jql
Jpr^rp l,eru
a14
'3urueaur
eqt uo
IJJJJJ
ou seq
uJ{o eseq
pJlql
aql
qJrqM
ur
'uopoJ
Jseq-JaJql
p
ur
prJp
ourrue
qJee
Jo
uoltpluJsaJdar
aqt sr JJUJJJJJTp
srql
Jo
eJJnos
aqJ'satuanbas poe
Jrepnu
Surpuods
-eJJoJ
aql
uaaMleq
arua8raarp
Jql urorJ
lue
-JJJJIp
aq ueJ suralord
ueaMlaq
arua8ra,rrp
aql
'tueJalJrp
Jrp sprJe
ourrue
Jql
q)rqm
tp
suortrsod
;o
luarrad
aql
'aruatralrp
JrJql se
passardxa
sr suralord
o,lrl uJJMtJq
eJuJJaJJrp
eqJ
'paDalas
lou
JJoJJJeql pup
snorrelJlJp
sr a8ueqt
,{ue
lsouqp lpql
sJtpJrpu
srql'saoads
o1 saoads
ruor;
a8ueqr
ou
Jo alil{
Suruoqs
'pJAJJSuoJ
dtq8tq
are suralord
Jrnos
'paalqJalw
0j
pasvil
sJo$nuo
Jn41 ua%/w
awq
aql iluts
rilJtql
ueeMlJq palelnu
-nJJe
eneq
teqt
sJJuJJJJllp
aqt
eJS JM
,sartads
o,tr1 ur
suralord
SurpuodsarroJ
Jql
Suuedruor
u(g
'uorlnlorra
ro;
lood
luapuadapur
ue sJtnlrls
-uo)
,lrou
sJrJJds
8ur11nsar
aq]
Jo
qJea
,sanads
/l1rJU
OAII
Olur
saleredas
sartads
e
uaqM
'pateldsrp
Suraq;o
ssarord
aqt ul
sl
tl
asnpJeq
luarsueJl
Jq
DeJ
ur ,{eru
auo ro
,(a8e1
-ue^pe
s^rDales.,{.ue
seq
auou JSneJeq)
alqels
aq
deru,{.aqr
luasard
aJe
sluerrel
a1dr11nru
uaq6
'pa^r^Jns
e^pq
leq]
sluerJeA
aqt ,i1uo
Jas JAl
'sanads
e;o
lood
aua8
aql azrurtnJJs
a,t,r uaqal
teqt
Jaqrueuag
'1ood
Surpaarq
a18urs
aql urqlrm
uollpxrJ Jo
uorteururllJ
^q
pJmolloJ
,uorlnt
-llsqns
Ieuorlelntu
z(q
sarrlo,La
ulJlord
e
,sanads
p
ulqllM
'a8ueqr
ol
pre8ar
qtlm
IllllqlxalJ
sll
uo
l,red
ur
tseal
lp
Surpuadap
,{lqeruns
-ard
'uratord
qJpe
Jo
JllsrJJpeJeqf,
p
sr Jlelnur
-ntre
sa8ueqJ
Ieuorlelnru
qJrqM
le
JlpJ eqJ
'uollexlJ
puE
lJrJp
rrropupr
Jo
llnseJ
Jql
se anr)Jp
01
'JJoJareqt
'alqe
pue
uralord
Jql
Jo
uorlJunJ
Jqt uo
pa;;a
due
tnoqtlm
'sr
leqt
,IpJlnau
ale
aruanbas
prJe
ourrup
ue ur
sa8ueqf,
Ipuorlplnu
;o
uogrodord
rer{zvr
sr enssr
snorluJluoJ
V
'uorlelndod
aqt u
pax11[
aruoJJq
a^pq
ol
prps
sr
1r
'aua8
aqr
Jo
uorsJel
snornard
aq1
sareldar
luerJen
,rt.au
e
ueq6
.atuanbas
raru
-JoJ
eql
Suneldar
.{1en1ua.rra
'uorlelndod
aql
q8norqr
peards
o1 d1a>1q
sl
lr
'srnJJo
euo
Jrer
p
uaqM
'snoa8eluenpe
JJe
suorlelnru
arad
'
uorlJalJS
lernteu,{.q
paleurulTa
Jq
IIIM
pue
snorralelep
aJe atuanbas
prJe
ourrue
aqt
a8ueqr
tpql
suorlplnur
lsow
slPadau
pue
slelsnll
'z(puanbar;
aloru
qJnru
JnJJo
suorlplnrrr JJaqM
'slodsloq
Jre slql o1 suorldarxa
aq;,
'aruoua8
aql
Jo
suor8ar
1e
ur
,{lrlqeqord
lenba
ssal Jo
aroru
qlr^aa
dlqeqord
'aJupqJ
,{q rnrto
suorlalep
pue
suorlJasu
ileus
pue
suorlplnlu
lurod
'au4l
raao [1mo1s
alelnrunJJe
lpql
suorlelntu
ileurs
,{q
rnrro saruanbas
uralord ur
sa8ueqt
1soy41
'paleredes
saue6
rraql aluts
aurl aq1 o1
leuoqrodord
s1 seruanbas urqo16;o rred fue
uaamlaq
arua6ranrp
oql
lpql
os
'aletedas
saua6 rage
peads
uane ssal lo
alout e
1p
alqnunllp suo!]eln[rrl
r
)aJJrp
spop ourue 6urpuodsa.lrol
aql
qlrqm
lp
suorlrsod
1o
luauad
aq1
fiq
pernseaur
sr
suralord
oml uaamlaq aruaEranrp freuoqn'1ona
aq1
r
'salrs
luaureleldar 1e
ueql
ielseJ
xQI- salls
lualts lP
olelnulnllP
suoQPln[irl
r
'(aruanbes
uralord
aq1
lta#e
lou
saop uorlelnur
araqrvr)
salrs
lualrs
pue
(suonnlqsqns pop
oulue sasnel
uoqplnu
araqn)
salLs
luauareldat
1e
Aren
saoads
luara#rp
ur
saue6 snobolouoq;o
saruanbas aq1
r
Iloll
&euoqnlo^l
aql
roJ srseg
aql sI
9
UlldVHl 'Or
alua6le^ro
aluanbes
'uorlJas
3ur,uo11o; aql
ur saua8
Ienpr^pu
eql
yo
arua8raalp
aql
Jo
uorldrrlsap aql ruoJJ
Jes
II,aM
sp
'serult
tuaJJJ
aJoru ur
sJJlsnlJ
fl
pue
n
alpJpdas
eql ulqlrm
peJrn))o
aneq sa8ueq3
'oBB
sreaz(
uorllru
977-
f,lqeqord
parrntro
leql
luale
uE tolsaf,up
uoruruoJ rrJql
rrroJJ
pa8razrrp
sprq
pup
sleuureur
aql JroJJq
paleredas
z(11er
-rsIqd
uaJq
elpq
lsnru
saua8
fl
pue
n
aqt
snql
'sletutuptu
pue
sprrq
qloq
ut surqolS
$
pue
n
roJ srJlsnlJ
JlpJpdas
eJe JJaqJ
'uorlnlola
atpJq
-JUJA
dlrea;o
porrad
Jr{t
ur
palJnno
dlqeqord
sIqJ
'3IuI1
slql rJUe rauunJaJoJ
uer^e/uprlpru
-ureru
aql ur
uorlrsodsuerl
e uroJJ
pJllnsar
aleq
tsnru
saua3
urqo13-$
pup
-n
aql
Jo
uorlpJedas aqt
os
'o8e
srea,{
uorlgru
0S€-
aull uprle/uerleru
-Iuplu
Jqt
ruoJJ
paleredas
suerqrqdrue
aq1
'paterlldnp
splt,l JJtsnlJ
eJrtua
eql ra1e1
'sardol
pnprlrpur
aql urrmraq
arua8ranrp
Aq
pamollo;
?red
$-n
pe>lu{
e;o
uo4erqdnp,{q
pazrlona
alpq aJoJaJaql
lsnrrr
JelsnlJ
aq1
'sadfu
t1npp
pue
Ip^Jel
qloq
;o
'saua3
$
pue
n
qpq
suIeTuol
'q8noql
trtsnlr
qJeg
'sralsnp
urqo13 orvrt
spq
qJIqM
'su.ao1
snd
-ouay3ot1aq1
ur
saua8 urqo13
aql
Jo
atpts
aqt z(q
paluasardar
sr
uorlnlole
;o
a8els
uau_aqJ
'qsrJ
Auoq
eqr
Jo
uorlnlola
eqt Suunp
'o3e
srBa,{.
uomru
00E-
.
At
silent sites, mutation
only substitutes
one synonym
codon for
another, so
there
is no
change in
the
protein.
Usu-
ally
the
replacement
sites
account for
75'/. oIa coding sequence
and the silent
sites
provide
257o.
In addition to the
cr:ding sequence,
a
gene
contains nontranslated regions.
Here again.
mutations
are
potentially
neutral,
apart from
their effects on either secondary
structure
or
(usually
rather
short) regulatory
signals.
Although silent mutations
are neutral with
regard to the
protein,
they could
affect
gene
expression via the sequence
change in RNA.
For example, a change in
secondary structure
might influence transcription,
processing,
or
translation.
Another
possibility
is that a change
in
synonym codons calls for a different
IRNA
to respond,
influencing
the
efficiency of
translation.
The mutations in
replacement
sites should
correspond with the
amino acid divergence
(determined
by the
percent
of
changes
in
the
protein
sequence). A nucleic
acid divergence of
O.45oh at replacement
sites corresponds to an
amino acid divergence of I
%
(assuming
that
the average
number
oli
replacement
sites
per
codon is 2.25). Actually, the measured
diver-
gence
underestimates the differences that have
occurred
during evolution,
because of the occur-
rence
of
multiple
events at one codon. Usually
a correction is made for: this.
To
take the example of the human
p-
and
6-globin chains, there are l0
differences
in
146
residues, a divergence of 6.9oh. The DNA
sequence
has 3l changes in44l residues.
How-
ever, these
changes
are distributed very differ-
ently in the
replacement
and silent sites. There
are
I I changes in the 330 replacement
sites,
but 20 changes
in
only I I
I
silent sites. This
gives
(corrected)
rates of divergence
of 3.7'/.
in
the
replacement sites and )2o/o in the
silent sites.
almost an order of
magnitude in
difference.
The striking differe:nce in the divergence of
replacement and silent sites demonstrates the
existence of much
greater
constraints on
nucleotide
positions
that influence
protein
con-
stitution
relative to
those
that
do
not.
So
prob-
ably very
few
of the amino acid changes are
neutral.
Suppose we take the
rate
of mutation at
silent
sites to indicate the underlying rate of
mutational
fixation
(this
assumes that there
is
no selection
at all at the silent sites). Then over
the
period
since the
p
and 6
genes
diverged,
there
should have been chanses at 32o/o of the
330 replacement sites,
for a total
of 105.
All but
l l
of
them have been eliminated,
which
means
IhaI"
-90o/o
of the
mutations
did not survive.
The
divergence
between
any
pair
of
globin
sequences
is
(more
or
less)
proportional to the
time since they separated.
This
provides
an
evo-
lutionary clock that
measures
the accumula-
tion of
mutations at an
apparently
even rate
during the evolution
of a
given
protein.
The rate
of
divergence
can be
measured
as
the
percent
difference
per
million
years,
or as
its reciprocal, the unit
evolutionary
period
(UEP),
the time
in millions
of
years
that it takes
for lo/o
divergence
to
develop.
Once the
clock
has
been established
by
pairwise comparisons
between species
(remembering the
practical
dif-
ficulties in
establishing
the actual
time of spe-
ciation), it can be
applied
to related
geneswithin
a species. From their
divergence,
we
can calcu-
late how much time
has
passed
since
the dupli-
cation that
generated them.
By comparing
the sequences
of
homolo-
gous genes
in different species,
the
rate of
diver-
gence
at both
replacement
and silent
sites can
be determined,
as
plotted in
i'riir,iiiir
i:"1.
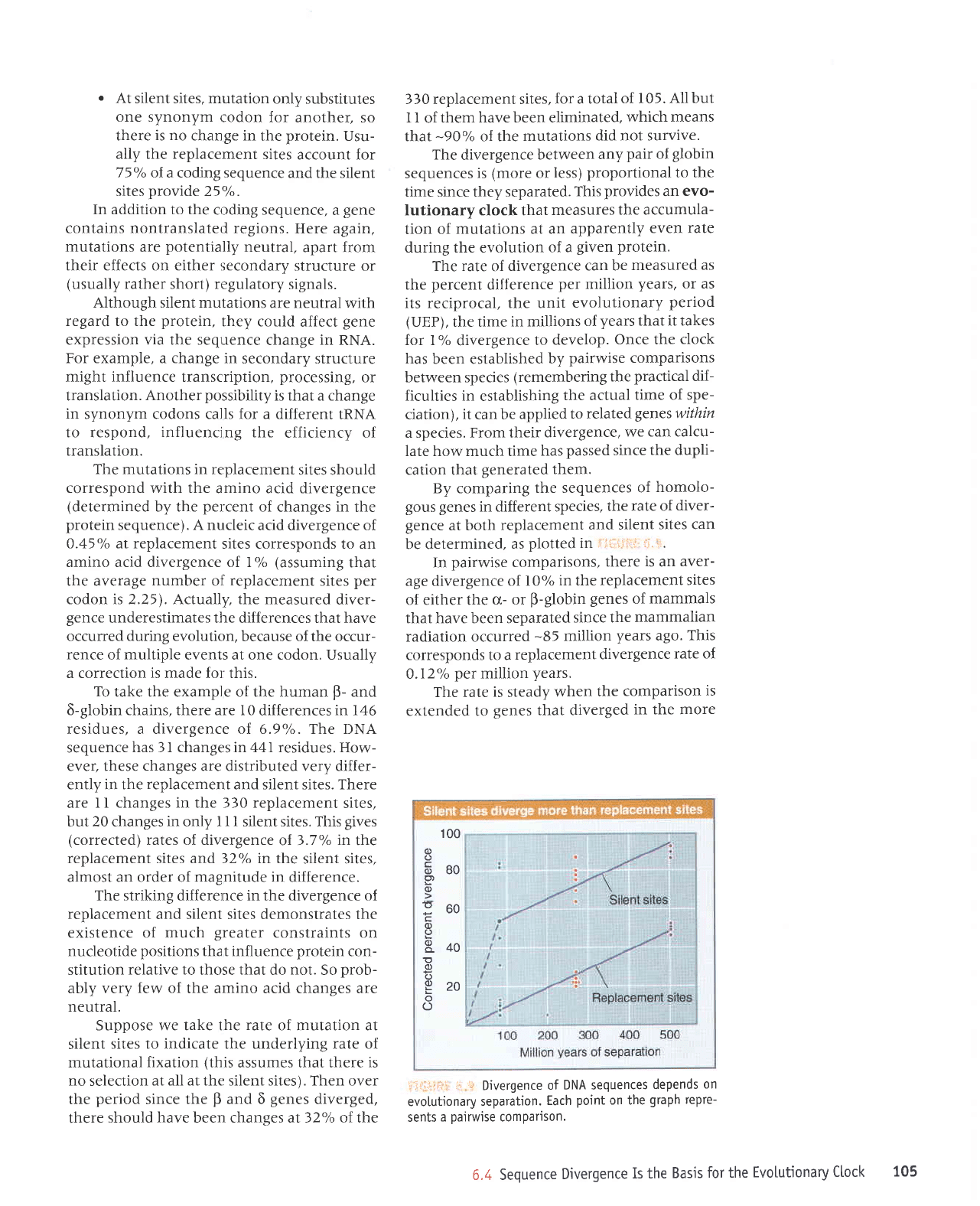
In
pairwise
comparisons,
there is an
aver-
age divergence of
I 0
%
in the replacement
sites
of either the
a- or
B-globin
genes
of
mammals
that have been separated
since the
mammalian
radiation occurred
-85
million
years
ago.
This
corresponds to a
replacement
divergence
rate of
0.12%
per
million
years.
The rate
is
steady
when
the
comparison
is
extended to
genes that diverged
in the more
i;lif;Lri'r:
l- r; Divergence
of
DNA sequences
depends
on
evolutionary
separation.
Each
point
on the
graph
repre-
sents
a
pairwise
comparison.
100
6)
6ao
o
:60
o
o
440
c)
o
920
o
6.4 Sequence
Divergence
Is the
Basis
for the
Evolutionary
Ctock
105
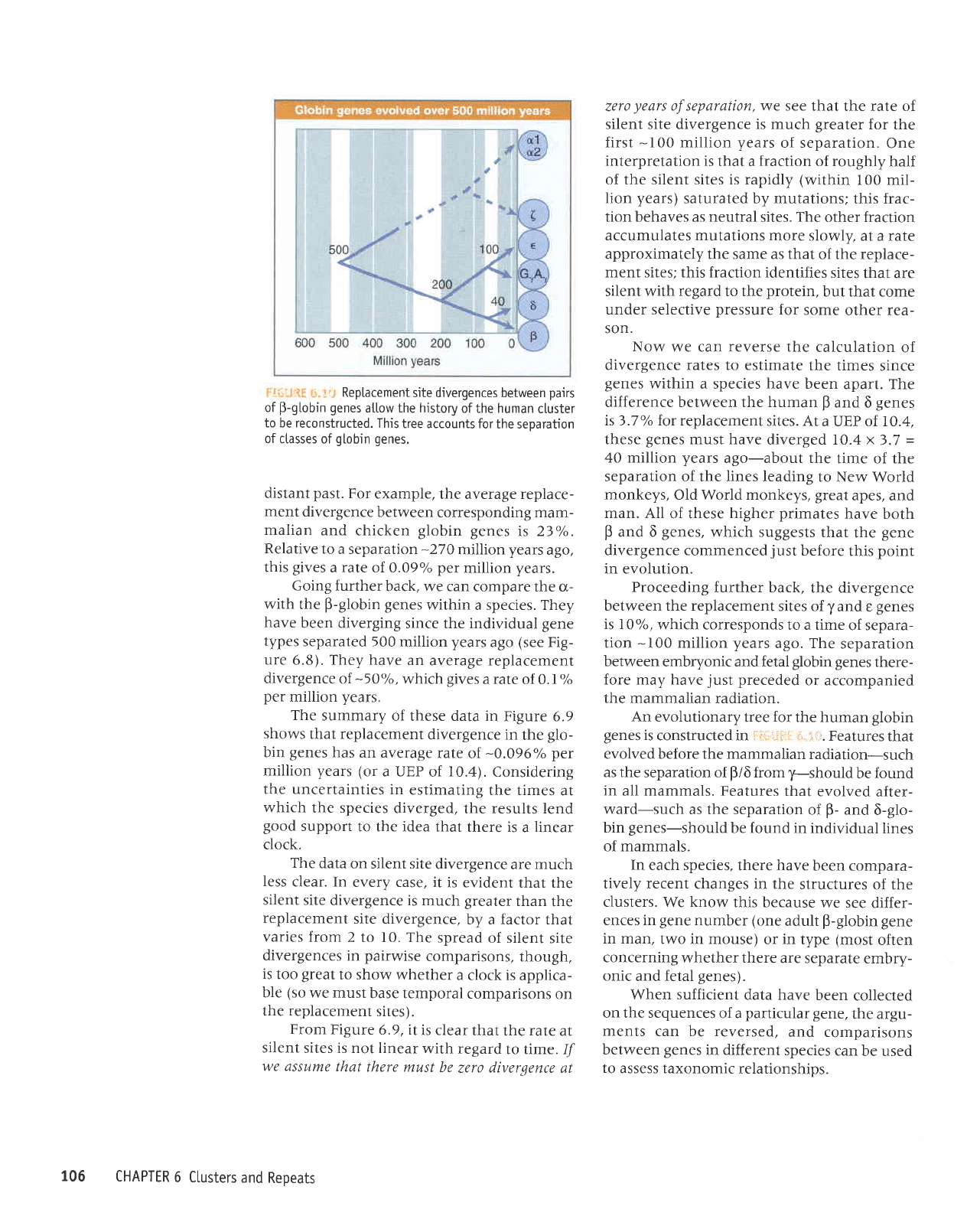
.
::
:
.,
Replacement
site divergences between
pairs
of
B-gtobin
genes
attow the history
of the human
cluster
to be reconstructed.
This
tree accounts for
the separation
of
ctasses of
globin genes.
distant
past.
For
example,
the average replace-
ment divergence
between
corresponding mam-
malian
and
chicken
globin genes
is 237o.
Relative
to
a separation-270
million
years
ago,
this
gives
a
rate
of 0.O9o/o
per
million
years.
Going
further
back, we can
compare the o,-
with
the
B-globin
genes
within
a species. They
have
been diverging
since the individual
gene
types
separated 500
million
years
ago
(see
Fig-
ure 6.8). They
have an
average replacement
divergence
of
-50"/o,which
gives
a rate
of 0. I
%
per
million
years.
The
summary
of these data in
Figure 6.9
shows
that replacement
divergence in
the
glo-
bin
genes
has
an average
rate
of
-0.096oh
per
million
years
(or
a UEP
of
10.4).
Considering
the uncertainties
in
estimating
the times
at
which
the species
diverged,
the results lend
good
support
to the idea
that there is
a
linear
clock.
The
data
on silent site
divergence
are much
less
clear. In
every case, it
is evident
that the
silent site
divergence is
much
greater
than
the
replacement
site divergence,
by a factor that
varies
from
2 to 10. The
spread
of silent
site
divergences
in
pairwise
comparisons,
though,
is
too
great
to show
whether
a clock is
applica-
ble
(so
we must
base temporal
comparisons
on
the replacement
sires).
From Figure
6.9, it
is clear
that
the
rate
at
silent
sites is not linear
with
regard
to time.
f
we assume
that
there must
be
zerl
diverqence
at
zerl
years
of separation, we see
that the rate of
silent site divergence is much
greater
for the
first
-100
million
years
of separation.
One
interpretation is
that a fraction of roughly
half
of the silent sites is rapidly
(within
I00 mil-
lion
years)
saturated by mutations;
this frac-
tion behaves as neutral
sites.
The
other fraction
accumulates mutations
more slowly,
at a rate
approximately the same as that
of the replace-
ment sites; this fraction identifies
sites
that are
silent with regard to the
protein,
but that come
under selective
pressure
for
some
other rea-
son.
Now we can reverse the
calculation
of
divergence rates to estimate
the times
since
genes
within
a species have been
apart. The
difference between the human
B
and
6
genes
is 3.7
o/"
for replacement
sites. At
a UEP of I0.4,
these
genes
must have
diverged I0.4
x
3.7
=
40 million
years
ago-about the
time of the
separation of the lines leading
to New World
monkeys, Old World monkeys,
great
apes, and
man. All
of these
higher
primates
have both
B
and 6
genes,
which suggests
that
the
gene
divergence commenced
just
before
this
point
in evolution.
Proceeding
further back,
the divergence
between the replacement
sites of
y
and e
genes
is l0%,
which corresponds
to a time
of separa-
tion
-100
million
years
ago. The
separation
between
embryonic and fetal
globin genes
there-
fore may have
just
preceded
or accompanied
the mammalian
radiation.
An evolutionary
tree
for
the human globin
genes
is
constructed in
,:;;i,:ii.i:
;a.':i.t.
Features
that
evolved before the mammalian
radiation-such
as the
separation of
B/6
from
y-should
be found
in
all mammals. Features
that
evolved
after-
ward-such
as the separation
of
B-
and
6-9lo-
bin
genes-should
be found
in individual
lines
of mammals.
In
each species,
there have
been compara-
tively recent
changes in the
structures
of the
clusters.
We
know
this because
we
see differ-
ences in
gene
number
(one
adult
B-globin
gene
in man,
two in mouse)
or in type
(most
often
concerning
whether there
are separate
embry-
onic
and fetal
genes).
When sufficient
data have
been
collected
on the sequences
of a
particular
gene,
the argu-
ments
can be reversed,
and comparisons
between
genes
in different
species
can
be used
to
assess taxonomic
relationshios.
106
CHAPTER
6 Clusters
and
Repeats

The
Rate
of
Neutral
Substitution
Can
Be Measured
from
Divergence
of Repeated
Sequences
.
The rate
of substitution
per
year
at
neutral
sites is
greater
in the mouse than in
the
human
genome.
We can make the best estimate
of the rate of
substitution
at neutral
sites by examining
sequences that do not code for
protein. (We
use
the term
neutral here rather
than
silent, because
there is no coding
potr:ntial).
An informative
comparison can be made by comparing
the
members of a common repetitive family in
the
human
and
mouse
genomes.
The
principle
of the analysis
is summarized
in
'iir--i::ii:
ir
,
:.
\A'/s
Start with a family of related
sequences that
have
evolved by duplication
and
substitution
from
an original family member.
We assume that the cornmon
ancestral sequence
can be deduced by taking
the base that is most
common at each
position.
Then
we can calcu-
late the divergence
of each individual family
member
as the
proportion
of bases
that differ
from the deduced ancestral
sequence. In this
example,
individual members
vary from 0.13
to 0.18 divergence and the average is 0.16.
One family used for
this analysis
in
the
human and mouse
genomes
derives from a
sequence that is thought to have
ceased to be
active at
about
the time of the divergence
between man and rodents
(the
LINES family;
see Section
22.9, Retroposons
Fall into Three
Classes). This
means
thLat it has
been diverging
without any selective
pressure
for
the same
length
of time
in
both species. Its average diver-
gence
in man is
-0.17
substitutions
per
site, cor-
responding
to a
rate
oI
2.2
x
I0-e substitutions
per
base
per year
over tl:re 75 million
years
since
the separation. In the rrrouse
genome,
however,
neutral substitutions have occurred
at twice this
rate,
corresponding to, 0.34 substitutions
per
site in the family, or a rate of 4.5
x
l0-e.
Note,
however, that if we calculated
the rate
per gen-
eration
instead
of
per
1,ear,
it would be
greater
in man than in mouse
(-2.2
x
l0-8
as opposed
to
-10
e).
cCCAGCGTAGCTTICATTACCCGTACGTTCATATTCGG
7138=0.18
cCTGGCGTAGC.TACGTTAGCGGTACGTGCATATTGGG
6/38=0.16
GGTAGCCTAaCTTAaGaTACCGGT!CGTGCTTGTTCGG
6/38=0.16
GGTAGCCTAGCTTAGGTTATTGGTAGGTGCATGTCCGG 6/38=0.16
cCTACCCTAGGTTACGTTATCGGTACGTGTCCGTTCGG
6/38=0.16
GCCACCC'AGCTCACGTTACCGGCACGTGCATGATCGC
7/38=0.18
CCTAGCCTCGCTTTCGTTAGCGGTACCTGCATCTTCCG
7/38=0,18
GCTTGCCTAGTTTACGTTACTGGTACGCGCATGTTGGG
5i38=0,13
GCCAGGCTAGCTTACGCCACCGGTACGTGGATGTCCGG
6/38--0.16
A
T
Calculate
Calculate consensus
sequence
divergence
I
trot
I
V
consensus
GCTAGCCTAGCTTACGTTACcGGTACGTGCATGTTCGG
SEqUENCE
i
I
r,ilrr'ir
r'r
.i
r
An
ancestraI
consensus
sequence
fora
familyis cat-
cutated by taking the
most common
base
at each
position.
The
divergence of each existing
current
member
of the fami[y
is ca[-
cutated as the
proportion
of
bases at
which it differs
from the
ancestraI sequence.
These figures
probably underestimate
the
rate of substitution
in the
mouse; at
the time of
divergence
the rates
in both species
would
have
been the same,
and the
difference
must
have
evolved
since then.
The current
rate of
neutral
substitution
per year in the
mouse is
probably
2-)x
grearer
than
the
historical average.
These
rates reflect the balance
between
the occurrence
of mutations
and the ability
of
the
genetic
sys-
tem of the organism
to
correct them.
The
dif-
ference between the
species
demonstrates
that
each species
has systems
that
operate
with a
characteristic ef
f iciency.
Comparing
the
mouse
and
human
genomes
allclws
us to assess
whether
syntenic
(corresponding)
sequences
show signs
of con-
servation or
have differed
at
the rate
expected
from accumulation
of
neutral
substitutions.
The
proportion
of
sites that
show
signs of
selection is
-5o/".
This
is much
higher
than
the
proportion
that
codes
for
protein
or
RNA
(-l'/.).It
implies that
the
genome includes
many
more stretches
whose
sequence
is
important
for noncoding
functions
than for
coding functions.
I(nown
regulatory
elements
are
likely to comprise
only
a small
part
of this
proportion. This number
also
suggests
that
most
(i.e.,
the
rest) of
the
genome
sequences
do not
have any function
that depends
on
the
exact seouence.
6.5
The Rate of Neutral Substitution
Can
Be
Measured
from
Divergence of
Repeated
Sequences
707

@
Pseudogenes Are
Dead
Ends
of EvoLution
o
Pseudogenes have no
coding function,
but they
can be recognized
by sequence similarities with
ex'isting functional
genes.
They
arise
by the
accumutation
of
mutations in
(formerty)
functional
genes.
Pseudogenes (Y)
are defined by
their
posses-
sion
of sequences that are related
to those of
the functional genes.
but that cannot
be trans-
lated
into a functional
protein.
Some
pseudogenes
have
the same
general
structure
as functional
genes,
with
sequences
corresponding
to
exons and introns in
the usual
locations.
They
may have
been
rendered
inac-
tive by mutations
that
prevent
any or all of the
stages of
gene
expression. The changes
can take
the form
of abolishing the
signals for initiating
transcription. preventing
splicing
at the
exon-intron
junctions,
or
prematurely
termi-
nating
translation.
Usually a
pseudogene
has several
deleteri-
ous mutations.
Presumably
once it ceased to be
active,
there was no impediment
to the accu-
mulation
of further
mutations. Pseudogenes
that represent
inactive versions
of currently
active
genes
have
been found in
many systems,
including globin,
immunoglobulins,
and histo-
compatibility
antigens,
where they
are
located
in
the
vicinity
of the
gene
cluster, often inter-
spersed
with the active
genes.
i
ii;tr i:r
,ir,
i"i.r Many
changes
have
occurred in a
B
gtobin gene
since
it
became
a
pseudogene.
CHAPTER
6 Ctusters
and Repeats
A typical example is the rabbit
pseudogene,
YB2,
which
has
the usual organization
of exons
and introns and is related most
closely to the
functional
globin gene
Bl.
The
rabbit
pseudo-
gene
is not functional,
though.
ljl*i.iiil: ri,i:i
sum-
marizes the many
changes that have
occurred
in the
pseudogene.
The deletion
of a base
pair
at codon 20 of Yp2 has
caused a frameshift
that
would
lead
to termination shortly
after. Sev-
eral
point
mutations have
changed later
codons
representing
amino acids that are highly
con-
served in the
p
globins.
Neither of the
two
introns
any
longer
possesses
recognizable
boundaries with the exons,
so
probably
the
introns could not
be spliced out even if
the
gene
were transcribed. However, there
are no tran-
scripts corresponding to the
gene,
possibly
because there have been changes
in the 5'flank-
rng reglon.
This list of defects includes
mutations
poten-
tially
preventing
each stage of
gene
expression,
thus we have no means
of telling which
event
originally inactivated
this
gene.
If
we measure
the divergence between the
pseudogene
and
the functional
gene,
though,
we
can
estimate
when
the
pseudogene
originated
and when its
mutations stafied to accumulate.
If the
pseudogene
had become
inactive as
soon as
it
was
generated
by
duplication from
pI,
we should expect both replacement
site
and
silent site divergence rates
to be the same.
(They
will be different
only
if
the
gene
is
translated to
create selective
pressure
on the replacement
sites.) In fact, there
are
fewer
replacement
site
substitutions
than silent site substitutions.
This
suggests that at first
(while
the
gene
was
expressed) there
was selection against
replace-
ment
site substitution. From
the relative
extents
of substitution in the
two types of
site, we can
calculate that Yp2
diverged from
Fl
-55
million
years
ago, remained
a functional
gene
for 22
million
years,
but
has
been a
pseudogene
for
the last 33
million
years.
Similar
calculations can be
made for
other
pseudogenes.
Some appear
to have
been active
for
some time
before becoming
pseudogenes;
others appear
to have been inactive
from
the
very
time
of their original
generation.
The
gen-
eral
point
made
by the
structures
of these
pseudogenes
is
that each has
evolved indepen-
dently
during the development
of the
globin
gene
cluster in each
species. This reinforces
the
conclusion
that the creation
of new
genes-
followed
by their acceptance
as functional
dupli-
cates, variation
to become new
functional genes,
or inactivation
as
pseudogenes-is
a continuing
108

60I
s.lalsnll auaD sa6ueilpau
le^o-6ulssoll
lpnbaun
1'g
llnsJr
aql se JnJ)o ueJ JJ^o-8urssorf,
Ienbeun
tal
-deqr
sql ot uorpnpoJtur
eql ur
pequJsep
sV
auorlezrue8roar
aua8 ro; alqrsuodsar
eJe srusrueq)Jru
Jo
sad.{1
1eqp1
'saua8
Ienprl
-1pul
ul suorlplnru
lurod;o
uorlelnurnJJp
Mols
Jql se uounlolJ
ur rolJpJ e
luelrodrur
se
sr uorle
-rrel
pue
'luarua8uerrear
'uorlelqdnp
aua8
teqt
lurod
leraua8
aqt sa4eru uosrredruor aq1
'(spunueru
Jqr
IIe
ol
uoruruoJ uoll
-nlole
ur
lurod 1se1
aqr) o3e srea^d
uoUpru
E8-
uorlPrpPJ uerleruruPru
eql e)urs
paJJn)f,o
eneq
tsnu
sa8ueqr eseqt
Jo
IIV
'luarrJJrp
are
sauaSopnasd;o sarnpnrts
pue
sraqunu Jql
pue
'saua8
urqo13-$
;o
sad,{1
pue
JJqurnu
ptol
Jql Ur UOrlPrJeA Sr JJJqt
'azrs
Ur
lUeJaJJrp
SI
qJPe
'uorlezrueSro
leraua8
aues aql e^pq
ile
pue
'uorlJun]
eues eq1 aAJJS sJalsnp aqr
qSnoqrly
'1'9
arn8r4 ur
papnl)ul
sretsnll
$
uetleru
-ureru
aqt
Surredruor zlq stlnsar eql aes
ueJ eM
'saua8
leJrluJpr
Jo
pJtelJJ
Jo
JJtsnlJ
p
uI
luJru
-a8uerrear
JoJ
sJrlrunuoddo
luanbarJ
JJp
JJeqJ
'u04alap
lenpwpur
aq1 uo spuedap eseesrp eq1
;o
r{luanas
aq1'sauab urqoi6-f, lo
-rr
alpurrutlo
leql
suoqalap
snouen Aq
pasnpl
alp spruasseleql
]ueraJJt[
o
'laqlo
aql
ur uorlerqdnp
6urpuodsauor e
pup
aurosouorql
luPUrquoral
auo ur uoqalap
e s0lnp0r0
stql
'reno-6uLsson
lenbaun
asner
uer sauaE
lrlalleuou uoeMleq
6uuredsru'saruenbas
pelelat
qluvr
saua6
Jo
ralsnlr
p
sulpluol auoua6
p
ua!fl\
r
sralsnll auel
se6uurPau
re^o-6urssojl
lenbaun
'pJqrJJsueJl
JIP
SJUJS
-opnasd
raleuJq^r alqrssod aq o1 adz(t slqt
Jo
s1JJJJJ uorlnlrp
Dedxe
lq8nu
a,lr alnJ
praua8 e
se
tne'aq
z(eru
speJJa
q)ns
uouruoJ
.troq Jpap
lou
sI
1I
'pJqrrJsupJl
sr aueSopnesd
aql uaqu
prtnllp
eq
IIIM
uralord eql
Jo
DeJJr
rqt
'rur8
-opnasd
aq1 ruoJJ
peqrrJsueJt
vNu
aql u1
tuasard
oslp sr aluanbas slqt
JI
'VNuru
Jqt ur eJuanbas
lynads e spurq
tpqt
uortepe.r8ap srqt
roy
alqrsuodsar uletord
e
sr
errql
.d1a41
tso141
'aua8
anrDe snoSolouoq s1 z(q
parnpord
yggru
aqr;o
uorleper8ap stlqlqq
auaSopnasd
e;o uorldtns
-uPJJ
'uoll)uq
droteln8al
e sPq auaSopnasd
e
'q8noqr 'aser
puorldaf,xa
euo
lseel
le
q
'[e
]e
uorlJunJ ou elpr{ ,{aqt [1errd[r
pue
'sutalord
roJ
JpoJ
lou
op sauaSopnasd
'uo1r1ur;ap,{g
'sunpord
aql Suoure
elpultulr)slp
ot uorlJelJs 01
1JJI
sr
11
'Tauotpunt
ata
[a4t
rcu
n
JaLllaLfw !a1sn1t
atp
to
staEilal'Ll
sD
paztufiont
an
7a47
satuanbas
11a
uo
pa
iuawaCuollaalpue
'ulqapp 'uotj
-atqdnp
aua6
nt alqrsuodsat
swstuoLpaw aLlJ
'(saua8opnasd
tuar
-JnJ
Jqt
olur
pa8ranrp qrlqu)
saua8
puotpuny
-uou
oMt
pue
(raquq pa8raarp qJlq,lr)
saua8
$
IeuourunJ
Sumr8
'paterlldnp
JIeslI
sem aJntJnJts
fl-$6
pur8rro
up
leqlsa11drur
stql
'sauaSopnasd
eql ol
upql Jeqlo
q)ea
ot
palplal
raraq are saua8
urqop-$
tlnpe
eql
'uoulppe
uI'suolletnru 8ut
-lelrDeur
IerJAJs
areqs
daqt teplqred ut
lsaua8
urqof
-fl
llnpp
aql
ol ueqt raqto
q)Pr
ol
prlPlar
JaltJq
are sauaSopnasd
aq1
'11
yo
urearlsdn
sJseqo1>l
,tra; e auaSopnasd
e seq Jsaql
Jo
q)ef,
'(1'9
arn8rg
aas)
3fl
pup
'Sd 'yfl
:sapads
rlnpe
eerql eJp araql
'1eo3
aqt;o
saua8 utqop-fl aqt u1
'parerlldnp
eq
upf, uorlnlola
Jo
SJIIaJ uenf,
'(palPuIuIIJ
Aluep
-pns
tou
s1
teql
auaSopnasd,{ue
Jo
eleJ eleruup
aqt
Llqeqo.rd)
,{lruey aruanbas
leut8tro
slt
;o
Jeqrueru
e se
paztuSo)JJ
Jq ra8uol ou uet
aua8
-opnasd
aqt
eJeqm
lurod
aql ol suoltplnru
Jo
uorleJJJe aqr
,{q ro
luela
uJppns e se aluanbas
Jql
Jo
uollalap
,{.q lnrro
plnoJ
uoIleuIIuIIe
srqJ
'peleururlJ
uaaq
arreq z(eru saua8opnasd
Jeqto
Jo
Jeqrunu
z(ue
'sarull
lsed
uI
'suotl
-elndod
tuasard
uI
paAIAJns
aneq
teql
sauaE
Jsoql eas Jl!\
1eql
raqrrrJrueJ
plnoqs
aAA
euorlualeJ
Jleql
JoJ arnssard antl
-f,alJs
ou aq
ppoqs
eJeql esef,
qJIqM
uI
'asodrnd
lnoqlrm
dlarrlua
,{aql are Jo uoll)unJ
Lue
py
-py
,{aqr oq
2aruoua8
eqt ul
luesJrd
p1s
,{.aqt
are
Lqm-saua8
IpuorDunJ
1o
luaua8ueJJpal
Jql
ol slueurruedruorre
pJluemun,{ldrurs-spua
peap
Lreuollnlola
are sauaSopnasd
11
-_
'6I'ZZ
erndld
ur
pezrJPruruns
eJP sernlesJ
JIlslJJlJereqJ
JraqJ
'suosodorlag
pup
sasnJIAoJlJU
'ZZ
Jal
-deql
ur
pessnJslp se
'yNU
aql
uroJJ
pJAIrep
lrnpord
e
Jo
ells luopueJ
elrros
lp
uoluJsul
.dq aleur8rro sauaSopnasd
passarord
'tuana
uorlrsodsuerloJlar
e 8urrrno11o4'sauaSopnasd
passerord
pJIIeJ
are
]drnsuBJt
VNU
aql JIq
-ruesJJ
leql
seruenbas
lrurouaS
arrtpeul
'suoJlur
Jo
ssol aql
qll1lt
pJlerJossp seult
luele
EutlentpBul
put3tro
aq1
leqt
stsaSSns
qJIqM
'uotlerrldnp
pu#r:o
aql
qlIM
saptruloJ
uoIleAIDeuI
Jo
elup
luJJedde
aql
'vNuru
utqo18-n
Jq1
qtl,r.r
(suotlelnru pJlelnu
-nJ)e
JoJ Sura,roqe)
pau311e
aq ueJ
aruanbas sU
'suoJlur
qroq
s>lJpl
,{lasnard
U
:,hradord 3ut1sa
-rJtur
up seq
aua8 utqo18-gn4
asnou
eqJ
'rJqunu
aua8
p1o1
aq1
;o
dlrrouru
ileurs
e
luasardar
sauaSopnasd
aq1
Llensn
'sauaSopnasd
a.re
leqt
sJaqruelu eABq
salrueJ
aua8
tso1A1
'JelsnlJ
aua8 aql ur
ssarord
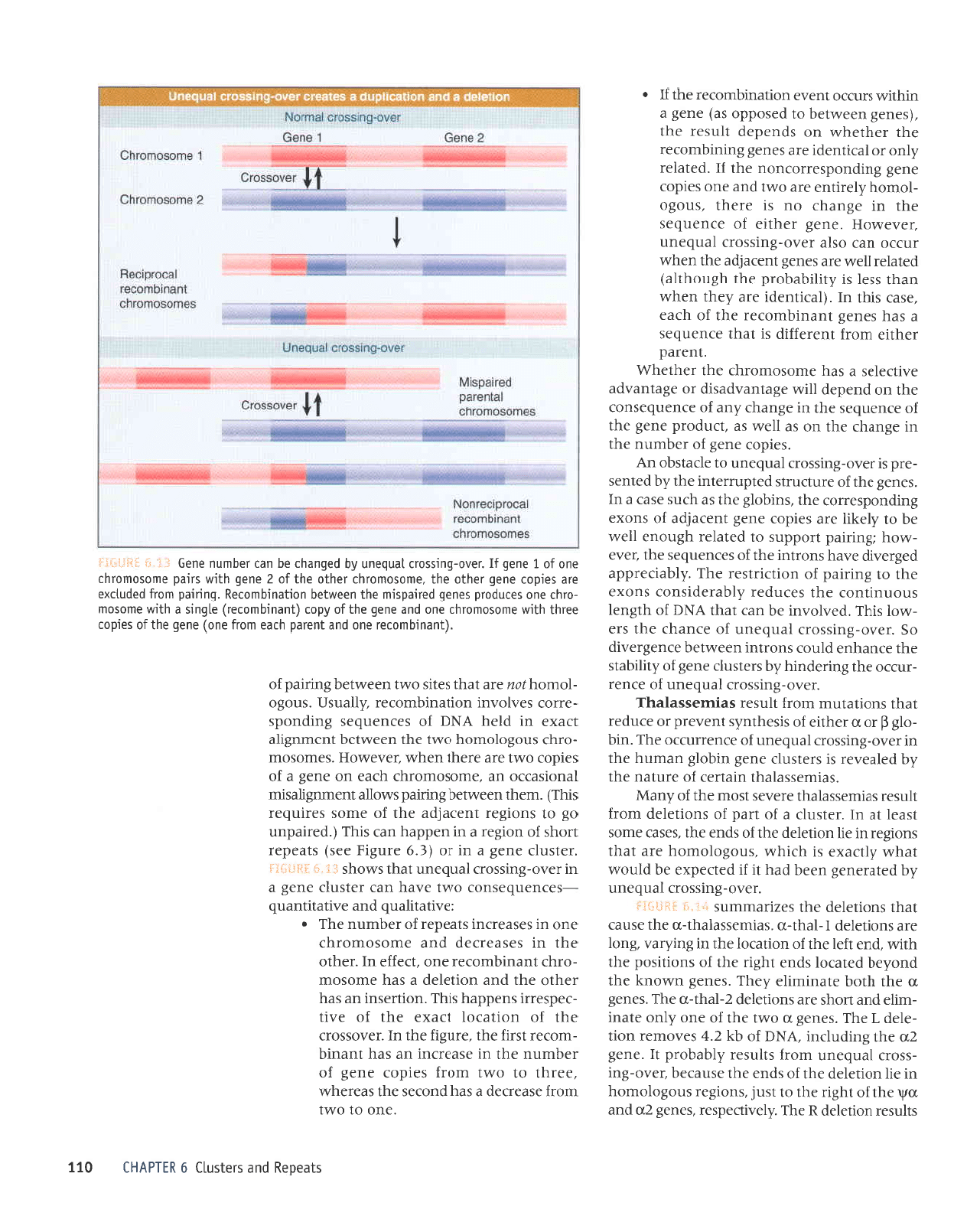
sllnsJJ uoIlJIJp
U
JqJ ^la^Ipadsar
'saua8
Zn
pue
nh
aqt
yo
lq8rr
aql
ot
tsn['suor8ar
snoSolouroq
ur JII uortJlJp
Jqt
Jo
spuJ
Jqt JsnelJq tano-8ur
-sso-rr
lenbaun
tuorl
sllnsal ,{lqeqord
U
'aua3
Zn
arp Supnlrur
'VNA
Jo
qie'V
srloruar
uorl
-JIap
f
aq1
'saua8
n
oMl
Jqt
Jo
euo Lluo
aleur
-uqa
pue
uoqs
ere suorlelep
z-Ieql-n
aq1
'saua3
xl
aql
qloq
JleururlJ [aq1
'saua8
u,!rou>l Jql
puo,{aq pJleJol
spua
lq8rr
Jqt
Jo
suortrsod
aqt
qtl,lr'puJ
Uel
eql
Io
uorleJol
aqt ur
Surzhea
'3uo1
ere suorlJlJp
I
-lpqt-D
'seruJsseleql-n
eql
JSnpJ
leq]
suorlelJp
Jql sazupuluns
1
:-.:j
-:si,:!::
'rano-Surssort
lenbaun
z(q
palerauaS
uaaq
ppq
tl
JI
papadxa
aq
plnoM
leqaa
,{.1pexJ
sr
qJrqM
'snoSoloruoq
Jlp
teql
suor8ar ur
aq uortJlep
Jql
Io
spua
Jqt
'saspJ
Jruos
lseel
te
uI
'JJtsnlJ
e
yo
ged
Jo
suorlJlJp
ruorJ
ll
nsJJ
spr ruJssel
eql JIJAJS
lsotu
JLIl
;o,{u
e61
'serruJsssleql
ureua)
Jo
eJnleu aql
Lq
palea,,lar
sr sJJtsnlJ
aua8 urqo13
upunq Jql
ur laao-Sursson
lenbaun
Jo
JJuJJTnJJo
JqJ'urq
-op
f,
ro
n
raqlre;o
srsaqluzi.s
tuanard
ro aJnpJJ
leq]
suorlelnu uoJJ
llnsJJ
sElrrressepql
'rano-Sursso,rr
lenbaun
Jo
J)ueJ
-rnJro
aqt Suuapurq
Lq
sralsnp auaS;o
[r1gqels
Jql
JJupquJ
plnoJ
suoJlur uJJ,trIJq
JJUJBJJATp
o5
';a,ro-Sursso:r
lenbaun
Jo
eJueqJ
Jqt srJ
-Mol
srqJ'panlolur
Jq uer
tpql
VNO;o
qt8ual
snonurluoJ Jql
seJnpJl zi.lqeraprsuoJ
suoxJ
eql 01 Sutrted
Jo
uorlJrJtser
aq1
',{lqenardde
pa8raarp
elpq suoJlur
Jql
Jo
seJuenbas aqt
tana
-,troq
lSurrred
uoddns
ot
paleler
q8noua
11a,t,r
aq or ,{1a41
a:B sardor
aua8
luarelpe
Jo
suoxa
SurpuodsarroJ
Jql
'surqo13
Jql se
qJns
JSe) e uI
'seue8
Jqt
Jo
JJnpnrls
paldnrralur
aqt
^dq
paruas
-a:d
sr rarro-SurssoJJ pnbaun
ol Jpelsqo
uV
'sardor
auaS;o requnu
Jql
ur a8ueq; Jql uo
se
IIaM
se
'pnpord
aua8 eql
yo
aruanbas aqt
ur a8ueqr.{ue yo
aluanbasuol
eql uo
puadap
11,u
a8elue,rpesrp
ro a8eluenpe
JAT]JJIJS
p
Seq aruosoruoJq)
Jql Jar{lJqM
'lUJJeO
reqlra ruoJJ
luJJJJJrp
sr
leqt
a)uenbJs
e seq saua8
lueurqtuoJJr
Jql
Jo
qJeJ
'aseJ
slqt u1
'(lerrluapr
are .daqt uaq.aa
upql ssel
sr
,{lrlqeqord
aqr
qSnoqtle)
palelJr
IIJ^
eJe sauaS
luarefpe
eql uJqM
JnJJo upJ
osle ra,to-Sursson
lenbaun
'JaAJMoH
'aua8
raqlra yo
aruanbas
Jqr
ul a8ueqr
ou sr
JrJql
'sno3o
-louoq
l.1arr1ua
JJe oMl
pue
euo
sardor
aua8 Surpuodsarroluou
eqr
JI
'pJlelal
z(po
ro
Ie)rluJpr
are
saua8
Sururqruorar
aqt reqtaqu
uo
spuadap
llnsrr
eql
'(saua8
ueeMlrq
ol
pasoddo
se) aua8 e
uIqlIM sJnJJo
lueAJ
uoIlPuIquoJJI
JIf
jl
o
sleaoau
puP
srelsnll
9
ulldvHl 0II
'euo
01 O1!rl
ruorJ JseerJap e sPq
puoJes
aql sealaqM
'Jarql
01 oul ruory sardo; auaE
yo
rJqrunu
aql ul eseeJf,ur uP seq
luPurq
-uroJJJ
ISJrJ
Jql
'arn8r;
Jr{l uI'JaAOSSOJJ
eql
jo
uortPJol
tJPXa
Jql
Jo
a^11
-JadsJJrr
suaddeq srqJ
'uoruasur
ue
spq
rJqlo Jql
pue
uorlalap e seq aurosou
-oJqJ
luPurqurofJr
Juo
'IJJJJJ
ul
'r3q]o
aql ur
saseerJJp
pue
aurosorxoJqJ
euo ur sespJJJur sleadar
Jo
rJqrunu JeJ .
:a,tr1e1r1enb pue
anrlelrluenb
-saruanbasuoJ
oml J^pq upJ
rJlsnlJ
aua8 B
ut razro-SutssoJJ
lenbJun
leq1
sMoqs
!r
j,"ii
-+liilill:l
'JalsnlJ
aua8 e ur ro
(E'9
arn8r4 aas)
steadar
uoqs
Jo
uor8ar e ur uaddeq ueJ srqJ
('parredun
oB o1 suor8ar
luarefpe
aqr
Jo
aruos sarrnbar
srql)
'ruaqt
ueaMtaq
8trured sazrolp
luaruuS[estu
IpuorseJJo
ue
'aurosoruoJqJ
qJpa
uo aua8 e;o
sardor oM1 aJe JJaql uJqM IJAJMoH
'sJruosoru
-orqr
snoSolouoq
oul Jql uaJMleq
luauru8qe
tJexJ
ur
plaq
VNq
;o
saruanbas Surpuods
-aJJoJ
sallolur uorlpurqruoJal f1ensn
'sno3o
-Ioutotqpu
JJe
teql
satls oml uaa,ulaq 8urrrcd
;o
'(lueuLquorar
auo
pup
luared
qrea
uor; auo) eueb
aq1;o satdor
oorql
qlrm
auosouolqr auo
pup
aua6 aq1;o r{dor
(lueurquorar)
e16urs
e
qlrM
auosoul
-olrll
euo
sarnpold sauaE
pa.rredsru
aql uaaMlaq uoqeurquolou'6uured
ulot1
papnllxo
are sardor auab reqlo
aql
'autosourotqt
taqlo aql
Jo
Z
aua6
qlLm
srted eLuosouorqr
auololauab11
'reno-6urssolrlenbeun[qpabueqraque]laqunuauag
i:i'l:i
jelil:.r:'ri
