Lewin Benjamin (ed.) Genes IX
Подождите немного. Документ загружается.

is essential for the
organism. Sometimes
muta-
tions
cause visible deleterious
effects
short of
lethality, in
which case we also
count them
as
identifying
an essential locus.
When the muta-
tions are
placed
into
complementation
groups,
the number
can be cornpared
with the number
of bands inthe region,
orindividual
complemen-
tation
groups
may even
be assigned to individ-
ual bands. The
purpose
of these
experiments has
been to determine whether
there is a consistent
relationship between
bands and
genes.
For
exam-
ple,
does
every band crlntain
a single
gene?
Totaling the analyses
that have
been car-
ried
out over the
past
l0
years,
the number of
lethal
complementation
groups
is
-70%
of the
number of bands. It is
an open
question
whether
there is any functional
significance
to this rela-
tionship. Irrespective
of the cause, the equiva-
Ience
gives
us a reasonable
estimate for the lethal
gene
number of
-3600.
By
any measure,
the
number of lethal loci
in Drosophila is
signifi-
cantly less than
the total number
of
genes.
If
the
proportion
of essential
human
genes
is similar to
other eukaryotes,
we would
pre-
dict a range of 4000
to 8000
genes,
in
which
mutations would
be lethal or
produce
evidently
damaging effects. At
present,
I300
genes
have
been identified in which mutations
cause evi-
dent defects. This is
a substantial
proportion
of
the expected total, especially in
view of the fact
that many lethal
genes
may act
so early that we
never see their effects. This
sort of bias may also
explain the results in
iii";i-:lii
* :r:.,
which show
that
the
majority
of known
genetic
defects are
due to
point
mutations
(where
there is more
Iikely
to be at
least
some residual function
of
the
gene).
How
do we explain the survival
of
genes
whose deletion appears to have no
effect? The
most likely explanation is
that the organism has
alternative ways of fulfilling
the same function.
The simplest
possibility
is that
there
is redun-
dancy, and that
some
genes
are
present
in mul-
tiple copies. This is certainly true in
some cases,
in which multiple
(r:elated)
genes
must be
knocked out in order
to
produce
an effect. In a
slightly
more
complex scenario, an organism
might have two separate
pathways
capable of
providing
some activity. Inactivation of either
pathway
by
itself
would not be damaging, but
the simultaneous occurrence of mutations in
genes
from both
pathways
would
be deleterious.
Such situations can be tested by combin-
ing mutations. In
principle,
deletions in two
genes,
neither
of which is lethal by itself, are
introduced
into
the same strain. If the double
mutant dies, the strain
is called a synthetic
lethal. This technique
has been used to
great
effect with
yeast,
where
the
isolation of double
mutants can be automated.
The
procedure
is
called
synthetic
genetic
array analysis
(SGA).
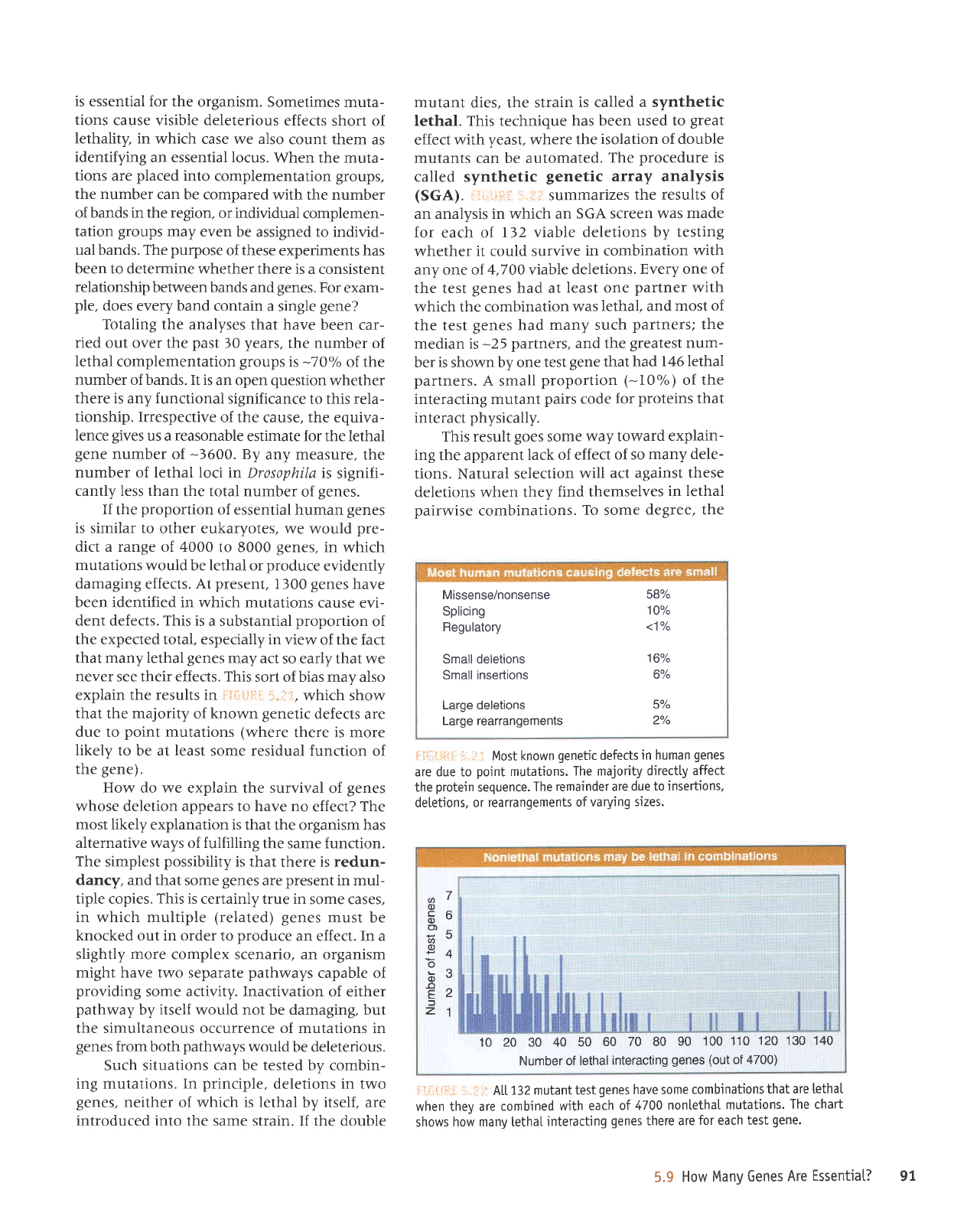
i.ii;;ri!il
ir.,i,i
summarizes
the results of
an analysis in which an
SGA screen was
made
for each oI 132
viable deletions
by testing
whether
it could survive
in combination
with
any one of
4,700
viable
deletions.
Every one
of
the test
genes
had at
least one
partner
with
which the combination
was
lethal, and
most of
the test
genes
had
many such
partners;
the
median is
-25
partners, and the
greatest
num-
ber is shown by one
test
gene
that had
146 lethal
partners.
A small
proportion
(-I0%)
of the
interacting mutant
pairs
code
for
proteins
that
interact
physically.
This result
goes
some
way toward
explain-
ing the apparent
lack of effect
of so
many dele-
tions.
Natural
selection
will
act against these
deletions when they
find themselves
in
lethal
pairwise
combinations.
To some
degree, the
f;i{l!ii,'-
11.;ii
Most known
genetic
defects
in human
genes
are due to
point
mutations.
The majority directty
affect
the
protein
sequence.
The
remainder are due
to insertions,
detetions, or
rearrangements
of varying
sizes.
,'ii*t.jFi:i:.,iir A[t132mutanttestgeneshavesomecombinationsthatareletha[
when they are
combined
with each
of 47Q0
nontethaI
mutations.
The chart
shows
how many lethal
interacting
genes
there are
for each test
gene.
Missense/nonsense
587"
Splicing
10%
Regulatory
<1o/o
Small deletions
16%
Small
insertions
6%
Large deletions
5%
Large rearrangements
2o/o
7
q)
66
o)-
6c
9n
Ez
z1
10 20 30
40 50
60 70 80
90 100
110
120 130 140
Number
of lethal
interacting
genes
(out
of 4700)
5.9
How
Manv Genes
Are
Essentiat? 91
organism has
protected
itself against the dam-
aging
effects of mutations by building in redun-
dancy. However, it
pays
a
price
in the form of
accumulating
the
"genetic
load"
of
mutations
that are not deleterious in
themselves, but that
may
cause serious
problems
when combined
with other
such
mutations
in future
genera-
tions. The
theory of natural
selection would
suggest
that the loss of the individual
genes
in
such
circumstances
produces
a sufficient disad-
vantage
to maintain the
active
gene
during the
course of evolution.
@
Genes Are Expressed
at
Widety Differing
Levels
.
In
any
given
cet[. most
genes
are expressed at a
low levet.
.
0nly
a smatl number of
genes,
whose
products
are
speciatized for
the ce[[ type,
are
high[y
expressed.
The
proportion
of DNA represented in
an nRNA
population
can be determined
by the amount
of the DNA
that can hybridize
with the RNA.
Such a
saturation analysis
typically identifies
-loh
of the DNA as
providing
a template for
nRNA.
From this
we can calculate
the
number
of
genes,
so long
as we know the average length
of an nRNA.
For a lower
eukaryote such as
yeast,
the total number
of expressed
genes
is
-4000.
For
somatic tissues
of
higher
eukary-
otes, the number
usually is 10,000
to I5,000.
The
value is
similar for
plants
and for
verte-
brates.
(The
only consistent
exception to this
type
of value is
presented
by mammalian
brain,
for
which much
larger numbers
of
genes
appear
to
be expressed,
although the exact
quantitation
is not
certain.)
I(inetic
analysis
of the reassociation
of an
RNA
population
can be used to
determine its
sequence
complexity.
This type
of analysis typ-
ically identifies
three
components in
a eukary-
otic
cell. Just as
with a DNA reassociation
curve,
a single
component hybridizes
over about two
decades
of Rot
(RNA
concentration
x
time) val-
ues,
and a reaction
extending
over a
greater
range
must
be resolved
by computer
curve-
fitting
into individual
componenrs.
Again,
this
represents
what is really
a continuous
spectrum
of sequences.
An
example
of an excess mRNA x
cDNA
reaction
that
generates
three components
is
given
in
i
il.ii.Jiitir
i',,i:i:
CHAPTER
5 Genome
Sequences
and
Gene Numbers
.
The first component has
the same char-
acteristics as a control reaction of oval-
bumin mRNA with its DNA
copy. This
suggests that the
first
component is in
fact
just
ovalbumin nRNA
(which
indeed occupies about half
of the mes-
senger mass in
oviduct tissue).
.
The next component
provides
15% of
the
reaction,
with a total complexity
of
l5 kb. This corresponds to7
ro 8 mRNA
species of average length 2000
bases.
.
The last
component
provides
35o/o
of
the
reaction,
which corresponds
to a
complexity of 26 Mb. This
corresponds
to
-13,000
mRNA species
of average
length 2000 bases.
From
this analysis, we can see that
about
half
of the mass of mRNA in the cell represents
a single mRNA,
-I5o/o
of the mass is
provided
by a
mere 7
to 8 mRNAs, and
-35oh
of the mass
is divided into the large number
of 13,000
nRNA
species.
It is
therefore obvious
that the
mRNAs
comprising each component
must be
present
in
very
different amounts.
The average number
of molecules
of each
mRNA
per
cell is called its abundance.
It can
be calculated
quite
simply if the total mass
of
RNA in
the cell is known. In the examole
shown
j'iii:ij*i:
1r.il; Hybridization between
excess mRNA
and cDNA
identifies
severaI
components in
chick oviduct
cetts, each
characterized
by the Rot172 of reaction.
First
Second Final
component component
component
50%
at
15Yo
at
35% at
Roto
0.0015
Roto
0.04 Rots
30
4zc
o
o
c
N
Eso
o
f25

in
Figure 5.2),Ihe
total mRNA
can be accounted
for
as
I00,000
copies
of the first
component
(ovalbumin
nRNA),
4000
copies of each
of the
7 to 8 mRNAs in
the second
component,
but
only
-5
copies of each
of the 13,000 mRNAs
that constitute the last
component.
We
can divide the nRNA
population
into
two
general
classes,
according
to their
abundance:
.
The oviduct is
an extreme
case, with so
much
of the mRNA represented
in only
one species, but most
cells do contain
a
small number
of RNAs
present
in
many
copies
each. This abundant
mRNA
component typically
consists
of
<I00
different mRNAs
present
in I000
to
10,000 copies
per
cell. It
often corre-
sponds
to a major
part
of the mass,
approaching
50'/" of
the total mRNA.
.
About
half of the mass
of the mRNA
con-
sists of a large
number
of sequences, of
the order of 10,000,
each represented
by only a small number
of copies in
the
mRNA-say. <10.
This is
the scarce
mRNA or
complex nRNA
class. It is
this class that drives
a saturation reaction.
How Many
Genes
Are Expressed?
mRNAs expressed
at low [evets overtap
extensivety
when different
cetl types are compared.
The
abundantty expressed mRNAs
are usualty
specific
for
the ce[[ type.
-10.000
expressed
genes
may
be common to most
cetl types of a
higher
eukaryote.
Many
somatic tissues of higher
eukaryotes have
an expressed
gene
number
in the range
of
10,000
to
20,000.
How much
overlap is there
between the
genes
expressed
in different tis-
sues? For example,
the expressed
gene
num-
ber of chick liver is
-l1,000
to
I7,000,
compared
with the value for
oviduct of
-l
1.000 to I 5.000.
How many of these two
sets of
genes
are iden-
tical? How many
are specific for each tissue?
These
questions
are usually addressed
by ana-
Iyzingthe transcriptome-the
set of sequences
represented in RNA.
We see immediately
that there are likely to
be substantial differences among the
genes
expressed in the aburrdant
class. Ovalbumin,
for
example,
is
synthesized only in
the oviduct,
and not at all in the liver. This means
lhat 50o/o
of the mass
of
mRNA in the oviduct
is specific
to that tissue.
The abundant mRNAs
represent only a
small
proportion
of the
number of expressed
genes,
though.
In terms of the total
number of
genes
of the organism,
and of the
number
of
changes in
transcription
that must be made
between different cell
types, we
need to know
the extent of overlap between
the
genes
repre-
sented in the scarce
mRNA classes
of different
cell
phenotypes.
Comparisons
between
different tissues show
that,
for
example,
-75'/o
of
the sequences
expressed in liver and
oviduct are the same.
In
other
words,
-I2,000
genes
are expressed
in
both liver and oviduct,
-5000
additional
genes
are
expressed only
in
liver, and
-3000
addi-
tional
genes
are expressed
only
in oviduct.
The scarce
mRNAs overlap
extensively.
Between mouse liver
and kidney,
-90%
of the
scarce
mRNAs
are
identical,
Ieaving a difference
between the tissues
of only
I000
to
2000 in
terms of the number of
expressed
genes.
The
general
result obtained
in several comparisons
of
this sort
is that only
-l}oh
of the
nRNA
sequences of a cell
are unique
to it. The major-
ity
of sequences
are
common to
many-
perhaps
even all-cell
types.
This suggests that
the common
set of
expressed
gene
functions,
numbering
perhaps
-10,000
in mammals,
comprise
functions that
are needed in all cell types.
Sometimes
this type
of function
is referred to as
a housekeeping
gene
or
constitutive
gene.
It contrasts
with
the
activities
represented
by specialized
func-
tions
(such
as ovalbumin
or
globin)
needed only
for
particular
cell
phenotypes. These are some-
times called luxury
genes.
Expressed Gene
Number
Can
Be Measured
En Masse
"Chip"
technotogy atlows
a snapshot
to be taken
of the expression of
the entire
genome
in
a
yeast
cetl.
-75olo
(-4500
genes)
of
the
yeast
genome
is
expressed under
normaI
growth
conditions.
Chip technotogy attows
detaited
comparisons of
retated anjmal celts to
determine
(for
exampte) the
differences
in expression
between
a normal cetl
and a cancer cett.
5.12
Exnressed
Gene
Number Can
Be Measured
En Masse 93
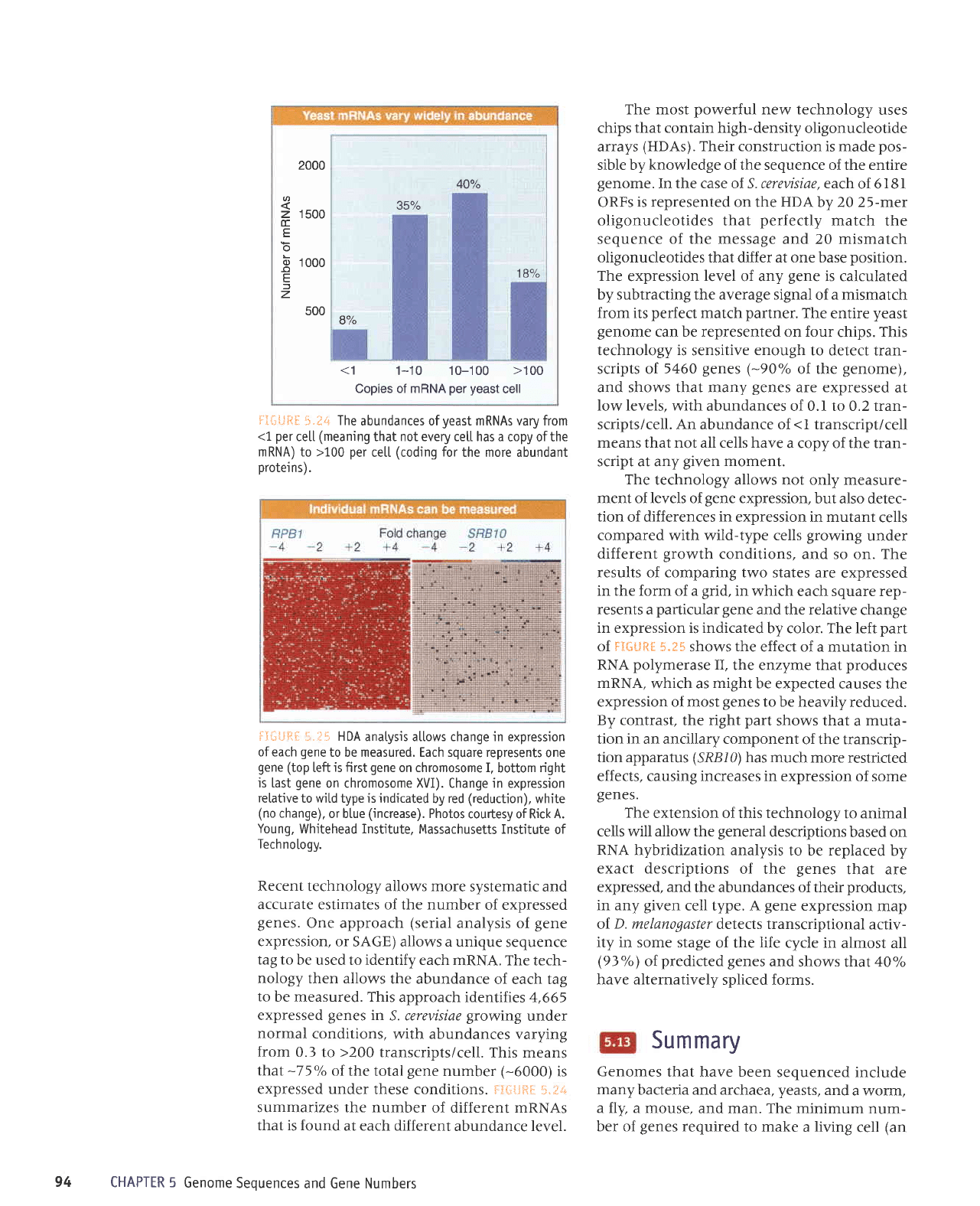
o
z
(E
E
q)
E
l
z
2000
1
500
1
000
500
4OTo
357o
18%
8%
<1
1-10 10-100
>100
Copies of mRNA
per yeast
cell
=:5;*fti.
1.t& The
abundances ofyeast mRNAs vary from
<1
per
cetl
(meaning
that not every cetl has a
copy of the
mRNA) to >100
per
cett
(coding
for
the more abundant
protei
ns).
i:**fti l:.=5 HDA
analysis attows change in
expression
of each
gene
to be measured. Each square represents
one
gene
(top
left
is
first
gene
on chromosome I,
bottom
right
is last
gene
on chromosome XVI).
Change
in
expression
retative
to wil.d type is indicated
by
red
(reduction),
white
(no
change),
or btue
(increase).
Photos
courtesy of Rick A.
Young,
Whitehead Institute,
Massachusetts Institute
of
Iechno[ogy.
Recent
technology allows more
systematic and
:il**riT,'#x',#i':idnrJ'"'Jffi
tag to
be used to identify each
mRNA. The tech-
nology
then allows
the abundance
of each tag
to
be measured. This
approach identifies
4,665
expressed
genes
in
S. cerevisiae
growing
under
normal
conditions,
with abundances varying
from
0.3
to
>200
transcripts/cell.
This means
that
-75o/o
of the
total
gene
number
(-6000)
is
expressed under
these conditions. i:irl{iS[:
$.irtri
summarizes the number
of different mRNAs
that is found
at each different
abundance level.
CHAPTER 5
Genome
Sequences
and Gene Numbers
The most
powerful
new technology
uses
chips
that contain high-density
oligonucleotide
arrays
(HDAs).
Their construction is made
pos-
sible by knowledge of the sequence of the entire
genome.
In the case of. S. cerevisiae, each
of 6
I
8 I
ORFs
is represented
on the
HDA
by 20 25-mer
oligonucleotides that
perfectly
match
the
sequence of the message and 20 mismatch
oligonucleotides that differ at one base
position.
The
expression
level
of any
gene
is calculated
by subtracting the average signal
of
a mismatch
from its
perfect
match
partner.
The entire
yeast
genome
can be represented on four
chips.
This
technology is
sensitive
enough
to detect tran-
scripts of
5460
genes (-90%
of the
genome),
and shows that many
genes
are expressed
at
low levels,
with abundances of 0.I to 0.2 tran-
scripts/cell. An abundance of
<l
transcript/cell
means that not all cells have
a copy of the tran-
script at any
given
moment.
The
technology allows not only measure-
ment of levels of
gene
expression,
but also detec-
tion
of differences
in
expression in mutant
cells
compared with wild-type cells
growing
under
different
growth
conditions, and so on. The
results of comparing two states
are expressed
in the form of a
grid,
in
which each square rep-
resents
a
particular gene
and the relative
change
in expression is indicated
by color. The left
part
of
fitil!flfl
5.?5
shows
the
effect of a mutation in
RNA
polymerase
II,
the enzyme that
produces
nRNA,
which as
might
be expected
causes the
expression of most
genes
to be heavily
reduced.
By contrast, the right
part
shows that
a
muta-
tion
in
an ancillary component
of the transcrip-
tion apparatus
(SRB10)
has
much more restdcted
effects, causing increases in expression
of some
genes.
The
extension of this technology
to animal
cells will allow the
general
descriptions
based on
RNA
hybridization analysis
to be replaced
by
exact descriptions
of the
genes
that are
expressed, and
the abundances of their
products,
in any
given
cell type. A
gene
expression
map
of.
D. melanogaster
detecls transcriptional
activ-
ity in some stage
of the life cycle in
almost all
(93"/")
of
predicted
genes
and shows
that
40'h
have alternatively
spliced forms.
Summary
Genomes
that have been
sequenced include
manybacteria
and archaea,
yeasts,
and
a worm,
atly, a mouse,
and man. The
minimum num-
ber of
genes
required
to make
a living
cell
(an
94

obligatory intracellular
parasite)
is
-470.
The
minimum number required
to make a free-liv-
ing cell is
-1700.
A typical
gram-negative
bac-
terium has
-1500
genes.
Strains of E. coli vary
from 4300
to
5400
genes.
The average bacter-
ial
gene
is
-1000
bp long
and
is
separated from
the
next
gene
by a space of
-
I 00 bp. The
yeasts
S.
pombe
and S. cerevisiae
have 5000 and 6000
genes,
respectively.
Although the fly D. melanogasler
is a more
complex organism and has a larger
genome
than
the worm C. elegans, the fly has fewer
genes
(13,600)
than
the worm
(18,500).
The
plant
Arabidopsis has 25,000
genes,
and the
lack
of a
clear
relationship
between
genome
size and
gene
number
is
shovm by the fact that the rice
genome
is 4x larger
but contains only a 507o increase in
gene
number, to
-40,000.
Mouse and man each
have 20,000 to 30,000
genes,which
is much less
than
had
been expected. The complexity of
development of an organism may
depend on
the nature of the interactions
between
genes
as
well as their total number.
About 8000
genes
are
common to
prokary-
otes and eukaryotes and are likely to
be
involved
in
basic
functions. A further 12,000
genes
are
found in multicellular
organisms. Another 8000
genes
are added to make an animal, and an addi-
tional8000
(largely
involved
with the
immune
and
nervous
systems) are found in vertebrates.
In each organism that has
been sequenced, only
-50%
of the
genes
have defined functions.
Analysis of lethal
genes
suggests that
only
a
minority
of
genes
is
essential
in
each organism.
The sequences comprising
a eukaryotic
genome
can be
classified in
three
groups:
non-
repetitive sequences are unique; moderately
repetitive
sequences are dispersed and
repeated
a small
number
of times
in
the form of
related,
but not identical, copies; and highly repetitive
sequences
are short and usually repeated as tan-
dem arrays.
The
proportions
of the types of
sequence are characteristic
for
each
genome,
although larger
genomes
tend to have a smaller
proportion
of nonrepetitive DNA. Almost 50%
of the
human
genome
consists of
repetitive
sequences, the vast
majority
corresponding to
transposon sequences. Most structural
genes
are
located in nonrepetitive DNA. The complex-
ity of nonrepetitive
DNA is
a better reflection of
the complexity of the organism than the total
genome
complexity; nonrepetitive DNA reaches
a maximum complexity of
-2
x
l0e
bp.
Genes are expressed at widely varying
lev-
els.
There may be I05 copies of nRNA for an
abundant
gene
whose
protein
is
the
principal
product
of the
cell,
I03 copies
of each
mRNA
for
<10
moderately
abundant
messages,
and
<10
copies of
each
nRNA for
>10,000
scarcely
expressed
genes.
Overlaps
between
the mRNA
populations
of cells
of different
phenotypes
are
extensive; the
majority
of
mRNAs are
present
in most cells.
Non-Mendelian
inheritance
is explained
by the
presence
of DNA
in organelles
in the
cytoplasm.
Mitochondria
and
chloroplasts
both
represent
membrane-bounded
systems
in which
some
proteins
are synthesized
within
the
organelle, whereas
others
are imported.
The
organelle
genome
is usually
a circular
DNA that
codes
for
all
of the
RNAs and
for some of
the
pro-
teins that are
required.
Mitochondrial
genomes
vary
greatly
in size,
from
the
l6 kb minimalist
mammalian
genome
to the 570
kb
genome of
higher
plants.
It is
assumed that
the
larger
genomes
code
for addi-
tional functions.
Chloroplast
genomes range
from 120 to 200
kb.
Those that
have been
sequenced
have a similar
organization
and cod-
ing functions.
In both
mitochondria
and chloro-
plasts,
many
of the
major
proteins
contain
some
subunits synthesized
in the organelle
and
some
subunits
imported
from
the cytosol.
Mammalian
mtDNAs
are transcribed
into a
single transcript
from
the
major coding
strand,
and
individual
products are
generated
by
RNA
processing.
Rearrangements
occur
in mitochon-
drial
DNA rather
frequently
in
yeast,
and
recom-
bination between
mitochondrial
or between
chloroplast
genomes has been
found.
Ttansfers
of DNA
have occurred
from
chloroplasts
or
mito-
chondria to
nuclear
genomes.
References
BacteriaI
Gene
Numbers
Range
0ver
an
0rder
of Magnitude
Reviews
Bentley, S.
D. and
Parkhill, J.
(2004).
Comparative
genomic
structure
of
prokaryotes. Annu.
Rev.
Genet. )8,
77
l-792.
Hacker, J.
and l(aper,
J.
B.
(2000). Pathogenicity
islands
and the
evolution
of
microbes.
Annu
Rev.
Microbiol. 54,
641-679.
Research
Blattner, F. R. et
al.
(1997).
The complete
genome
sequence
of.
Escherichia
coli
I(12. Science
277
,
t453-1474.
Deckert, G.
et al.
(I99S).
The complete
genome of
the hyperthermophilic
bacterium
Aquifex aeoli'
cus. Nature
392,
353-358.
References
95

Galibert, F. et
al.
(2001).
The
composite
genome
oi
the legume
symbiont Sinorhizobium
meliloti
Science 29),
668-672.

analysis of
human
chromosome
18. Nature
4)7, 55t-555.
The Y
Chromosome Has
Several
Mate-Specific
Genes
Resea rc h
Skaletsky, H. et al.
(2003).
The
male-specific
region of the human Y
chromosome is
a
mosaic of discrete
sequence classes. Nature
42), 825-837.
More
Comptex Species Evotve
by
Adding
New Gene Functions
Reference
The
Chimpanzee Sequencing and Analysis
Con-
sortium
(2005).
Initial sequence
of the chim-
panzee genome
and
comparison with the
human
genome.
Nature 4)7, 69-87.
How Many
Genes
Are Essentia[?
Resea rc h
Giaever et al.
(2002).
Functional profiling
of the
S cerevisiae
genome.
Nature 418,
)87-391.
Goebl,
M.
G. and Petes, T. D.
(1986).
Most
of the
yeast
genomic
sequences
are not essential for
cell
growth
and
division. Cell 46,98)-992.
Hutchison,
C. A. et al.
(1999).
Global transposon
mutagenesis and a rninimal mycoplasma
genome.
Science 286, 2165-2169.
Kamath, R.
S.,
Fraser, A.
G., Dong, Y., Poulin, G.,
Durbin, R.,
Gotta, M., Kanapin, A., Le Bot,
N.,
Moreno, S., Sohrmann, M.,
Welchman,
D. P.,
Zipperlen, P., and Ahringer,
J.
(2001).
System-
atic functional analysis of
the C. elegans
genome
using RNAi. Nature 421,231-2]7.
Tong, A. H. et al.
(2OO4l.
Global
mapping
of the
yeast
genetic
interaction
network. Science )0),
808-8
r
3.
Genes
Are Expressed
at Widel.y
Diftering
Leve[s
Resea
rc h
Hastie,
N.
B. and
Bishop, J. O.
(1976\.
The expres-
sion of three abundance
classes
of mRNA
in
mouse tissues.
Cell 9,761-774.
Expressed Gene
Number Can
Be
Measured
En Masse
Reviews
Mikos,
c.
L. c. and
Rubin, G.
M.
(I996).
The role
of the
genome
project
in determining
gene
function:
insights from
model organisms.
Cel/
86,52t-529.
Young, R. A.
(2000). Biomedical
discovery
with
DNA
arrays.
Cell
102, 9-L5.
Resea rch
Holstege, F.
C.
P. et al.
(1998). Dissecting the
regu-
latory
circuitry
of a
eukaryotic
genome.
Cell
95
,
7 17
-7
28.
Hughes, T. R., Marton,
M. J., Jones,
A.
R., Roberts,
C. J., Stoughton,
R., Armour,
C. D.,
Bennett,
H.
A., Coffey,
E.,
Dai, H.,
He, Y. D., Kidd,
M. J.,
Ifing, A,
M., Meyer,
M. R., Slade,
D., Lum,
P. Y., Stepaniants,
S.
B.,
Shoemaker,
D. D.
et al.
(2000).
Functional
discovery
via
a com-
pendium
of
expression
profiles.
Cell
102,
t09-t26.
Stolc, V. et al.
(2004).
A
gene
expression
map
for
the
euchromatic
genome
of. Drosophila
melanogaster. Science
306,
65 5-660.
Velculescu,
V.
E. et al.
(1997).
Characterization
of
the
yeast
transcriptosome.
Cell 88,
243-251.
References
97
Clusters
and Repeats
CHAPTER
OUTLINE
Introduction
Gene Duptication
Is
a Major Force
in Evolution
e
Duplicated genes
may
diverge
to
generate
different
genes
or
one copy may
become inactive.
GE
Globin
Ctusters
Are Formed
by
Duplication
and
Divergence
.
Att
gtobin
genes
are
descended
by duptication
and
mutation
from
an
ancestral
gene
that had
three exons.
.
The ancestral
gene
gave
rise to myogtobin,
leghemogtobin,
and
cr and
B
gtobins.
o
The
c- and
B-gtobin
genes
separated
in
the
period
of early
vertebrate
evotution,
after which
duptications
generated
the
individuaI
ctusters
of separate
cx-
and
B-tike
genes.
o
0nce
a
gene
has
been inactivated
by mutation. it may
accu-
mutate
further
mutations
and become
a
pseudogene,
which is
homotogous
to the
active
gene(s)
but has
no functional rote.
Sequence
Divergence
Is
the Basis for
the Evo[utionary
Clock
r
The
sequences
of homologous genes
in
different species
vary
at reptacement
sites
(where
mutation
causes
amino
acid
substitutions)
and silent
sites
(where
mutation does
not affect
the
protein
sequence).
r
Mutations
accumutate
at silent
sites
-10x
faster
than
at
reptacement
sites.
o
The
evolutionary
divergence
between
two
proteins.is
mea-
sured
by the
percent
of
positions
at which
the correspon-
ding amino
acids
differ.
o
Mutations
accumutate
at
a
more
or
less even speed
after
genes
separate,
so
that the
divergence
between any
pair
of
gtobin
sequences
is
proportionaI
to
the time since
their
genes
separated.
The
Rate
of Neutral
Substitution
Can Be
Measured from
Divergence
of
Repeated
Sequences
o
The rate
of
substjtution
per
year
at neutral
sites is
greater
in
the mouse
than in
the human
genome.
Pseudogenes
Are Dead
Ends
of Evolution
r
Pseudogenes
have no
coding function,
but they
can be rec-
ognized
by sequence
similarities
with
existing functional
genes.
They
arise
by the
accumutation
of mutations
in
(for-
merly) functionaI genes.
UnequaI
Crossing-over
Rearranges
Gene
Ctusters
o
When
a
genome
contains
a cluster
of
genes
with
retated
sequences,
mispairing
between nonatlelic
genes
can cause
unequal
crossing-over.
This
produces
a detetion in
one
recombinant
chromosome
and
a conesponding
duptication
in
the other.
98
r
Different thatassemias
are caused by
various
detetions
that
eliminate o- or
B-globin
genes.
The
severity of the
disease
depends on the individual deletion.
Genes
for rRNA
Form Tandem Repeats
o
Ribosomal
RNA
is
coded by a large number
of
identical
genes
that are tandemly
repeated
to form
one or
more
clusters.
r
Each
rDNA
ctuster is organized
so that transcription
units
giving
a
joint
precursor
to the major rRNAs
atternate with
nontranscribed
spacers.
The Repeated
Genes
for
rRNA Maintain
Constant
Seouence
r
The
genes
in
an
rDNA
ctuster a[[ have an identical
sequence.
r
The nontranscribed
spacers consist of
shorter repeat'ing
un'its whose number
varies so that the
lengths of individuaI
spacers are different.
Crossover Fixation
Coutd Maintain IdenticaI
Repeats
.
Unequal crossing-over
changes the size
of a cluster of tan-
dem reDeats.
.
IndividuaI repeating
units can be etiminated
or can spread
through the cluster.
Satellite
DNAs
Often Lie in Heterochromatin
.
Highty repetitive DNA
has a very short repeating
sequence
and no
coding
function.
.
It
occurs
in
large blocks
that can
have
distinct
physical
properties.
I
It is
often the major
constituent of centromeric
heteroch ro mati
n.
Arthropod
Satettites
Have
Very Short IdenticaI
Repeats
r
The repeating
un'its
of arthropod satetlite DNAs
are
on[y a
few
nucleotides
long.
Most
of the copies
of the sequence
are
identical.
Mammatian
Satetlites
Consist of
HierarchicaI
Repeats
r
Mouse
satettite DNA has
evotved by duplication
and muta-
tion of
a short
repeating
unit to
give
a basic repeating
unit
of 234
bp
in
which the
originaI
ha[f.
quarter,
and eighth
repeats
can be recognized.
Minisatetlites
Are
UsefuI for
Genetic Mapping
o
The variation
between microsatettites
or minisateltites
in
individuaI
genomes
can be used to identify
heredity
unequivocatly
by showing
that 50%
of the bands in
an
indi-
vidual
are derived from
a
particular parent.
Summarv
Introduction
A
set of
genes
descended
by
duplication and
variation
from
some ancestral
gene
is called
a
gene
family. Its mernbers
may
be clustered
together or dispersed
on different
chromosomes
(or
a combination
of both). Genome
analysis
shows that many
genes
belong
to families; the
25,000
genes
identified
in the human
genome
fall into
-15,000
families,
so the average
gene
has
a couple of relatives
in the
genome (see
Figure 5.7).
Gene fam;ilies
vary enormously in
the degree of relatedness
between members,
from
those consisting of multiple
identical mem-
bers to those for which the relationship
is
quite
distant. Genes are usu;llly related
only by their
exons, with introns
having diverged
(see
Sec-
tion 3.5, Exon Sequences Are
Conserved but
Introns Vary).
Genes may also
be
related
by only
some of their exons, w.hereas
others are unique
(see
Section
3.9,
Some Exons
Can Be
Equated
with
Protein Functions).
The initial event tl:rat
allows related exons
or
genes
to develop is
a duplication,
when a
copy is
generated
of some
sequence within the
genome.
Tandem
duplication
(when
the dupli-
cates
remain
together) may
arise through errors
in replication
or recombination.
Separation of
the duplicates can
occur by a translocation
that transfers material from
one chromosonre
to another. A
duplicate at a new location may
also be
produced
directly
by a transposition
event that
is
associated with
copying a
region
of
DNA from
the vicinity
of the transposon.
Duplications may appl.g
either to intact
genes
or
to collections of exons or even individual
exons.
When an intact
gene
is involved,
the act of dupli-
cation
generates
two copies of a
gene
whose
activities are
indistinguishable,
but then usu-
ally the copies diverge
as each accumulates dif-
ferent mutations.
The members of a well-related
structural
gene
family
usually
h;rve
related or even iden-
tical functions, although they may
be expressed
at different times or
in
different cell types. As a
result, different
globin proteins
are expressed
in
embryonic and adult red blood cells, whereas
different actins are utilized in muscle
and
non-
muscle
cells.
When
genes
have diverged signif-
icantly, or when only some
exons are
related,
the
proteins
may have different functions.
Some
gene
families
consist of
identical
members. Clustering
is
a
prerequisite
for main-
taining identity between
genes,
although clus-
tered
genes
are
not necessarily
identical.
Gene
clusters range from extremes in
which
a dupli-
cation
has
generated
two adjacent
related
genes
to cases where
hundreds of
identical
genes lie
in a tandem array. Extensive
tandem
repetition
of a
gene
may occur
when the
product
is
needed
in unusually
large
amounts.
Examples are the
genes
for rRNA or
histone
proteins.
This cre-
ates a special situation
with
regard to the
main-
tenance of
identity and
the effects of
selective
pressure.
Gene clusters offer
us an opportunity
to
examine the
forces involved
in evolution
of the
genome
over
larger
regions than
single
genes.
Duplicated sequences,
especially
those
that
remain in the same
vicinity.
provide
the sub-
strate for further evolution
by
recombination.
A
population
evolves
by the classical
recombi-
nation illustrated
in
; l'iiil'"iI::
ir.
i
and
ii.;1,
in which
an
exact
crossing-over
occurs.
The recombinant
chromosomes
have the same
organization
as
the
parental
chromosome.
They contain
pre-
cisely the same
loci in the
same order,
but con-
tain different combinations
of alleles,
providing
the raw material
for natural
selection.
How-
ever, the existence
of
duplicated
sequences
allows aberrant events
to
occur occasionally,
which changes
the content
of
genes
and
not
just
the combination
of
alleles.
Unequal crossing-over
(also
known
as
nonreciprocal
recombination)
describes
a
recombination
event occurring
between two
sites that
are not
homologous.
The
feature that
makes such events
possible
is the
existence
of
repeated
sequences.
r
r,rilillir :
"i
shows
that this
allows one
copy of a
repeat
in one chromosome
to misalign
for recombination
with
a different
of
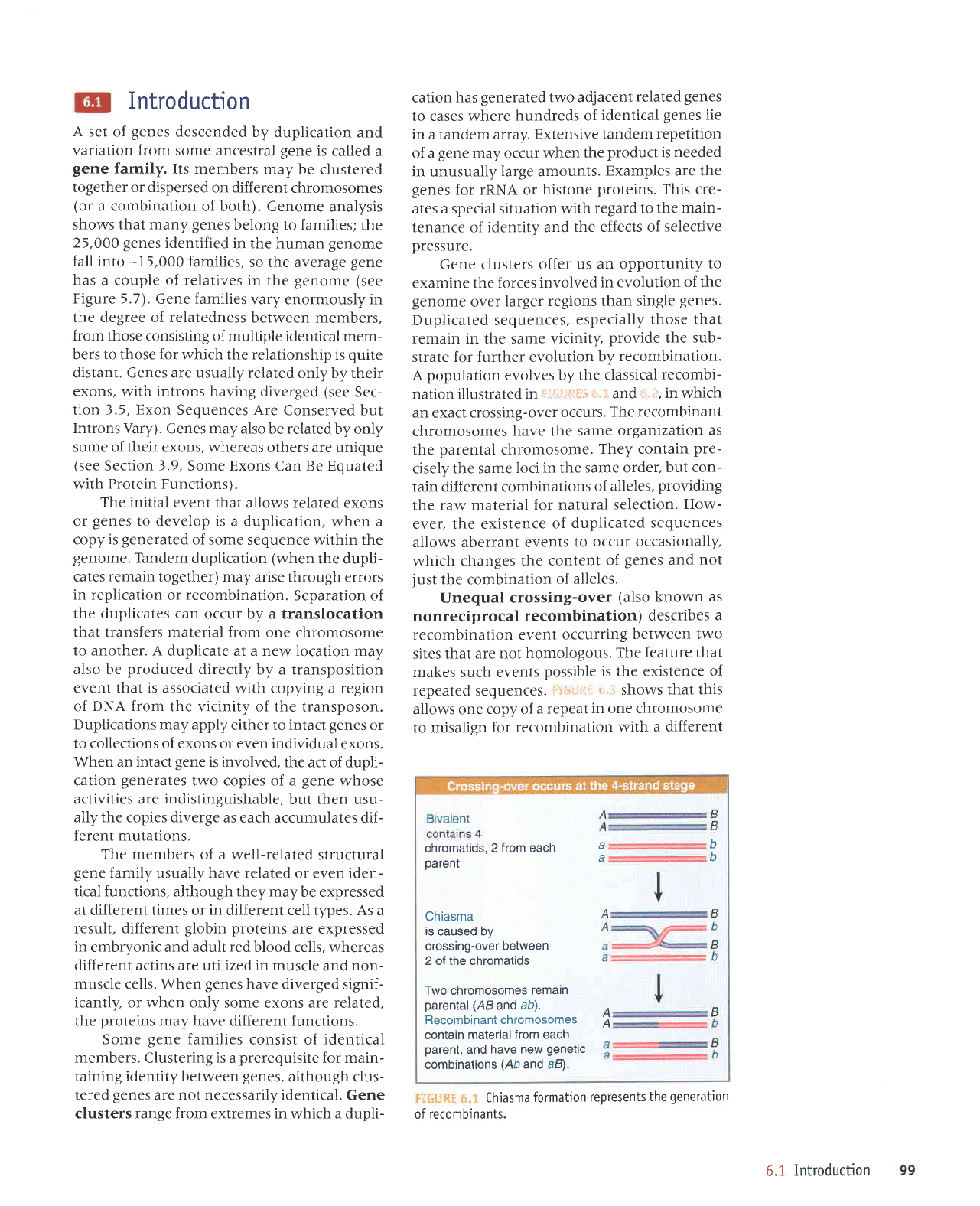
recombinants.
chromatids, 2
from each
a
D
^h
parent
d
u
Chiasma
is
caused by
crossing-over between
2 of the chromatids
Two chromosomes
remain
parental (ABand
ab).
Recombinant
chromosomes
contain material
from each
parent,
and
have new
genetic
combinations
(Ab
and aB).
6.1
Introduction
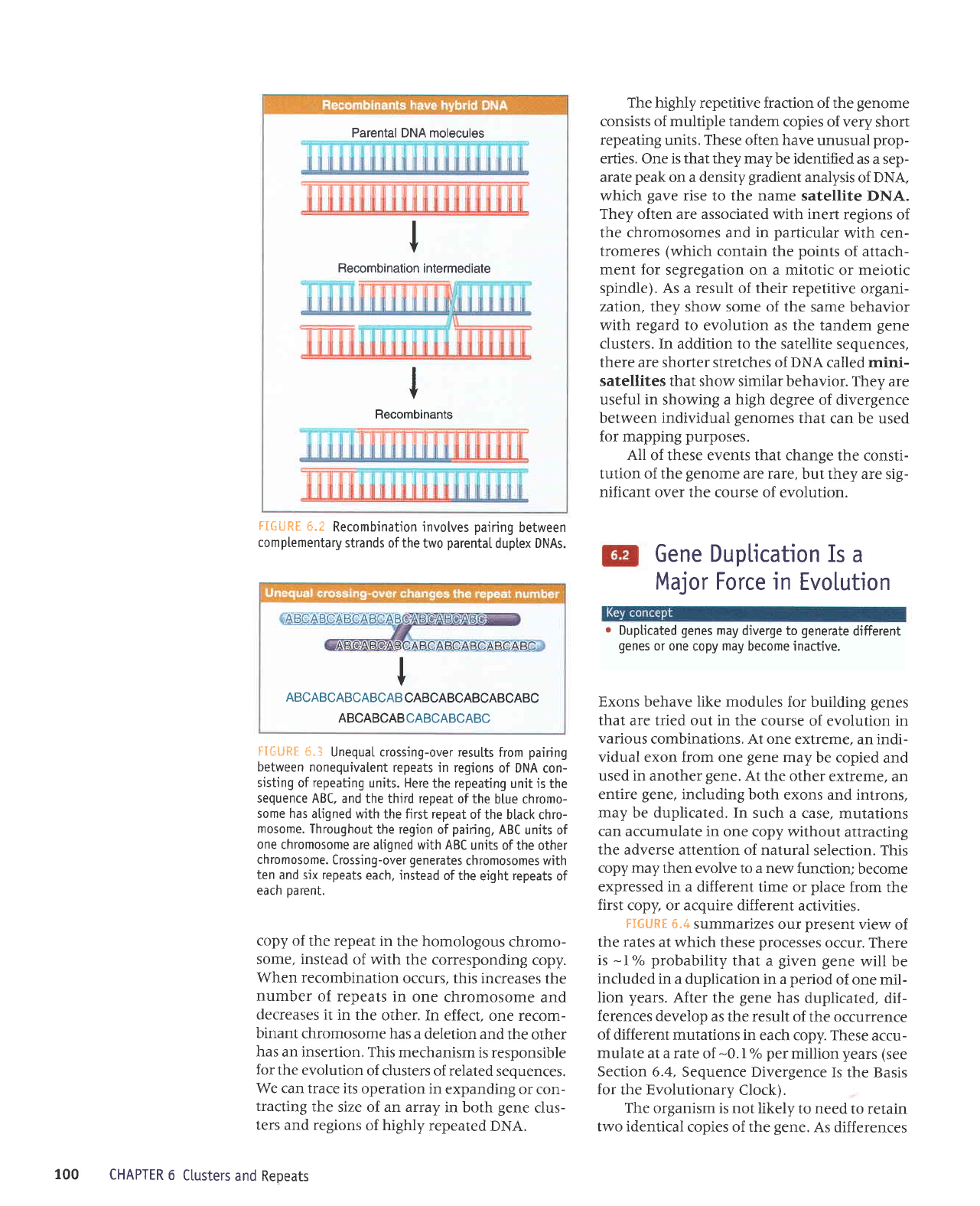
Parental DNA molecules
I
Becombination
intermediate
I
Recombinants
FI6URE
6.t
Recombination invotves
pairing
between
comptementary
strands of the
two
parentaI
duptex DNAs.
I
ABCABCABCABCAB
CABCABCABCABCABC
ABCABCABCABCABCABC
fIfiUFf
*"3
UnequaI crossing-over results
from
pairing
between
nonequivatent
repeats in regions
of DNA
con-
sisting
of
repeating
units. Here
the repeating
unjt is the
sequence ABC.
and the third repeat
of the
blue chromo-
some has
atigned with the first repeat
of
the black chro-
mosome.
Throughout
the region
of
pairing,
ABC
units of
one
chromosome
are atigned with ABC
units ofthe
other
chromosome.
Crossing-over
generates
chromosomes with
ten and six repeats
each, instead
of the
eight
repeats
of
eacn
Darent.
copy of
the repeat in
the homologous
chromo-
some, instead
of
with the corresponding
copy.
When recombination
occurs, this increases
the
number
of repeats
in one
chromosome
and
decreases
it in the
other. In
effect, one recom-
binant
chromosome
has a
deletion and
the other
has
an insertion.
This mechanism
is responsible
for
the
evolution
of clusters of related
sequences.
We can trace
its operation in
expanding
or con-
tracting
the
size of an array
in both
gene
clus-
ters and regions
of
highly
repeated
DNA.
Reoeats
The highly repetitive fraction of the
genome
consists of multiple tandem
copies of very short
repeating
units. These often have
unusual
prop-
erties. One is that they may
be
identified
as a sep-
arate
peak
on a density
gradient
analysis
of DNA,
which
gave
rise to the name satellite
DNA.
They often are associated
with
inert
regions
of
the chromosomes and in
particular
with cen-
tromeres
(which
contain
the
points
of attach-
ment for
segregation on a mitotic
or
meiotic
spindle). As a result of their repetitive
organi-
zation, they show some of the
same behavior
with regard to
evolution as the tandem
gene
clusters. In addition to the satellite
sequences,
there are shorter stretches of DNA
called mini-
satellites that
show similar behavior. They are
useful
in
showing a high degree
of divergence
between individual
genomes
that
can be used
for mapping
purposes.
AII
of these events that change
the consti-
tution of the
genome
are rare,
but they are
sig-
nificant
over the course of evolution.
Major
Gene
Duplication
Is
a
Force in
Evolution
r
Dupticated
genes
may diverge to
generate
different
genes
or one
copy
may
become inactive.
Exons behave like modules
for building
genes
that are tried out in the course
of evolution in
various
combinations. At one
extreme, an indi-
vidual exon from one
gene
may
be copied
and
used in
another
gene.
At the other
extreme,
an
entire
gene,
including both
exons and introns,
may be duplicated. In
such a case,
mutations
can accumulate
in one copy without
attracting
the
adverse attention of natural
selection. This
copy
may
then evolve to a new
function;
become
expressed in a
different time or
place
from the
first
copy, or acquire different
activities.
FISUSE
S"4 summarizes
our
present
view
of
the
rates
at which these
processes
occur. There
is
-l%
probability
that a
given
gene
will
be
included
in a
duplication in a
period
of one
mil-
Iion
years.
After the
gene
has
duplicated,
dif-
ferences develop
as the result
of the occurrence
of
different mutations in
each copy. These
accu-
mulate
at a rate
of
-0.1"/o
per
million
years (see
Section
6.4, Sequence Divergence
Is
the Basis
for
the Evolutionary
Clock).
The organism is
not likely
to nee d to retain
two identical
copies of the
gene.
As differences
100
CHAPTER
6
Clusters
and
