Knapp J.S., Cabrera W.L. Metabolomics: Metabolites, Metabonomics, and Analytical Technologies
Подождите немного. Документ загружается.


Metabolomics 237
A useful approach to unraveling and understanding complex biological networks is to
decompose networks into basic functional and structural units [48]. Recent application of
convex analysis to metabolic networks leads to the development of network-based metabolic
pathway analysis and the decomposition of metabolic networks into metabolic extreme
pathways that are true functional units of metabolic systems [48]. Metabolic extreme
pathways are derived from limited knowledge of the metabolic networks, but provide an
integrated predictive description of metabolic networks [48]. Some interesting reports on
usefulness of pathway analysis are presented in Table 1.
D. Pathway Analysis Tool
There are also many pathway analysis tools at present. These tools are very useful.
Details of some important tools will be hereby presented.
1. Pathway Miner [53]
Pathway Miner catalogs genes based on their role in metabolic, cellular and regulatory
pathways [53]. A Fisher exact test is provided as an option to rank pathways [53]. The genes
are mapped onto pathways and gene product association networks are extracted for genes that
co-occur in pathways [53]. Pathway Miner is a freely available web accessible tool at
http://www.biorag.org/pathway.html [53].
2. WholePathwayScope [54]
WholePathwayScope (WPS) is for deriving biological insights from analysis of High
Throughput data [54]. WPS extracts gene lists with shared biological themes through color
cue templates [54]. WPS statistically evaluates global functional category enrichment of gene
lists and pathway-level pattern enrichment of data [54]. WPS incorporates well-known
biological pathways from KEGG (Kyoto Encyclopedia of Genes and Genomes) and Biocarta,
GO (Gene Ontology) terms as well as user-defined pathways or relevant gene clusters or
groups, and explores gene-term relationships within the derived gene-term association
networks (GTANs) [54]. WPS simultaneously compares multiple datasets within biological
contexts either as pathways or as association networks. WPS also integrates Genetic
Association Database and Partial MedGene Database for disease-association information. We
have used this program to analyze and compare microarray and proteomics datasets derived
from a variety of biological systems [54]. The tool is freely available at
http://www.abcc.ncifcrf.gov/wps/wps_index.php [54].+
3. ArrayxPath [55]
ArrayXPath (http://www.snubi.org/software/ArrayXPath/) is a web-based service for
mapping and visualizing microarray gene-expression data for integrated biological pathway
resources using Scalable Vector Graphics [55]. By integrating major bio-databases and
searching pathway resources, ArrayXPath automatically maps different types of identifiers
from microarray probes and pathway elements [55]. When one inputs gene-expression
clusters, ArrayXPath produces a list of the best matching pathways for each cluster [55].

Viroj Wiwanikit 238
4. Genome Expression Pathway Analysis Tool [56]
Genome Expression Pathway Analysis Tool (GEPAT) offers an analysis of gene
expression data under genomic, proteomic and metabolic context [56]. GEPAT offers various
statistical data analysis methods, as hierarchical, k-means and PCA clustering, a linear model
based t-test or chromosomal profile comparison [56]. GEPAT offers no linear work flow, but
allows the usage of any subset of probes and samples as a start for a new data analysis [56].
GEPAT relies on established data analysis packages, offers a modular approach for an easy
extension, and can be run on a computer grid to allow a large number of users [56]. It is freely
available under the LGPL open source license for academic and commercial users at http://
gepat.sourceforge.net [56].
Introduction to Metabolonics
Metabolonics is a science to study cellular metabolic activities. The aim of metabolonics
is to study the complete metabolic response of living things to genetic modifications or
environmental stimuli. The knowledge in this new “omic” science is still limited but carries
great hope in science and medicine.
References
[1] Fridman, E; Pichersky, E. Metabolomics, genomics, proteomics, and the identification
of enzymes and their substrates and products. Curr Opin Plant Biol., 2005, 8(3), 242-8.
[2] German, JB; Bauman, DE; Burrin, DG; Failla, ML; Freake, HC; King, JC; Klein, S;
Milner, JA; Pelto, GH; Rasmussen, KM; Zeisel, SH. Metabolomics in the opening
decade of the 21st century: building the roads to individualized health. J Nutr., 2004,
134(10), 2729-32.
[3] Arita, M. Additional paper: computational resources for metabolomics. Brief Funct
Genomic Proteomic., 2004, 3(1), 84-93.
[4] Mendes, P. Emerging bioinformatics for the metabolome. Brief Bioinform., 2002, 3(2),
134-45.
[5] Lange, BM; Ghassemian, M. Comprehensive post-genomic data analysis approaches
integrating biochemical pathway maps. Phytochemistry., 2005, 66(4), 413-51.
[6] Hunter, P; Nielsen, P. A strategy for integrative computational physiology. Physiology
(Bethesda)., 2005, Oct; 20, 316-25.
[7] Eungdamrong, NJ; Iyengar, R. Computational approaches for modeling regulatory
cellular networks. Trends Cell Biol., 2004, Dec;14(12), 661-9.
[8] Hofestädt, R; Thelen, S. Quantitative modeling of biochemical networks. In Silico Biol.,
1998, 1(1), 39-53.
[9] Cabrera, ME; Saidel, GM; Kalhan, SC. Modeling metabolic dynamics. From cellular
processes to organ and whole body responses. Prog Biophys Mol Biol., 1998, 69(2-3),
539-57.
[10] Arita, M. Computer modeling of metabolic networks. Tanpakushitsu Kakusan Koso.,
2003, Jun; 48(7), 823-8

Metabolomics 239
[11] Wiechert, W. Modeling and simulation: tools for metabolic engineering. J Biotechnol.,
2002, Mar, 14, 94(1), 37-63.
[12] van Helden, J; Wernisch, L; Gilbert, D; Wodak, SJ. Graph-based analysis of metabolic
networks. Ernst Schering Res Found Workshop., 2002, (38), 245-74.
[13] Duran, AL; Yang, J; Wang, L; Sumner, LW. Metabolomics spectral formatting,
alignment and conversion tools (MSFACTs). Bioinformatics., 2003, 19(17), 2283-93.
[14] Shinoda, K; Yachie, N; Masuda, T; Sugiyama, N; Sugimoto, M; Soga, T; Tomita, M.
HybGFS: a hybrid method for genome-fingerprint scanning. BMC Bioinformatics.,
2006, Oct 29, 7, 479.
[15] Wishart, DS; Tzur, D; Knox, C; Eisner, R; Guo, AC; Young, N; Cheng, D; Jewell, K;
Arndt, D; Sawhney, S; Fung, C; Nikolai, L; Lewis, M; Coutouly, MA; Forsythe, I;
Tang, P; Shrivastava, S; Jeroncic, K; Stothard, P; Amegbey, G; Block, D; Hau, DD;
Wagner, J; Miniaci, J; Clements, M; Gebremedhin, M; Guo, N; Zhang, Y; Duggan, GE;
Macinnis, GD; Weljie, AM; Dowlatabadi, R; Bamforth, F; Clive, D; Greiner, R; Li, L;
Marrie, T; Sykes, BD; Vogel, HJ; Querengesser, L. HMDB: the Human Metabolome
Database. Nucleic Acids Res. 2007 Jan, 35, (Database issue), D521-6.
[16] Lemer, C; Antezana, E; Couche, F; Fays, F; Santolaria, X; Janky, R; Deville, Y;
Richelle, J; Wodak, SJ. The aMAZE LightBench: a web interface to a relational
database of cellular processes. Nucleic Acids Res., 2004, Jan 1, 32(Database
issue):D443-8.
[17] Hou, BK; Kim, JS; Jun, JH; Lee, DY; Kim, YW; Chae, S; Roh, M; In, YH; Lee, SY.
BioSilico: an integrated metabolic database system. Bioinformatics., 2004, Nov 22,
20(17), 3270-2.
[18] Karp, PD; Riley, M; Paley, SM; Pellegrini-Toole, A; Krummenacker, M. EcoCyc:
Enyclopedia of Escherichia coli Genes and Metabolism. Nucleic Acids Res., 1997, Jan
1, 25(1), 43-51.
[19] Karp, PD; Riley, M; Paley, SM; Pellegrini-Toole, A; Krummenacker, M. Eco Cyc:
encyclopedia of Escherichia coli genes and metabolism. Nucleic Acids Res., 1999, Jan
1, 27(1), 55-8.
[20] Karp, PD; Riley, M; Paley, SM; Pellegrini-Toole, A; Krummenacker, M. yc:
Encyclopedia of Escherichia coli genes and metabolism. Nucleic Acids Res., 1998, Jan
1, 26(1), 50-3.
[21] Karp, PD; Riley, M; Paley, SM; Pelligrini-Toole, A. EcoCyc: an encyclopedia of
Escherichia coli genes and metabolism. Nucleic Acids Res., 1996, Jan 1, 24(1), 32-9.
[22] Dogrusoz, U; Erson, EZ; Giral, E; Demir, E; Babur, O; Cetintas, A; Colak, R.
PATIKAweb: a Web interface for analyzing biological pathways through advanced
querying and visualization. Bioinformatics., 2006, Feb 1, 22(3), 374-5.
[23] Chen, M; Hofestädt, R. PathAligner: metabolic pathway retrieval and alignment. Appl
Bioinformatics., 2004, 3(4), 241-52.
[24] Caspi, R; Foerster, H; Fulcher, CA; Hopkinson, R; Ingraham, J; Kaipa, P;
Krummenacker, M; Paley, S; Pick, J; Rhee, SY; Tissier, C; Zhang, P; Karp, PD.
MetaCyc: a multiorganism database of metabolic pathways and enzymes.
Nucleic Acids
Res. 2006, Jan 1, 34(Database issue):D511-6.
[25] The MetaCyc Database. Karp, PD; Riley, M; Paley, SM; Pellegrini-Toole, A. Nucleic
Acids Res., 2002, Jan 1, 30(1), 59-61.

Viroj Wiwanikit 240
[26] Zhang, P; Foerster, H; Tissier, CP; Mueller, L; Paley, S; Karp, PD; Rhee, SY. MetaCyc
and AraCyc. Metabolic pathway databases for plant research. Plant Physiol., 2005,
May;138(1), 27-37.
[27] Kopka, J; Schauer, N; Krueger, S; Birkemeyer, C; Usadel, B; Bergmüller, E; Dörmann,
P; Weckwerth, W; Gibon, Y; Stitt, M; Willmitzer, L; Fernie, AR; Steinhauser, D.
GMD@CSB.DB: the Golm Metabolome Database. Bioinformatics., 2005, Apr 15,
21(8), 1635-8.
[28] Mazurek, S; Eigenbrodt, E. The tumor metabolome. Anticancer Res., 2003, Mar-Apr,
23(2A), 1149-54.
[29] Kolch, W; Mischak, H; Pitt, AR. The molecular make-up of a tumour: proteomics in
cancer research. Clin Sci (Lond)., 2005, May, 108(5), 369-83.
[30] Claudino, WM; Quattrone, A; Biganzoli, L; Pestrin, M; Bertini, I; Di Leo, A.
Metabolomics: available results, current research projects in breast cancer, and future
applications. J Clin Oncol., 2007, Jul 1, 25(19), 2840-6
[31] Rochfort, S. Metabolomics reviewed: a new "omics" platform technology for systems
biology and implications for natural products research. J Nat Prod., 2005, Dec, 68(12),
1813-20.
[32] Xu, M; Lin, DH; Liu, CX. Current status and prospect of metabonomics. Yao Xue Xue
Bao., 2005, Sep, 40(9), 769-74.
[33] Sivachenko, A; Kalinin, A; Yuryev, A. Pathway analysis for design of promiscuous
drugs and selective drug mixtures. Curr Drug Discov Technol., 2006, Dec, 3(4),
269-77.
[34] Goodacre, R. Metabolomics of a superorganism. J Nutr., 2007, Jan; 137(1 Suppl),
259S-266S.
[35] van der Greef, J; Hankemeier, T; McBurney, RN. Metabolomics-based systems biology
and personalized medicine: moving towards n = 1 clinical trials? Pharmacogenomics.,
2006, Oct;7(7), 1087-94.
[36] Schuster, D; Steindl, TM; Langer, T. Predicting drug metabolism induction in silico.
Curr Top Med Chem., 2006, 6(15), 1627-40.
[37] Fox, T; Kriegl, JM. Machine learning techniques for in silico modeling of drug
metabolism. Curr Top Med Chem., 2006, 6(15), 1579-91.
[38] Mankowski, DC; Ekins, S. Prediction of human drug metabolizing enzyme induction.
Curr Drug Metab., 2003, Oct; 4(5), 381-91.
[39] Lindon, JC; Holmes, E; Nicholson, JK. Metabonomics in pharmaceutical R&D. FEBS
J., 2007, Mar; 274(5), 1140-51.
[40] Fu, J; Swertz, MA; Keurentjes, JJ; Jansen, RC. MetaNetwork: a computational protocol
for the genetic study of metabolic networks. Nat Protoc., 2007, 2(3), 685-94.
[41]
Rom
ero, R; Espinoza, J; Gotsch, F; Kusanovic, JP; Friel, LA; Erez, O; Mazaki-Tovi, S;
Than, NG; Hassan, S; Tromp, G. The use of high-dimensional biology (genomics,
transcriptomics, proteomics, and metabolomics) to understand the preterm parturition
syndrome. BJOG., 2006, Dec;113, Suppl, 3, 118-35.
[42] Goesmann, A; Haubrock, M; Meyer, F; Kalinowski, J; Giegerich, R. PathFinder:
reconstruction and dynamic visualization of metabolic pathways. Bioinformatics., 2002,
Jan;18(1), 124-9.

Metabolomics 241
[43] Bourqui, R; Cottret, L; Lacroix, V; Auber, D; Mary, P; Sagot, MF; Jourdan, F.
Metabolic network visualization eliminating node redundance and preserving metabolic
pathways. BMC Syst Biol., 2007, Jul 3, 1, 29.
[44] Klamt, S; Stelling, J; Ginkel, M; Gilles, ED. FluxAnalyzer: exploring structure,
pathways, and flux distributions in metabolic networks on interactive flux maps.
Bioinformatics., 2003, Jan 22, 19(2):261-9.
[45] Papin, JA; Price, ND; Wiback, SJ; Fell, DA; Palsson, BO. Metabolic pathways in the
post-genome era. Trends Biochem Sci., 2003, May;28(5), 250-8.
[46] Becker, MY; Rojas, I. A graph layout algorithm for drawing metabolic pathways.
Bioinformatics., 2001, May; 17(5), 461-7.
[47] Karp, PD; Paley, SM. Representations of metabolic knowledge: pathways. Proc Int
Conf Intell Syst Mol Biol., 1994, 2, 203-11.
[48] Xiong, M; Zhao, J; Xiong, H. Network-based regulatory pathways analysis.
Bioinformatics., 2004, Sep 1, 20(13), 2056-66.
[49] Papin, JA; Price, ND; Edwards, JS; Palsson, B. BØ. The genome-scale metabolic
extreme pathway structure in Haemophilus influenzae shows significant network
redundancy. J Theor Biol., 2002, Mar 7, 215(1), 67-82.
[50] Price, ND; Papin, JA; Palsson, BØ. Determination of redundancy and systems
properties of the metabolic network of Helicobacter pylori using genome-scale extreme
pathway analysis. Genome Res., 2002, May, 12(5), 760-9.
[51] Wiback, SJ; Palsson, BO. Extreme pathway analysis of human red blood cell
metabolism. Biophys J., 2002, Aug, 83(2), 808-18.
[52] Papin, JA; Palsson, BO. The JAK-STAT signaling network in the human B-cell: an
extreme signaling pathway analysis. Biophys J., 2004, Jul, 87(1), 37-46.
[53] Pandey, R; Guru, RK; Mount, DW. Pathway Miner: extracting gene association
networks from molecular pathways for predicting the biological significance of gene
expression microarray data. Bioinformatics., 2004, Sep, 1, 20(13), 2156-8.
[54] Yi, M; Horton, JD; Cohen, JC; Hobbs, HH; Stephens, RM. WholePathwayScope: a
comprehensive pathway-based analysis tool for high-throughput data. BMC
Bioinformatics., 2006, Jan 19, 7, 30.
[55] Chung, HJ; Kim, M; Park, CH; Kim, J; Kim, JH. ArrayXPath: mapping and visualizing
microarray gene-expression data with integrated biological pathway resources using
Scalable Vector Graphics. Nucleic Acids Res., 2004, Jul 1, 32(Web Server
issue):W460-4.
[56] Weniger, M; Engelmann, JC; Schultz, J. Genome Expression Pathway Analysis Tool--
analysis and visualization of microarray gene expression data under genomic,
proteomic and metabolic context. BMC Bioinformatics., 2007, Jun 2, 8, 179.

In: Metabolomics: Metabolites, Metabonomics… ISBN: 978-1-61668-006-0
Editors: J.S. Knapp and W.L. Cabrera, pp. 243-251 © 2011 Nova Science Publishers, Inc.
Chapter 9
THE ROLE OF SPECIFIC ESTROGEN METABOLITES
IN THE INITIATION OF BREAST AND OTHER
H
UMAN CANCERS
Eleanor G. Rogan
*
and Ercole L. Cavalieri
Eppley Institute for Research in Cancer and Allied Diseases,
University of Nebraska Medical Center, 986805 Nebraska Medical Center,
Omaha, NE, USA
Keywords: Breast cancer, cancer initiation, catechol estrogen quinones, depurinating DNA
adducts, estrogens.
Introduction
Various types of evidence have implicated estrogens in the etiology of human breast
cancer [1-8]. They are generally thought to cause proliferation of breast epithelial cells
through estrogen receptor-mediated processes [4]. Rapidly proliferating cells are susceptible
to genetic errors during DNA replication, which, if uncorrected, can ultimately lead to
malignancy. While receptor-mediated processes may play an important role in the
development and growth of tumors, accumulating evidence suggests that specific oxidative
metabolites of estrogens, if formed, can be endogenous ultimate carcinogens that react with
DNA to cause the mutations leading to initiation of cancer [6-9]. Thus, estrogen metabolites,
specifically catechol estrogen-3,4-quinones, are hypothesized to be endogenous initiators of
breast, prostate and other human cancers.
Several lines of evidence, including metabolism and carcinogenicity studies by Liehr and
coworkers, led to the recognition that the 4-hydroxylated estrogens play a major role in the
genotoxic properties of estrogens [1-3]. We have hypothesized that the estrogens estrone (E
1
)
and estradiol (E
2
) initiate breast and other human cancers by reaction of their electrophilic
*
E-mail address: egrogan@unmc.edu. Tel: 402-559-4095, Fax: 402-559-8068. (Corresponding author)

Eleanor G. Rogan and Ercole L. Cavalieri 244
metabolites, catechol estrogen-3,4-quinones [E
1
(E
2
)-3,4-Q], with DNA to form depurinating
adducts [5-8]. These adducts generate apurinic sites leading to mutations that may initiate
breast, prostate and other human cancers [6-9].
The estrogens, E
1
and E
2
, are obtained via aromatization of 4-androstene-3,17-dione and
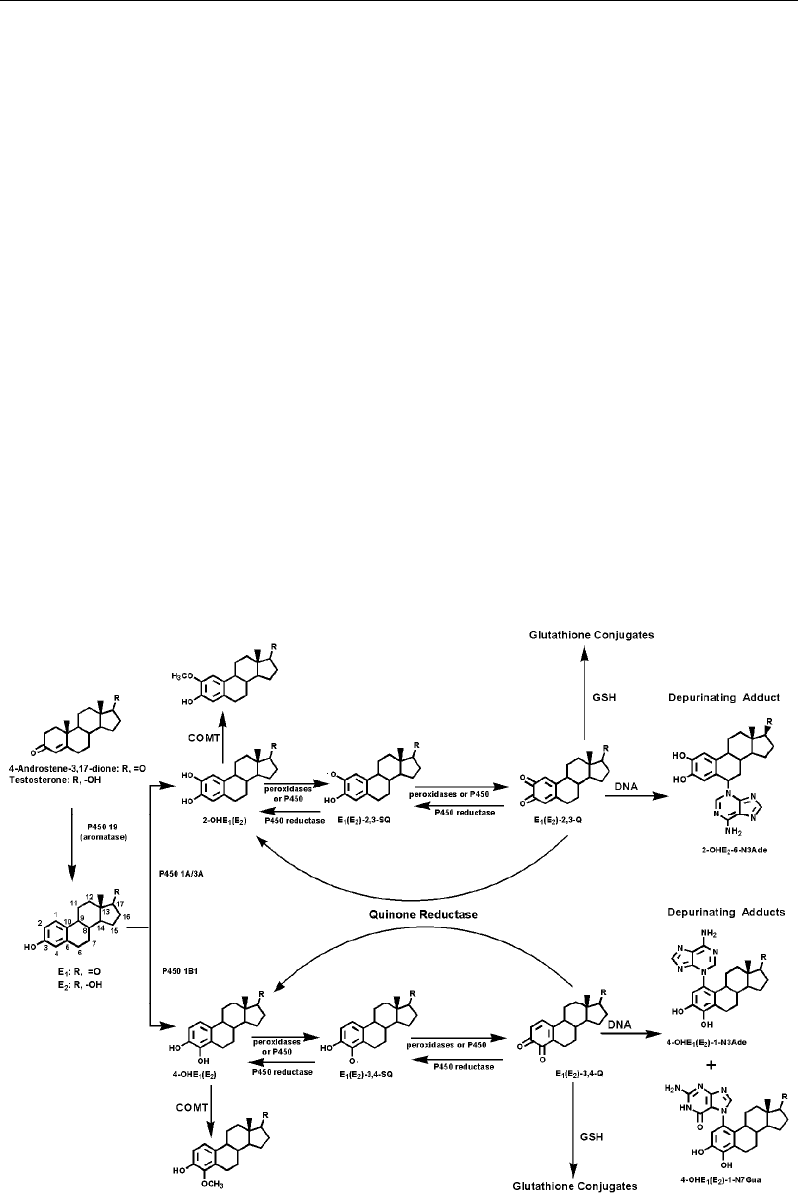
testosterone, respectively, catalyzed by cytochrome P450(CYP)19, aromatase (Figure 1). E
1
and E
2
, which are biochemically interconvertible by the enzyme 17β-estradiol dehydrogenase,
are metabolized to the 2-catechol estrogens, 2-OHE
1
(E
2
), and 4-OHE
1
(E
2
), predominantly
catalyzed by the activating enzymes CYP1A1 [10] and 1B1 [10-13], respectively, in
extrahepatic tissues. The estrogens are also metabolized, to a lesser extent, to 16α-hydroxy
derivatives (not shown). The catechol estrogens are further easily oxidized to the catechol
estrogen quinones, E
1
(E
2
)-2,3-Q and E
1
(E
2
)-3,4-Q (Figure 1) by metal ions, peroxidases and
cytochrome P450. In general, the catechol estrogens are inactivated by conjugating reactions,
such as glucuronidation and sulfation. A common pathway of inactivation in extrahepatic
tissues, however, occurs by O-methylation catalyzed by the ubiquitous catechol-O-
methyltransferase (COMT) [14]. If formation of E
1
or E
2
is excessive, due to overexpression
of aromatase and/or the presence of excess sulfatase that converts the stored E
1
sulfate to E
1
,
increased formation of catechol estrogens is expected. In particular, the presence and/or
induction of CYP1B1 and other 4-hydroxylases could render the 4-OHE
1
(E
2
), which are
usually minor metabolites, as the major metabolites [15-17]. Thus, conjugation of 4-
OHE
1
(E
2
) via methylation in extrahepatic tissues might become insufficient, and competitive
catalytic oxidation of 4-OHE
1
(E
2
) to E
1
(E
2
)-3,4-Q could occur. (Figure 1)
Protection at the quinone level can occur by conjugation of E
1
(E
2
)-Q with glutathione
(GSH, Figure 1). A second inactivating process for E
1
(E
2
)-Q is their reduction to catechol
estrogens by quinone reductase. If these two inactivating processes are not effective, E
1
(E
2
)-Q
may react with DNA to form stable and depurinating adducts [5-8,18-20]. Imbalances in
estrogen homeostasis [17,20], that is the equilibrium between activating and protective
enzymes with the scope of avoiding formation of catechol estrogen semiquinones and
quinones, could lead to initiation of cancer by estrogens.
Catechol Estrogen Quinones as Mutagens Initiating Breast,
Prostate and Other Human Cancers
Experiments on estrogen metabolism [17,19-22], formation of DNA adducts [5-8],
carcinogenicity [23-25], and mutagenicity [9,26,27] provide a basis for the hypothesis that
reaction of certain estrogen metabolites, predominantly catechol estrogen-3,4-quinones, with
DNA can generate the critical mutations initiating breast, prostate and other cancers [7-9,28].
Imbalance in Estrogen Homeostasis
Estrogen metabolism involves a balance between activating and deactivating (protective)
pathways. There are several factors that can unbalance estrogen homeostasis, that is, the
equilibrium between activating and deactivating pathways, to limit formation and/or reaction
of the endogenous carcinogenic E
1
(E
2
)-Q with DNA. The first imbalancing factor could be
excessive synthesis of E
2
by high expression of aromatase (CYP19) in target tissues [29-31]

The Role of Specific Estrogen Metabolites… 245
and/or the presence of sulfatase that excessively converts stored E
1
sulfate to E
1
[32,33]. A
striking result of in situ production of E
2
in human breast tissue is the similar levels of E
2
in
breast tissue in pre-menopausal and post-menopausal women, even though plasma levels are
10-50 fold higher in pre-menopausal than post-menopausal women [34]. Both aromatase and
sulfatase contribute to in situ estrogen production [32,33].
A second critical factor leading to imbalances in estrogen homeostasis might be high
levels of 4-OHE
1
(E
2
) due to high expression of CYP1B1, which metabolizes E
2
predominantly to form 4-OHE
2
[10,12,35]. This could result in relatively large amounts of 4-
OHE
1
(E
2
) that, in turn, can lead to more extensive oxidation to the carcinogenic E
1
(E
2
)-3,4-Q.
A third factor could be a lack or a low level of activity of the protective COMT enzyme. If
this enzyme is insufficient, 4-OHE
1
(E
2
) will not be effectively methylated in extrahepatic
tissues, but will be oxidized to the ultimate carcinogenic metabolites E
1
(E
2
)-3,4-Q. A fourth
factor could be a low level of GSH and/or low levels of quinone oxidoreductase and/or CYP
reductase, which could leave available higher levels of E
1
(E
2
)-3,4-Q that may react with
DNA.
Imbalances in estrogen homeostasis have been observed in laboratory animals and in
breast tissue from women with breast cancer:
The Kidney of Syrian Golden Hamsters
The hamster provides an excellent model for studying estrogen homeostasis because
implantation of E
1
or E
2
in male Syrian golden hamsters induces 100% of renal carcinomas,
but does not induce liver tumors [36]. Therefore, comparison of the profile of estrogen
metabolites, conjugates and DNA adducts in the two organs, after treatment of hamsters with
E
2
, should provide information on the relative imbalance in estrogen homeostasis in the two
tissues [20]. In the liver, more O-methylation of 2-OHE
1
(E
2
) was observed, whereas more
formation of E
1
(E
2
)-Q was detected in the kidney. These results suggest greater oxidation of
catechol estrogens to E
1
(E
2
)-Q and less protective methylation of 2-OHE
1
(E
2
) in the kidney.
When normal levels of GSH were depleted before hamsters were treated with E
2
, very low
levels of catechol estrogens and methoxy catechol estrogens were observed in the kidney
compared to the liver, suggesting little protective reduction of E
1
(E
2
)-Q to catechol estrogens
in the kidney. More importantly, the 4-OHE
1
(E
2
)-1-N7Gua depurinating adduct arising from
reaction of E
1
(E
2
)-3,4-Q with DNA was detected in the kidney, but not in the liver [20].
These results suggest that tumor initiation in the kidney occurs because of poor
methylation of catechol estrogens, rendering more likely competitive oxidation of catechol
estrogens to E
1
(E
2
)-Q, as well as poor quinone reductase activity to remove the E
1
(E
2
)-Q.
These two effects produce a large amount of E
1
(E
2
)-Q, which can react with the nucleophilic
groups of DNA.
The Mammary Gland of ERKO/Wnt-1 Mice
Mammary tumors develop in female estrogen receptor-α knock-out (ERKO)/Wnt-1 mice
despite their lack of functional estrogen receptor-α [37]. Extracts of hyperplastic mammary
tissue and mammary tumors from these mice were analyzed by HPLC interfaced with an
Eleanor G. Rogan and Ercole L. Cavalieri 246
electrochemical detector [21]. Picomole amounts of the 4-catechol estrogens were detected,
but their methoxy conjugates were not. Neither the 2-catechol estrogens nor 2-methoxy
catechol estrogens were detected. 4-OHE
1
(E
2
)-GSH conjugates or their hydrolytic products
(conjugates of cysteine and N-acetylcysteine) were detected in picomole amounts in both
tumors and hyperplastic mammary tissue, demonstrating the formation of E
1
(E
2
)-3,4-Q.
These preliminary findings indicate that estrogen homeostasis is unbalanced in the mammary
tissue, in that the normally minor 4-catechol estrogen metabolites were detected in the
mammary tissue, but not the normally predominant 2-catechol estrogens. Furthermore,
methylation of catechol estrogens was not detected, whereas formation of 4-OHE
1
(E
2
)-GSH
conjugates was. These results are consistent with the hypothesis that mammary tumor
development is primarily initiated by metabolism of estrogens to E
1
(E
2
)-3,4-Q, which may
react with DNA to induce oncogenic mutations.
The Prostate of Noble Rats
Estrogen metabolites and conjugates were analyzed in the ventral and anterior lobes of
the rat prostate, which are not susceptible to estrogen-induced carcinogenesis, and in the
susceptible dorsolateral and periurethral prostate of rats treated with 4-OHE
2
or E
2
-3,4-Q
[22]. The analyses revealed that the areas of the prostate susceptible to induction of
carcinomas have less protection by COMT, quinone reductase and GSH, thereby favoring
reaction of E
1
(E
2
)-3,4-Q with DNA.
Figure 1. Formation, metabolism, conjugation and DNA adducts of estrogens.
