Knapp J.S., Cabrera W.L. Metabolomics: Metabolites, Metabonomics, and Analytical Technologies
Подождите немного. Документ загружается.


Microbial Metagenomics: Concept, Methodology and Prospects… 197
Flint, H. J., Bayer, E. A., Rincon, M. T., Lamed, R. & White, B. A. (2008). Polysaccharide
utilization by gut bacteria: potential for new insights from genomic analysis. Nature Rev
Microbiol., 6, 121-131.
Friedrich, M. W. (2006). Stable-isotope probing of DNA-insights into the function of
uncultivated microorganisms from isotopically labeled metagenomes. Curr Opin
Biotechnol., 17, 59-66.
Galbraith, E. A., Antonopoulos, D. A. & White, B. A. (2004). Suppressive subtractive
hybridization as a tool for identifying genetic diversity in an environmental metagenome:
the rumen as a model. Environ Microbiol., 6, 928-937.
Gillespie, D. E., Brady, S. F., Bettermann, A. D., Cianciotto, N. P., Liles, M. R. & Rondon,
M. R. (2002). Isolation of antibiotics turbomycin A and B from a metagenome library of
soil microbial DNA. Appl Environ Microbiol., 68, 4301-4306.
Guo, H., Feng, Y., Mo, X., Duan, C., Tang, J. & Feng, J. (2008). Cloning and expression of
beta-glucosidase gene umcel3G from metagenome of buffalo rumen and characterization
of the translated product. Sheng Wu Gong Cheng Xue Bao. 24, 232-38. (Article in
Chinese, abstract in English)
Hall, N. (2007). Advanced sequencing technologies and their wider impact in microbiology.
J Exp Biol., 210, 1518-1525.
Handelsman, J., Rondon, M. R., Brady, S. F., Clardy, J. & Goodman, R. M. (1998).
Molecular biological access to the chemistry of unknown soil microbes: a new frontier
for natural products. Chem Biol., 5, R245-R249.
Henne, A., Schmitz, R. A., Bomeke, M., Gottschalk, G. & Daniel R. (2000). Screening of
environmental DNA libraries for the presence of genes conferring lipolytic activity on
Escherichia coli. Appl Environ Microbiol., 66, 3113-3116.
Huson, D. H., Auch, A. F., Qi J. & Schuster, S. C. (2007). MEGAN analysis of metagenomic
data. Genome Res., 17, 377-386.
Jones, B. V. & Marchesi, J. R. (2007). Transposon-aided capture (TRACA) of plasmids
resident in the human gut mobile metagenome. Nature Methods., 4, 55-61.
Kang, S., Denman, S. E., Morrison, M., Yu, Z. & McSweeny, C. S. (2009). An efficient RNA
extraction method for estimating gut microbial diversity by polymerase chain reaction.
Curr Microbiol., 58, 464-471.
Kobayashi, K. S., Chamaillard, M., Ogura, Y., Henegariu, O., Inohara, N. & Nunez, G.
(2005). Nod2-dependent regulation of innate and adaptive immunity in the intestinal
tract. Science., 307, 731-734.
Kowalchuk, G. A., Speksnijder, A. G. C., Zhang, K., Goodman, R. M. & van Veen, J. A.
(2007) Finding the needles in metagenome haystack. Microbial Ecol., 53, 475-485
Lammle, K., Zipper, H., Breuer, M., Hauer, B., Buta , C., Brunner, H. & Rupp, S. (2007).
Identification of novel enzymes with different hydrolytic activities by metagenome
expression cloning. J Biotechnol., 127, 575-592.
Lan, P. T., Sakamoto, M., Sakata, S. & Benno, Y. (2006).
Bacteroides barnesiae sp
. nov.,
Bacteroides salanitronis sp. nov., and Bacteroides gallinarum sp. nov., isolated from
chicken caecum. Int J Syst Evol Microbiol., 56, 2853-9.
Lima, J. R., Ribin, A. O., Russell, J. B. & Mantovani, H. C. (2009). Bovicin HC5 inhibits
wasteful amino acid degradation by mixed ruminal bacteria in vitro. FEMS Microbiol
Lett., 292, 78-84.

B. Singh, T.K. Bhat, O.P. Sharma et al. 198
Lissens, G., Verstraete, W., Albrecht, T., Brunner, G., Creuly, C., Seon, J., Dussup, G. &
Lasseur C. (2004). Advanced anaerobic bioconversion of lignocellulosic waste for
bioregenerative life support following thermal water treatment and biodegradation by
Fibrobacter succinogenes. Biodegradation., 15, 173-83.
Liu, K., Wang, J., Bu, D., Zhao, S., McSweeny, C. S., Yu, P. & Li, D. (2009). Isolation and
biochemical characterization of two lipases from a metagenomic library of China
Holstein cow rumen. Biochem. Biophys Res Commun., 385, 605-611.
Liu, L., Tang, J. & Feng, J. (2009). Bacterial diversity in Guangxi buffalo rumen. Wei Sheng
Wu Xue Bao., 49, 251-256. (article in Chinese, abstract in English)
Lopez-Cortes, N., Reyes-Duarte, D., Beloqui, A., Polaina, J., Ghazi, I. & Golyshina, O. V.
(2007). Catalytic role of conserved HQGE motif in the CE6 carbohydrate esterase family.
FEBS Lett., 581, 4657-4662.
Lopez-Garcia, P. & Moreira D. (2008). Tracking microbial diversity through molecular and
genomic ecology. Review. Res Micrbiol., 159, 67-73.
Lu, J., Santo Domingo, J. & Shanks, O. C. (2007). Identification of chicken-specific fecal
microbial sequences using a metagenomic approach. Water Res., 41, 3561-3574.
Margulies, M., Egholm, M., Altman, W. E., Attiya, S., Bader, J. S. & Bemben, L. A., (2005).
Genome sequencing in microfabricated high-density picolitre reactions. Nature., 437,
376-380.
Martinez, A., Kolvek, S. J., Yip, C. L., Hopke, J., Brown, K. A., MacNeil, I. A. & Osburne,
M. S. (2004). Genetically modified bacterial strains and novel bacterial artificial
chromosome shuttle vectors for constructing environmental libraries and detecting
heterologous natural products in multiple expression hosts. Appl Environ Microbiol., 70,
2452-2463.
Morrison, M., Pope, P. B., Denman, S. E. & McSweeney, C. S. (2009). Plant biomass
degradation by gut microbiomes: more of the same or something new? Curr Opin
Biotechnol., 20, 358-363.
Myrick, K. V. & Gelbart, W. M. (2002). Universal fast walking for direct and versatile
determination of flanking sequence. Gene., 284, 125-131.
Nelson, K. E, Zinder, S. H., Hance, I., Burr. P., Odongo, D., Wasawo, D., Odenyo, A. &
Bishop R. (2003). Phylogenetic analysis of the microbial populations in the wild
herbivore gastrointestinal tract: insights into an unexplored niche. Environ Microbiol., 5,
1212-1220.
Nicholson, M. J., Evans, P. N. & Joblin, K. N. (2007). Analysis of methanogens diversity in
the rumen using temporal temperature gradient gel electrophoresis: identification of
uncultured methanogens. Microb Ecol., 54, 141-50.
Noguchi, H., Park, J. & Takagi, T. (2006). MetaGene: prokaryotic gene finding from
environmental genome shotgun sequences. Nucleic Acid Res., 34, 5623-5630.
Nordgard, L., Traavik, T. & Nielsen, K. M. (2005). Nucleic acid isolation from ecological
samples—vertebrate gut flora. Methods Enzymol
., 39
5, 38-48.
Ochman, H., Ayala, F. J. & Hartl, D. L. (1993). Use of polymerase chain reaction to amplify
segments outside boundaries of known sequences. Methods Enzymol., 218, 309-321.
Pace, N. R., Stahal, D. A., Lane, D. J. & Olsen, G. J. (1986). The analysis of natural microbial
populations by ribosomal RNA sequences. Adv Microb Ecol., 9, 1-55.

Microbial Metagenomics: Concept, Methodology and Prospects… 199
Palackal, N., Lyon, C. S., Zaidi, S., Luginbuhl, P., Dupree, P. & Goubet, F. (2007). A
multifunctional hybrid glycosyl hydrolase discovered in an uncultured microbial
consortium from ruminant gut. Appl Micobiol Biotechnol., 74, 113-124.
Qi, M., Nelson, K. E., Daugherty, S. C., Nelson, W. C., Hance, I. R , Morrison, M. &
Forsberg, C. W. (2005). Novel molecular features of the fibrolytic intestinal bacterium
Fibrobacter intestinalis not shared with Fibrobacter succinogens as determined by
suppressive subtractive hybridization. J Bacteriol., 187, 3739-3751.
Radajewski, S., Ineson, P., Parekh, N. R. & Murell, J. C. (2000). Stable-isotope probing as a
tool in microbial ecology. Nature., 403, 646-649.
Schloss, P. D. & Handelsman, J. (2005). Introducing DOTUR, a computer program for
defining operational taxonomic units and estimating species richness. Appl Environ
Microbiol.,1501-1506.
Schmeisser, C., Steele, H. & Streit, W. R. (2007) Metagenomics, biotechnology with non-
culturable microbes. Appl Microbiol Biotechnol., 75, 955-962
Schmidt, T. M., DeLong, E. F. & Pace, N. R. (1991). Analysis of marine plankton community
by 16S rRNA gene cloning and sequencing. J Bacteriol, 173, 4371-4378.
Selinger, L. B., Forsberg, C. W. & Cheng. K. J. (1996). The rumen: a unique source of
enzymes for enhancing livestock production. Anaerobe., 2, 263-284.
Shanks, O. C., Santo Domingo, J. W., Lamendella, R., Kelty, C. A. & Graham, J. E. (2006).
Competitive metagenomic DNA hybridization identifies host-specific microbial genetic
markers in cow fecal samples. Appl Environ Microbiol., 72, 4054-4060.
Sharma, R., John, S. J., Damgaard, M. & McAllister, T. A. (2003). Extraction of PCR quality
plant and microbial DNA from total rumen contents. Biotechniques., 34, 92-94, 96-97.
Shedova, E. N., Lunina, N. A., Berezina, O. V., Zverlov, V. V., Schwarz, V. &
Velikodvorskaia, G. A. (2009). Expression of the genes celA and XylA isolated from a
fragment of metagenomic DNA in Escherichia coli. Mol Gen Mikrobiol Virol., 2,
28-32 (Article in Russian, abstract in English)
Short, J. M. & Mathur, E. J. (1999). Production and use of normalized DNA libraries. US
Patent No. 6001574.
Singh, B., Bhat, T. K. & Singh, B. (2001). Exploiting gastrointestinal microbes for livestock
and industrial development. Asian-Aust J Anim Sci., 14, 567-586.
Singh, B., Gautam S. K. & Mukesh, M. (2009). Rumen ecosystem to boost productivity- a
metagenomic overview. Indian Dairyman. 61 (9), 50-55.
Singh, B., Gautam, S. K., Verma, V., Kumar, M. & Singh, B. (2008a). Metagenomics in
animals gastrointestinal tract- potential biotechnological prospects. Anaerobe
., 14,
13
8-144.
Singh, B., Bhat, T. K., Sharma, O. P. & Kurade, N. P. (2008b). Metagenomics in animal
gastrointestinal tract- a microbiological and biotechnological perspective. Indian J
Microbiol., 48, 216-227.
Toyoda, A., Iio, W., Mitsumori, M. & Minato, H. (2009). Isolation and identification of
cellulose binding proteins from sheep rumen contents. Appl Environ Microbiol., 75,
1667-1673.
Tuohy, K. M., Gougoulias, C., Shen, Q., Walton, G., Fava, F. & Ramani, P. (2009). Studying
the human gut microbiota in the trans-omics era- focus on metagenomics and
metabolomics. Curr Pharmaceut Des., 15, 1415-1427.

B. Singh, T.K. Bhat, O.P. Sharma et al. 200
Whitman, W. B., Coleman, C. D. & Wiebe, W. J. (1998). Prokaryotes: the unseen majority.
Proc Natl Acad Sci., USA. 95, 6578-6583.
Xu, J. (2006). Microbial ecology in the age of genomics and metagenomics: concepts, tools,
and recent advances. Mol Ecol., 15, 1713-1731.
Venter, J. C., Remington, K., Heidelberg, J. F., Halpern, A. L., Rusch, D. & Eisen, J. A.
(2004). Environmental genome shotgun sequencing of the Sargasso Sea. Science., 304,
66-74.

In: Metabolomics: Metabolites, Metabonomics… ISBN: 978-1-61668-006-0
Editors: J.S. Knapp and W.L. Cabrera, pp. 201-213 © 2011 Nova Science Publishers, Inc.
Chapter 6
NUTRIGENOMICS, METABOLOMICS AND
METABONOMICS: EMERGING FACES OF
MOLECULAR GENOMICS AND NUTRITION
B. Singh
1,*
, M. Mukesh
2
, M. Sodhi
2
, S.K. Gautam
3
,
M. Kumar
4
and P.S. Yadav
5
1
Regional Station, IVRI, Palampur-176 061, India
2
Animal Genetics Division, NBAGR Karnal-132 001, India
3
Department of Biotechnology, Kurukshetra University, Kurukshetra-136 119, India
4
Dairy Microbiology Division, NDRI Karnal-132 001, India.
5
Department of Buffalo Physiology and Reproduction, CIRB, Hisar-125 001, India
Abstract
Nutrition exhibits the most important life-long environmental impact on health. Nutrients, gut
microbial metabolites and other bioactive food constituents interact with body at system,
organ, cellular and molecular levels, and affect the expression of genome at several levels, and
subsequently, the overall production of metabolites. Direct measurement of cellular
metabolites is essential for the study of biological processes, and may allow causes of disease,
toxicological progression, and novel disease-biomarkers to be identified. Advances in
analytical techniques and the algorithms for management of the data has allowed a precise and
global analysis of biological substances such as DNA (genomics), RNA (transcriptomics),
proteins (proteomics) and smaller molecules (metabolomics). Holistic “omics” approaches are
indispensable to cover the complex nutrient-cell and gut microbial-host interactions. This
chapter presents an overview of nutrigenomics and metabolomics tools with reference to their
perspective in livestock health and production.
*
E-mail address: bsbpalampur@yahoo.co.in; Fax; +91 1894 233063; Phone +91 1894 230526. (Corresponding
author)

B. Singh, M. Mukesh, M. Sodhi et al. 202
Introduction
Imbalanced nutrition in terms of deficits of critical nutrients or excessive intake of certain
anti-nutritional compounds may cause a number of diseases and metabolic disorders. The
classic or traditional research related to human or animal nutrition, deals mainly with studying
the interactions of host-dietary components directly or using biomarker approaches (van
Ommen, 2004) or aims to study either deficiency or excess of nutrients in relation to health
ailments. Recent advances in genomics have propelled the development of new technologies
that have provided the researchers with methods to quickly analyze genes and their products
en masse.
The advent of modern analytical tools has led to realization that not only are certain
nutrients essential, but also that specific quantities of each were necessary for optimal health,
thereby leading to such notions as dietary recommendations, nutritional epidemiology, and
realization that food can directly contribute to disease onset (Mutch et al., 2005). Further, the
availability of human genome sequence information and large set of single nucleotide
polymorphisms (SNPs) in candidate genes and their correlation with metabolic imbalances
have pioneered a new era in modern genomics, and added new parameters to the molecular
nutrition panel. There is wide support to the theory that genetic variation in selected SNPs,
haplotypes, and copy number variants can have remarkable effect not only in an individual’s
response to dietary components, but also for their optimal utilization (Ferguson, 2006).
Nowadays it is recognized that understanding the effect of diet on health requires the
study of mechanisms of nutrients and other bioactive food constituents at cellar as well as
molecular levels. This is supported by the increasingly growing number of studies in humans,
animals and cell culture studies revealing that nutrients and other bioactive dietary ingredients
have crucial role in regulating gene-expression in diverse ways (Mead, 2007). Also, there are
enough scientific evidences illustrating that micronutrients and certain plant secondary
metabolites (PSMs) can interact with the genome, modify gene-expression and regulation,
alter protein and metabolite composition within the cells and tissues, and even participate in
genome repair (Fig. 1) (Singh et al., 2003; Marambaud et al., 2005; Zheng and Chen, 2006;
Fenech 2008). This concept puts nutritional genomic area at the food/ gene interphase
creating opportunities for the industry to develop commercially viable supplements or
nutraceuticals that can modify expression of the genes of interest (Subbiah, 2008).
Nutrigenomics Concept
Nutritional genomics is a recent offshoot of genetic revolution that was experienced over
the past decade. The term nutritional genomics, or nutrigenomics, appears to have its origin in
the context of plant biology, wherein it refers to work in interface of plant biochemistry
(specifically, secondary metabolism), genomics, and human nutrition (DellaPenna, 1999).
Muller and Kersten (2003) define nutrigenomics as “the applications of high throughput
genomics in nutrition research, and studying the genome wide influences of nutrition”. From
a nutrigenomics perspective, nutrients and dietary signals are detected by cellular sensor or
receptor systems that in turn, influence gene and protein expression, and subsequently the
metabolite production by the cell (Muller and Kersten, 2003; Mariman, 2006; Fenech, 2008).
Hence, pattern of gene expression, protein synthesis and modification and production of

Nutrigenomics, Metabolomics and Metabonomics 203
metabolites in response to the nutrients or nutritional regimes can be viewed as “dietary
signatures”. The nutrigenomics is primarily aimed at examining these “dietary signatures” in
a target cell, tissue or even entire organism, and thereby elucidating the impact of nutrition on
host homeostasis. According to Kaput et al. (2005) nutrigenomics is the study of how
constituents of the diet interact with genes, and their products, to alter phenotype and,
conversely, how genes and their products metabolize these constituents into nutrients,
antinutrients, and bioactive compounds. This new era of nutrition recognizes the complex
relation and interaction between the health of individual, its genome, and the life-long dietary
exposure, and has led to realization that nutrition is essentially a gene-environment interaction
science.
The nutritional genomics encompasses two broad areas namely, nutrigenomics, which
deals with interaction between dietary components and the genome as well as the resulting
changes in proteins and other metabolites, and the nutrigenetics, which aims at understanding
gene-based differences in response to dietary components and to develop novel nutraceuticals
that are most compatible with the health status of individuals based on their genetic makeup
(Kaput, 2008). Nutrigenetics and nutrigenomics have emerged as nascent areas that are
evolving quickly and riding on the wave of “personalized medicine” that is providing
opportunities in the discovery and development of nutraceutical compounds (Panagiotou and
Nielsen, 2009).
Nutrient-Gene Interaction
It is evident that diet is a complex mixture of substances and gut microbial metabolites
(Fig. 2) that supply both energy and building blocks to develop and sustain the cells or
organisms. The nutrients exhibit a variety of biological activities, ranging from protection
against diseases and acting as signaling molecules (Muller and Kersten, 2003). At molecular
levels, the nutrients relay signals and communicate a specific cell about the dietary
components. Therefore, the cellular processes including every step of genetic information
from gene-expression to the protein synthesis and degradation might be affected by the diet
and environmental factors.
In some ways, the nutrigenomics can be compared with pharmacogenomics
(personalizing drug therapy based on individual SNPs) which has made tremendous headway
in recent years as a tool to reduce individual drug toxicity and designing personalized
medicine (Giacomini et al., 2007; Relling and Hoffman, 2007). However, the important
difference is that pharmacogenomics is concerned with the effects of drugs that are pure
compounds administered in precise and small doses, whereas nutrigenomics encompasses
complexity and variability of nutrients (Muller and Kersten, 2003).
Advanced technologies for genomic analysis have led to identification of genes or
markers associated with genes of interest. Microarray technology for high-throughput
screening of changes in gene-expression has enormously advanced the nutrigenomic studies.
At the moment, the gene-expression microarrays have become a de facto golden standard to
evaluate changes in genome wide gene-expression under different conditions.
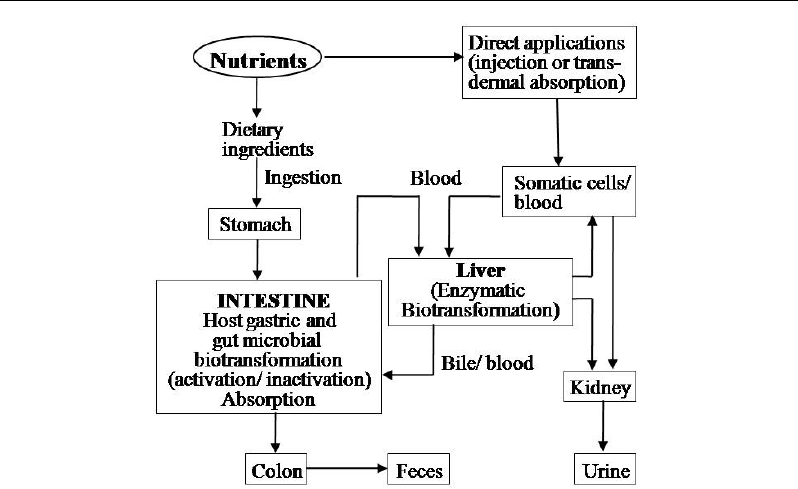
B. Singh, M. Mukesh, M. Sodhi et al. 204
Figure 1. A diagrammatic illustration of applications and effects of nutrients and bioactive PSMs. In
addition to oral intake, certain compounds can reach blood circulation via absorption or inhalation. Gut
and liver are the major sites for metabolism of these components. The gut microbes have ability to
detoxify as well as to generate bioactive compounds from dietary components.
Nutrigenomics in Livestock Perspective
Application of modern molecular biological techniques has potential to revolutionize the
animal nutrition. Presently, many of these technologies are used in research on searching and
identifying candidate genes and disease diagnosis. Veterinary nutritionists have begun
applying animal genomics to the field of nutrition. Integrating the information encoded in the
genome to applied nutrition and ultimately augmenting livestock production is the goal.
Further, when genomics is combined with metabolomics (discussed below), the whole animal
assessment may be achieved and may provide the opportunity for corrective interaction via
specific nutrients like, retinoic acid, fatty acids, vitamins and other compounds. The ability to
thoroughly understand the role of nutrients will be significantly enhanced by using
nutrigenomics and metabolomic approaches. In addition, nutrigenomics tools are being used
to link expression data with gene-function in the bovines, such as in vitro models of bovine
adipogenesis and bioinformatics tools to map gene network (Lehnert et al., 2006).
In the changing scenario of ruminant nutrition, the nutritional genomics has several
applications (Zdunczyk and Pareek, 2008). However, at present the nutrigenomics studies in
livestock sciences are rare, but they are likely to become more important, as we develop an
understanding of the relationship between nutrition, genetics, fertility and tissue growth
(Dawson, 2006). Molecular nutrition will serve as a new tool for nutritional research in
mitigating the problems related to animal health and production (Table 1).

Nutrigenomics, Metabolomics and Metabonomics 205
Table 1. A summary of challenging areas in augmenting livestock
production using nutrigenomics.
1. Gaining insight into the mechanisms or pathways by which dietary components affect animal
growth, tissue structure and overall performance by “up- or down-regulation” of target genes
2. Identifying the novel strategies for controlling the key metabolic processes by managing
gene expression rather than looking at animal performance based on traditional nutritional
responses
3. Evaluating the diet-mediated differential gene expression, identifying diet-induced
alterations in gene expression profile (Reverter et al., 2003)
4. Using proteomics tools (e. g. 2-D electrophoresis) to reveal information concerning
composition of egg and poultry meat proteins, and effect of dietary methionine on breast-meat
accretion
5. Evaluating the effects of use of transgenic crops on animal nutrition and health
6. Identification of the genes involved in carcinogenesis and anti-carcinogenesis process in
response to dietary toxins or PSMs
7. Elucidating the role of gut microbiota in host immunity and nutrient utilization
8. Analysis of regulation of myogenesis and its regulatory pathways in meat producing animals
What is Metabolomics?
With the advent of functional genomics during the last two decades and recent advances
in sequencing technologies, a substantial progress has been made in biological investigations.
With the evolution of second generation sequencing it is possible to sequence the entire
genome of an organism, and precisely analyze the huge data produced. The “omics”
applications now enable us to understand various aspects of cellular physiology and/ or
biology of an organism as affected or influenced by environmental stimuli or genetic
perturbations.
The current rise in diet-related diseases continues to be one of the most significant health
problems. The technologies are developed that have enormous impact on disease
investigation by studying the metabolic profile of a cell or an organism. The
1
H-nuclear
magnetic resonance (NMR) and mass spectrometry (MS)-based technologies to generate
profiles of metabolites in biofluids permit profiling of the entire metabolome, which provides
a sensitive intermediate phenotype linking the genotype, gut microbial composition and
personal health status (Oresic, 2009). The metabolomics combines strategies to identify and
quantify the cellular metabolites using sophisticated analytical technologies along with
applications of statistical and multivariate methods for deriving information and data
interpretation (Roessner and Bowne, 2009). The assessment of both essential nutrient status
and the more comprehensive systemic metabolic response to dietary, lifestyle and
environmental influences are necessary for the evaluation of physiological status in
individuals that can identify multiple targets of interventions needed to address metabolic
diseases (Zivkovik and German, 2009). As the cellular metabolites are considered to act as
“spoken language, or broadcasting signals” from the genetic architecture and the
environment, the metabolomics is considered to provide a direct “functional readout of the
physiological state” of an organisms (Gieger et al., 2008). Salient applications of
metabolomics have been summarized in Table 2.
B. Singh, M. Mukesh, M. Sodhi et al. 206
Techniques and Data Analysis in Metabolomics
Unlike transctriptomics and proteomics, which intend to determine a single or unique
class of end products (mRNA and proteins, respectively), the metabolomics has to deal with
components of very diverse physiochemical properties. Moreover, concentration of these
metabolites in biofluids varies from millimoler level (or higher) to picomoler, making it to
exceed the linear range of conventially employed analytical techniques (Garcia-Canas et al.,
2010). Hence, metabolomics relies on additional technologies to isolate and characterize
biological metabolites which can combine automation and miniaturization as for nucleic
acids. This includes techniques for tissue sampling, extraction of specific classes of
molecules, their storage, sample preparation and analyses. A combination of methods (Table
3) based on gas chromatography/mass spectrometry (GC/MS) and liquid chromatography/
mass spectrometry (LC/MS) has attained a high technical robustness, which makes them
more comparable to microarrays used for nucleic acids or protein studies (Hocquette, 2005).
The process of metabolomics comprises of four broad conceptual approaches, namely, i)
target analysis, ii) metabolic profiling, iii) metabolomics, and iv) metabolic fingerprinting.
Therefore, the specific application depends on the subject and requirement. Innovative
experimental designs combined with novel computational tools (Table 4) for handling
metabolomics data offer new opportunities for early disease detection as well as
characterization of dietary and therapeutic interventions in the context of human physiology
(Oresic, 2009). MS-based small molecular metabolite analysis is rapidly becoming a method
of choice and enables multiple biological paths discovering and validating functional
assignments (Baran et al., 2009). The combination of MS-based metabolic profiling with
genome-scale models of metabolisms and other –‘omics’ approaches provide opportunities to
expand our understanding of microbial metabolic-networks, stress-responses, and to identify
genes associated with specific enzymatic and regulatory activities (Baran et al., 2009). NMR
however, still remains most important instrument in metabolomic studies, though initially it
had certain limitations which were later overcome by incorporating software called as Eclipse
Version 3.0.
Table 2. Applications of metabolomic tools.
1. Predicting the physiological status of a cell or organism, detection of drug residues through
global profiling of metabolites in blood or body fluids
2. Developing actionable metabolic diagnostics and more comprehensive systemic metabolic
response to dietary, lifestyle and environmental influences (Zivkovik and German, 2009)
3. Using ‘metabolome fingerprints’ for predicting embryonic development through dynamic
changes in its metabolome (Hayashi et al., 2009)
4. Rapidly assessing the disturbances in metabolic profiles following administration of drugs,
and identifying the biomarkers of toxicity to assess the health risk of specific toxins
Comprehensive multidimensional techniques, such as GCxGC or LCxLC, are also a
revolutionary improvement in separation techniques that will be applicable in nutritional
metabolomics in near future. They provide enhanced resolution and a huge increase in
selectivity and sensitivity in comparison with conventional separation techniques (Garcia-
Canas et al., 2010). A number of commercially available software can be used for quantitative
