Elder K. Human preimplantation embryo selection
Подождите немного. Документ загружается.

HPE_Chapter22.qxp 7/13/2007 4:53 PM Page 286

OVARIAN STIMULATION AND HIGH-ORDER
MULTIPLE PREGNANCY
The use of ovarian stimulation has provided a means
of obtaining multiple oocytes per assisted reproduc-
tive technology (ART) treatment cycle. In most
cases the majority of oocytes retrieved will be suc-
cessfully fertilized, and as a result, several embryos
are typically available for transfer. Because the prob-
ability of an individual embryo forming a viable
pregnancy is relatively low (approximately 15%) it
has long been standard practice to transfer more
than one embryo per cycle. This approach has been
highly successful at increasing take-home-baby rates,
thus reducing the likelihood that a patient will need
to face the emotional, physical, and financial burden
of multiple treatment cycles.
Unfortunately, the transfer of multiple embryos
has also led to an explosion in the incidence of high-
order multiple gestations. Since the introduction of
in vitro fertilization (IVF) more than 20 years ago
the incidence of such pregnancies has increased
400% for women in their thirties and 1000% for
women in their forties (National Center for Health
Statistics). Although some patients initially welcome
the prospect of a ‘ready made family’, the medical
reality is that pregnancies of this type carry a sig-
nificantly elevated risk of serious complications
for both the mother and children. For the mother
there is increased risk of problems such as pre-
eclampsia, gestational diabetes, and vaginal and
uterine hemorrhaging. Fetal complications include
a high-risk of prematurity and very low birth weight,
as well as significantly elevated risks of miscarriage,
infant mortality, and cerebral palsy. High-order
multiple pregnancies also have financial implications
for both the parents and the health care provider.
1–3
For example, a triplet pregnancy will typically cost a
health care provider in the United States approxi-
mately $300 000.
EMBRYO VIABILITY ASSESSMENT –
MORPHOLOGY
The problem of multiple gestations in IVF stems
from the fact that current methods for predicting
embryo viability and implantation potential are
relatively poor. Multiple embryos are transferred
because this is the only way to maximize the proba-
bility that at least one viable embryo has been
selected. It is inevitable that in some cases all of
the embryos transferred will have the potential to
implant.
There is an increasing desire within the field of
reproductive medicine to overcome the problem of
multiple gestations. Additionally, in some countries,
legislation to limit the number of embryos trans-
ferred has come into force.
4
Although it may be
generally accepted that high-order multiple preg-
nancies are not a desirable outcome of IVF treatment,
many laboratories remain concerned about the
impact of a reduction in the number of embryos
transferred on pregnancy rates. This is particularly
true in countries where IVF success rates are pub-
lished and influence the number of patients seeking
treatment at a particular center.
If pregnancy rates are to be maintained while
the number of embryos transferred is reduced,
ultimately culminating in the routine use of single
embryo transfer at some point in the future, it will
be necessary to dramatically improve our ability to
assess embryo viability. One of the most important
challenges encountered each day in IVF laboratories
23. Future genetic and other technologies for
assessing embryos
Dagan Wells
HPE_Chapter23.qxp 7/13/2007 5:31 PM Page 287

HUMAN PREIMPLANTATION EMBRYO SELECTION
is the determination of which embryos to select for
transfer to the uterus. Which embryos have the
greatest probability of providing the patient with a
successful pregnancy?
In the majority of laboratories, microscopic
assessment of developmental stage and morphology
of the embryo is the most important guide to future
viability. Many different systems for scoring zygotes
and embryos based on their morphology have been
described, and in most cases these schemes have
been demonstrated to assist in distinguishing viable
embryos from non-viable to some extent.
5–11
However, morphological analysis remains a rela-
tively inefficient means of assessing embryos. All
physicians and embryologists working in IVF can
recount stories of patients who received two
embryos of extremely poor morphology and became
pregnant with twins. Similarly, we are all familiar
with cases in which embryo morphology was
perfect and yet no pregnancy ensued.
EMBRYO VIABILITY ASSESSMENT –
PREIMPLANTATION GENETIC
SCREENING
One reason why morphologically normal, appar-
ently healthy, IVF embryos may fail to form a preg-
nancy is the presence of aneuploidy – an incorrect
number of chromosomes. During the early 1990s,
research using fluorescence in situ hybridization
(FISH) revealed that chromosome anomalies in
human preimplantation embryos are startlingly
common. Analysis of between three and six of the
24 types of chromosome (22 pairs of autosomes
plus the X and Y), revealed that more than 50%
of cleavage stage embryos contained at least one
abnormal cell.
12–15
Later investigations using
comparative genomic hybridization (CGH), a more
comprehensive cytogenetic method that permits the
entire chromosomal set to be screened in a single
cell, suggested that between two-thirds and three-
quarters of human embryos contain abnormal cells
on day 3 postfertilization.
16,17
Most of the anom-
alies detected have been shown to be lethal,
18
and
their high prevalence in embryos produced during
IVF treatment is thought to explain many cases of
embryonic arrest, implantation failure, and early
spontaneous abortion. A critical point of note is
that most chromosomally abnormal embryos are
morphologically indistinguishable from their
euploid counterparts.
A major breakthrough in embryo assessment
came with the development of preimplantation
genetic diagnosis (PGD) for the detection of chro-
mosome abnormalities, known as PGD-AS (PGD-
aneuploidy screening) or PGS (preimplantation
genetic screening). The most widely applied PGS
strategy involves the biopsy of a single blastomere
on day 3 postfertilization. The blastomere can be
subjected to a wide range of genetic analyses. In the
case of chromosomal screening, the cell is spread
onto a microscope slide, fixed, and then analyzed
using FISH. At present, the most experienced labo-
ratories assess up to ten chromosomes per cell. This
is achieved by employing two sequential rounds
of hybridization using chromosome-specific probes
labeled with distinct fluorescent dyes.
The use of PGS as a guide to selecting embryos
to be considered for transfer, supplementing (not
replacing) morphological analysis, has been shown
to significantly improve several IVF outcomes. Not
surprisingly, the screening of chromosomes 13, 18,
21, X, and Y has led to a reduction in the number of
pregnancies affected by aneuploid syndromes (i.e.
Patau, Edward, Down, Turner and Klinefelter).
19
This is particularly evident in IVF patients where
the female partner is over 35 years of age, an age
after which the risk of producing a chromosomally
abnormal oocyte begins to increase rapidly. It is also
clear that certain groups of IVF patient display a
decline in the incidence of spontaneous abortion
following PGS. This is not surprising, given that
chromosome imbalance is known to be the major
cause of early miscarriage in natural cycles. Of first
trimester pregnancy losses, 60–70% display chromo-
some anomalies.
20–22
Of most significance to the issue of multiple
pregnancies is the impact of PGS on embryo implan-
tation rates. Although some controversy remains,
there is mounting evidence that PGS provides spe-
cific groups of IVF patient with a significant increase
HPE_Chapter23.qxp 7/13/2007 5:31 PM Page 288

FUTURE GENETIC AND OTHER TECHNOLOGIES FOR ASSESSING EMBRYOS
in implantation rate per embryo transferred.
23,24
Apparent contradictions in the literature regarding
the impact of PGS on implantation rate seem to be
a consequence of methodological differences between
studies.
25
Data suggest that increased implantation
is only conveyed by PGS strategies employing biopsy
of a single blastomere (rather than two) and analy-
sis of at least seven chromosomes. If PGS truly has
the ability to identify embryos with high implanta-
tion potential, then it is likely that this type of
screening can help to reduce the incidence of multiple
pregnancy, while maintaining an acceptable preg-
nancy rate. A recent study performed by Munné
et al has provided preliminary data in support of
this suggestion.
The patients taking part in the study were divided
into two groups: (1) embryos assessed using PGS
and transfer limited to one to two embryos per cycle;
and (2) a control group in which embryos were
transferred without any genetic screening and no
restriction was placed on the number of embryos
transferred to the patient. Data from over 100 cycles
were documented in each group, with patients
matched for a variety of factors including maternal
age. As with earlier studies, embryos transferred
after PGS displayed an approximate doubling of
implantation rate compared with the control group.
However, despite the increased implantation rate
for the PGS embryos, pregnancy rates for the two
groups were essentially identical (30% for the PGS
group, 33% for the control). This is because fewer
embryos were transferred per cycle in the PGS group
(average of 1.5 per cycle for PGS versus 3.6 for con-
trol). Importantly, no high-order multiple pregnan-
cies were seen in the PGS group, whereas two triplet
pregnancies and more twins were present in the con-
trol group (Munne et al, personal communication).
PREIMPLANTATION GENETIC SCREENING –
FUTURE DIRECTIONS
Although PGS has provided significant benefits in
terms of embryo screening and IVF outcome, exist-
ing protocols assess less than half of the chromoso-
mal complement. Examination of the chromosomes
not usually tested during PGS in a research context,
has shown that any chromosome can display aneu-
ploidy during preimplantation development. Thus,
it is inevitable that some aneuploid embryos are
incorrectly classified ‘normal’ using current PGS
methods, and may be inadvertently transferred.
Such embryos are believed to have little potential for
forming a viable pregnancy and most likely fail to
implant or spontaneously abort at an early stage of
gestation.
It seems logical that an expanded chromosomal
screen, permitting assessment of every chromo-
some, would improve the ability of PGS to identify
viable embryos. However, a comprehensive chro-
mosomal screen cannot be achieved using FISH due
to the restricted number of spectrally distinct fluo-
rochromes available for probe labeling. Although
this problem can be partially overcome by perform-
ing sequential FISH experiments on the same cell,
accuracy declines with each additional hybridiza-
tion and consequently most PGS laboratories limit
the number of rounds of FISH to two.
Although methods such as G-banding, multi-
plex-FISH (M-FISH), and spectral karyotyping
(SKY) provide information on every chromosome,
they are dependent on the presence of cells in
metaphase. Unfortunately the vast majority of blas-
tomeres sampled from day 3 embryos are found to
be in interphase. During this phase of the cell cycle
chromosomes are contained within the nucleus and
cannot be distinguished from one another. On the
rare occasions when a blastomere in metaphase is
biopsied, artifacts of the method used for fixation
and spreading, most notably loss of chromosomes,
are often observed.
Comparative genomic hybridization (CGH) is a
method related to FISH that permits analysis of the
entire chromosome complement. Aneuploidy detec-
tion is achieved via a competitive hybridization of
differentially labeled DNA samples to normal meta-
phase chromosomes
26
(Figure 23.1). In the case of
PGS, DNA from the sample (blastomere) is labeled
with a green fluorochrome, while chromosomally
normal reference DNA (usually from 46 XY lym-
phocytes) is labeled with a red fluorochrome. The
two labeled samples are simultaneously hybridized
HPE_Chapter23.qxp 7/13/2007 5:31 PM Page 289
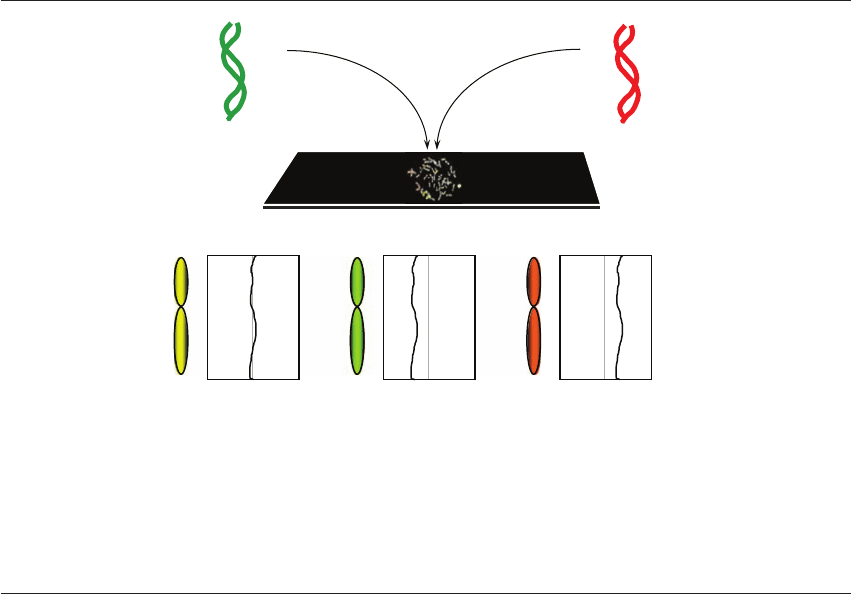
HUMAN PREIMPLANTATION EMBRYO SELECTION
to normal metaphase chromosomes on a microscope
slide. Red and green DNA fragments compete for
hybridization to their complementary sequences
on the chromosomes, causing each chromosome to
adopt a coloration related to its copy number in
each of the samples. For example, if the green (blas-
tomere) DNA was from an embryo with an extra
copy of chromosome 16, there would be relatively
more green copies of chromosome 16 DNA frag-
ments than red. This will cause green DNA to out-
compete red DNA for hybridization to chromosome
16. The overall effect is that chromosome 16 appears
more green than the other chromosomes. Chromo-
some imbalances of this type are most clearly
indicated by analyzing the ratio of green : red fluo-
rescence along the length of each chromosome.
CGH represents an excellent method for the simul-
taneous assessment of all chromosomes, and can
even reveal errors involving fragments of chromo-
somes, provided they are at least 5 Mb in size.
Unfortunately, CGH requires ~1 g of DNA
whereas a single blastomere contains only 5–10 pg.
For this reason it is necessary to perform whole
genome amplification (WGA) prior to CGH analy-
sis of individual cells. The method of choice for this
purpose is known as degenerate oligonucleotide
primed (DOP) PCR.
27,28
DOP-PCR utilizes a semi-
degenerate oligonucleotide primer that has the abil-
ity to anneal at many sites throughout the genome.
With the addition of a thermostable DNA poly-
merase, DNA synthesis is initiated at each site of
primer annealing. Thermal cycling using the princi-
ples of the polymerase chain reaction (denaturation,
annealing, synthesis) permits a dramatic amplifica-
tion of DNA from the original sample and provides
enough material for subsequent CGH analysis.
Normal
1 : 1
Trisomy
3 : 2
Monosomy
1 : 2
Normal DNA
46 XY
Test DNA
Ratio red : green
Figure 23.1 Comparative genomic hybridization (CGH) involves the simultaneous hybridization of differentially labeled DNA
samples – a chromosomally normal DNA (labeled with a red fluorochrome) and sample DNA (labeled with a green fluorochrome).
The relative number of red and green DNA fragments hybridizing to a given chromosome is related to the number of copies of the
chromosome in question in the sample compared with the control. An excess of chromosomal material in the sample causes a green
coloration, while a deficiency leads to a red coloration. Identical copy numbers of sample (green) and control (red) chromosomes
results in equal hybridization of red and green fragments and a yellow coloration. In most cases a computer is used to assist in the
calculation of red : green fluorescence intensity along the length of each chromosome.
HPE_Chapter23.qxp 7/13/2007 5:32 PM Page 290
FUTURE GENETIC AND OTHER TECHNOLOGIES FOR ASSESSING EMBRYOS
Studies applying CGH to the analysis of cleavage
stage embryos of good morphology confirmed that
aneuploidy could affect any chromosome during
human preimplantation development. Abnormalities
included chromosome breakage and varieties of
aneuploidy that are not seen in prenatal samples or
in material from spontaneous abortions (e.g. imbal-
ance affecting the larger chromosomes of groups A
and B).
16,17
These unfamiliar forms of abnormality
are presumed to be lethal during early stages of
development.
Analysis of the published CGH data reveals that
25–30% of embryos carry chromosome abnormali-
ties that would not be detectable using the nine-
chromosome PGS examination employed by most
PGD laboratories.
29,30
The fact that current PGS
protocols are not 100% successful in preventing the
transfer of aneuploid embryos argues for the devel-
opment of methods that permit a comprehensive
analysis of blastomere chromosomes. However, it
may be possible to significantly improve embryo
selection simply by adjusting the combination of
chromosomes screened.
In most cases the chromosomes selected for
screening are included because they are frequently
found to be abnormal in prenatal samples and mis-
carriages. However, these chromosomes are not
necessarily the most relevant in terms of implanta-
tion failure.
31
Unfortunately, there are insufficient
published CGH data to allow the incidence of
aneuploidy to be precisely calculated for individual
chromosomes, and consequently it is difficult to
determine precisely which chromosomes should be
prioritized for screening. However, pooling all exist-
ing CGH data can provide a rough indication of the
most common aneuploidies at the cleavage stage. In
most cases this analysis confirms the importance
of the chromosomes currently assessed by PGS.
However, it also suggests that screening might be
improved if some chromosomes were substituted by
others with higher preimplantation aneuploidy rates.
Chromosomes that have not been extensively
studied in human embryos, but appear to show above
average aneuploidy rates include chromosomes 2, 4,
7, 9, 12, and 20. This observation is based on a small
number of samples and requires confirmation using
CGH or FISH on a much larger series of embryos
before adjustment of clinical PGS strategies. In
addition to the CGH data, some FISH studies have
also indicated that a revision of the chromosomes
assessed by PGS may be necessary.
32
From the pooled CGH data, one can predict that
the proportion of aneuploid embryos detected could
be increased from 70–75% to ⬎95% if the number
of chromosomes screened by FISH was expanded
from nine to 15. However, although it may be possi-
ble to achieve high detection rates for aneuploid
embryos without screening the entire chromosome
complement, it is inevitable that maximum accu-
racy will only be achieved if all the chromosomes
are evaluated. To date CGH has been the most
promising method for this purpose. However, as
with FISH there are limitations to this technology.
The principal difficulties in applying CGH clini-
cally are the complexity of the technique and the
length of time required (approximately 5 days),
which is incompatible with the restricted timeframe
available for PGS. One strategy for overcoming the
problem of timeframe involves cryopreservation of
embryos after biopsy, with embryo transfer occur-
ring in a subsequent cycle.
29,30,33
The main drawback
of this approach is that freezing and thawing can
lead to a reduction in embryo implantation poten-
tial, a problem that is exacerbated by embryo biopsy.
An alternative strategy for the clinical applica-
tion of CGH is based upon assessment of the first
polar body (PB).
34
The first PB is available for analy-
sis 3 days earlier than biopsied blastomeres, provid-
ing sufficient time to perform CGH without the
need for cryopreservation. However, confirmatory
FISH analysis of blastomeres is advisable since chro-
matid anomalies detected in meiosis I have only a
50% chance of leading to an aneuploid embryo.
There is also the potential for misdiagnosis due to a
meiosis II error or a chromosome abnormality of
paternal origin.
Whatever the CGH strategy employed for PGS,
the complexity of the technique remains a major
problem. The protocol is labor intensive and neces-
sitates expertise in both molecular genetic and cyto-
genetic methods that are not generally available to
fertility clinics. A less complex approach will be
HPE_Chapter23.qxp 7/13/2007 5:32 PM Page 291

HUMAN PREIMPLANTATION EMBRYO SELECTION
necessary if CGH is to be widely applied. At the
present time, the best hope for a simplified method-
ology is microarray CGH. As with conventional CGH,
this method involves the competitive hybridization
of differentially labeled test and reference DNA
samples. However, in this case the labeled DNAs are
hybridized to DNA probes fixed to a microscope
slide rather than metaphase chromosomes. Each
probe is specific to a different chromosomal region
and occupies a discrete spot on the slide. Chromo-
somal loss or gain is revealed by the color adopted
by each spot after hybridization (ratio of red : green
fluorescence). Importantly, the evaluation of red :
green fluorescence is easily automated, circumvent-
ing the need for an experienced cytogeneticist.
Microarray CGH has been successfully applied
for the detection of aneuploidies in single cells after
whole genome amplification using DOP-PCR or an
alternative method known as multiple displacement
amplification (MDA).
35–37
Using this approach, com-
prehensive chromosome analysis has been achieved
in less than 48 hours (i.e. within the timeframe nec-
essary for PGS) and consequently the future appli-
cation of microarray CGH for PGS appears to be
extremely encouraging.
Although comprehensive chromosomal analy-
sis would seem to be advantageous for PGS, some
physicians have expressed concern that a full-
chromosome screening will increase the number of
embryos excluded due to aneuploidy, leading to
more IVF cycles in which there are no embryos
eligible for transfer. Even though current protocols
only assess nine or ten chromosomes, a small but
significant number of PGS cycles end without
embryo transfer due to all embryos receiving an
unfavorable diagnosis.
Fortunately, the small amount of data currently
available indicate that the proportion of embryos
diagnosed as normal may actually increase follow-
ing CGH analysis. In a small study comparing FISH
and CGH for the purposes of PGS, 40% of embryos
analyzed by CGH were found to be chromosomally
normal, while the proportion of embryos diag-
nosed normal by FISH was 33%.
30
The most likely
explanation for this counterintuitive observation is
inaccuracy in the FISH methodology caused by the
presence of micronuclei. Micronuclei, which are a
relatively common phenomenon in human blas-
tomeres, contain chromosomal material and are
prone to loss during fixation and spreading prior to
FISH. This may explain why an excess of mono-
somies (relative to trisomies) have been detected
following FISH analysis of human embryos. CGH
does not involve the spreading of the sample and
consequently avoids this source of error.
RELATIONSHIP BETWEEN GENE EXPRESSION
AND EMBRYO VIABILITY
The application of PGS has been shown to benefit
several groups of IVF patients and proves that chro-
mosome abnormality is an important factor affect-
ing embryo implantation potential. However, when
considering implantation failure it is also clear that
chromosome anomaly is only part of the story.
Many IVF-PGS cycles fail to provide a pregnancy,
despite the transfer of embryos classified as chro-
mosomally and morphologically normal. Further-
more, there are significant subgroups of IVF patients
that show little or no improvement in embryo
implantation following PGS (e.g. patients under 35
years of age). Although some of these failures may
be attributed to uterine or endocrine problems, it is
likely that many (perhaps most) are the conse-
quence of nonchromosomal embryological issues.
A host of vital, yet largely invisible, processes
occur during the first few days of life. These include
the transition away from a reliance on maternal
protein and mRNA that accompanies activation of
the embryo genome, and the first types of cellular
differentiation. Given the complex and fundamen-
tal nature of events the embryo must successfully
undertake at this time, it is perhaps unsurprising
that developmental arrest during the preimplanta-
tion phase is common. One of the great challenges
now facing scientists involved in translational IVF
research is to characterize the cellular pathways
perturbed in embryos destined for implantation
failure. An improved understanding in this area
may lead to new viability markers, revealing embryos
that are under pressure and heading for arrest, and
HPE_Chapter23.qxp 7/13/2007 5:32 PM Page 292
FUTURE GENETIC AND OTHER TECHNOLOGIES FOR ASSESSING EMBRYOS
assist in the optimization of ovarian stimulation
and embryo culture methods.
An extremely useful approach for investigating
the processes occurring in a biological sample (e.g. a
cell, a tissue, an embryo, etc.) is to examine gene
expression. The activity of individual genes is con-
stantly changing, fluctuating in response to the
changing needs of the cell. Variation in environ-
mental conditions will induce changes in gene
expression, as will factors such as altered metabolic
requirements, the presence of aneuploidy or DNA
damage, progress through the cell cycle, and processes
such as differentiation and apoptosis.
Quantification of the number of mRNA tran-
scripts derived from a given gene provides an indi-
cation of how actively expressed it is. This in turn
provides information on the activity of the cellular
pathways that require the product of that gene.
Embryonic gene expression at different preimplan-
tation stages has been assessed using real-time poly-
merase chain reaction (PCR). This method involves
extraction of RNA from the sample, reverse tran-
scription in order to produce cDNA and PCR
amplification of a cDNA fragment from the gene(s)
of interest. The accumulation of amplified DNA is
measured in each sample tube in real-time (i.e. once
per PCR cycle) by monitoring the fluorescence
emitted by sequence specific probes or generic DNA
stains. All samples are assessed relative to standards
containing known numbers of template molecules.
Measurements taken while amplification is still
proceeding in an exponential fashion, allow the
number of templates in each sample at the begin-
ning of the PCR to be calculated by reference to the
standards.
38
A recent gene expression study focused on genes
with roles in cell cycle regulation, DNA repair, sig-
naling pathways, and apoptosis, functions of great
importance during early development.
39
Although
transcripts from every gene assessed were detectable
at every stage, from mature oocyte to hatched blas-
tocyst, the number of transcripts at different stages
varied considerably. The quantity of mRNA tran-
scripts was generally high in oocytes, but decreased
dramatically after fertilization, in agreement with
earlier observations.
40–42
The data obtained clearly
indicate that transcripts from many genes reach extre-
mely low levels in 2- and 3-cell embryos, in some
cases scarcely above the threshold of detection.
39
Depletion of maternal mRNA after fertilization
appears to be a normal occurrence and yet may
have great significance for the embryo. Most cellular
pathways are controlled to some degree by the tran-
scriptional activation or repression of specific genes.
In other words, gene expression allows the subtle
control of cellular mechanisms in response to changes
in the intracellular and extracellular environment. It
is likely that prior to the activation of the embryonic
genome many important cellular pathways exist in a
rigid form, controlled by a reservoir of proteins
inherited from the oocyte. Consequently, the embryo
may have limited ability to respond to environ-
mental challenges during this period. This may have
important implications for the practice of in vitro
fertilization and embryo culture methods.
No embryonic gene expression was detected
until the 4-cell stage, consistent with previous data
demonstrating the onset of gene activation.
43,44
In
most cases the increase in transcript number upon
genome activation was modest, however, in a subset
of embryos dramatic increases in expression were
seen for the BRCA1 gene (several hundred fold).
39
BRCA1 is a multifunctional protein with roles in
cell cycle regulation and DNA repair. The increased
expression of BRCA1 in cleavage stage embryos may
indicate the presence of DNA damage. Such genetic
damage could be derived from the sperm (as indi-
cated by studies employing the sperm chromatin
structure assay)
45
or the oocyte. It is possible that
DNA repair is relatively inefficient until the embry-
onic genome is activated and fresh gene expression
permits the stimulation of DNA repair pathways.
From the 10-cell to morula stages the activity of
most genes examined stabilized or even underwent
a small decline. Thereafter, expression levels rose
proportionately with increasing cell number, and as
a result, most genes displayed greater transcript
numbers in blastocysts than at any other stage. The
high gene activity in blastocysts is not surprising, as
most of the genes assessed produce proteins that
interact with DNA or chromosomes (i.e. the quan-
tity of protein required is likely to be closely related
HPE_Chapter23.qxp 7/13/2007 5:32 PM Page 293
HUMAN PREIMPLANTATION EMBRYO SELECTION
to the number of nuclei). Gene expression was
clearly linked to developmental stage, with embryos
at similar stages having very similar patterns of gene
expression.
The gene expression data were also analyzed
using a hierarchical cluster algorithm, a mathemati-
cal method for identifying samples with similar
characteristics. The results are displayed graphically
as a dendogram, embryos with similar gene expres-
sion being placed on closely associated branches of a
tree diagram. Cluster analysis grouped embryos of
specific stages together, confirming statistically that
patterns of gene expression and developmental
stage are closely associated. Only 20% of embryos
failed to cluster with counterparts of similar stage.
Interestingly, more than half of these embryos dis-
played morphological abnormalities, suggesting
that such aberrations may cause (or be caused by)
perturbations of gene expression
46
(Figure 23.2).
As well as separating embryos by developmental
stage, cluster analysis was able to identify subgroups
of identically staged embryos that had distinct
expression profiles. Analysis of the morphological
notes for the embryos within these subgroups
revealed that one group was associated with the
presence of granular cytoplasm, condensed orga-
nelles, and multiple nuclei – features that are nega-
tively correlated with embryo implantation. A
second group of 4–10-cell stage embryos was found
to contain a preponderance of embryos with uneven
cleavage divisions.
46
Interestingly, not all the groups of embryos
displaying a distinct pattern of gene activity were
associated with a characteristic morphology. This
demonstrates that differences in expression are
frequently related to non-morphological factors,
which could include factors such as the cell cycle,
cellular stress, chromosomal status (e.g. aneuploid
0
2
4
6
8
10
ABCDEFGH I J ABCDEFGH I J
ABCDEFGH I J
ABCDEFGH I J
0
2
4
6
8
10
0
2
4
6
8
10
0
2
4
6
8
10
1
34
2
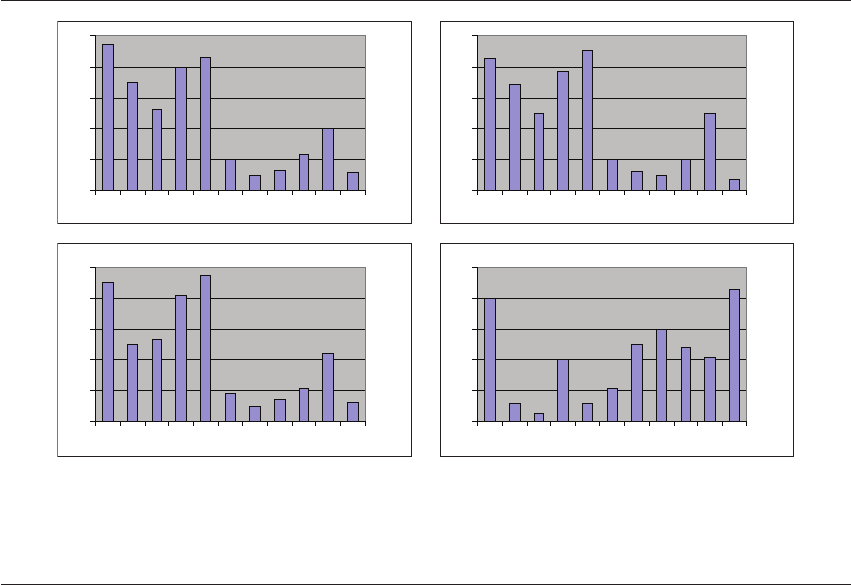
Figure 23.2 Quantification of gene expression reveals that most embryos show an expression profile characteristic of their develop-
mental stage. However, abnormalities can lead to disturbances in the normal pattern of gene expression. This figure shows the results
of gene expression analysis for ten genes (A–J) in four blastocysts (1–4). Blastocysts 1, 2, and 3 displayed very similar patterns of gene
expression and appeared to be morphologically identical (good morphology). However, analysis of blastocyst 4 revealed altered
expression of several genes. This blastocyst had a poorly defined inner cell mass and minor growth retardation.
HPE_Chapter23.qxp 7/13/2007 5:32 PM Page 294

FUTURE GENETIC AND OTHER TECHNOLOGIES FOR ASSESSING EMBRYOS
versus euploid), or microenvironment of individual
embryos. The potential of gene expression analysis
to reveal ‘hidden’ information concerning processes
occurring within the embryo may be of clinical
value, as some of these processes are likely to have a
bearing on viability.
Further studies will continue to improve our
understanding of the cellular processes occurring
during the preimplantation phase. A recent advance
has been the development of in vitro transcription
techniques that permit the mRNA content of single
oocytes to be amplified to levels sufficient for
microarray analysis.
47
Microarrays allow the simul-
taneous analysis of ⬎29 000 genes in a single exper-
iment and have allowed us to catalogue over 9000
genes that are expressed in human oocytes (Wells
et al, unpublished data). It is anticipated that the
application of such methods to the study of human
preimplantation development will greatly accelerate
the progress of discovery in this area. Our analysis of
human oocytes has already revealed several key cel-
lular pathways that are differentially expressed when
a chromosome abnormality is present. Not only
does this open up new possibilities for the detection
of aneuploidy, but also it reveals the cellular mecha-
nisms underlying this problem. The knowledge
gained from such studies may lead to the develop-
ment of preventative interventions, such as improved
ovarian stimulation regimens or optimized culture
media.
PROTEOMIC ANALYSIS OF HUMAN EMBRYOS
Investigation of proteins may prove to be even more
useful for assessing embryo viability than either
cytogenetic or gene expression analysis. Although
the gene expression data obtained provide an inter-
esting insight into the genetic activity of the early
embryo, a change in the number of mRNA transcripts
derived from a given gene does not necessarily indi-
cate altered utilization of the pathway in which it
functions. Most genes experience some degree of
regulation at the post-translational level, through
protein modification, degradation, or sequestra-
tion. Consequently, there may be occasions when a
change in the concentration of active protein is not
mirrored by an alteration in gene activity. Further-
more, individual genes usually produce more than
one type of protein, accomplished by utilizing
mechanisms such as alternative splicing and post-
translational modification. It is not possible to detect
post-translational modifications by looking at gene
expression.
Advances in mass spectrometry have led to the
development of techniques with sufficient sensitiv-
ity to permit the analysis of individual embryos. A
recent study by Katz-Jaffe and colleagues utilized
surface-enhanced laser desorption and ionization
time-of-flight mass spectrometry (SELDI-TOF MS)
to produce proteomic profiles for individual human
blastocysts.
48
The method involved the lysis of whole
blastocysts and binding their proteins to chips.
Bound proteins were released from the surface of
the chip and ionized by laser activation and the mass :
charge ratio of the liberated ions determined by
time-of-flight mass spectrometry. The mass : charge
ratios of the many different protein fragments pro-
duced during this process produce a unique pro-
teomic fingerprint for each sample assessed.
Analysis of individual blastocysts revealed differ-
ences in proteomic profiles related to morphology
and developmental stage. For example, degenerat-
ing embryos displayed numerous alterations in
protein expression when compared with developing
blastocysts.
48
Proteomic analysis also identified dif-
ferences between embryos that were not correlated
with either developmental stage or morphology. In
some cases such changes may be related to altered
utilization of specific cellular pathways that influ-
ence or reflect embryo viability. Analysis of the pro-
teins involved may allow morphologically identical
embryos to be distinguished in terms of their
implantation potential.
ASSESSMENT OF OOCYTES AND EMBRYOS
WITHOUT BLASTOMERE BIOPSY
Analyses of genes and proteins have demonstrated
that dynamic fluctuations in expression occur
throughout preimplantation development and that
HPE_Chapter23.qxp 7/13/2007 5:32 PM Page 295
