Elder K. Human preimplantation embryo selection
Подождите немного. Документ загружается.

HUMAN PREIMPLANTATION EMBRYO SELECTION
pregnancy groups in both day 3 and day 5 assays.
Significantly, the 50% group had values that fell
between the 0 and 100% groups (Figure 20.6). Using
the data obtained in the pilot studies, Bayesian prob-
ability statistics were used to establish a predictive
index of embryo viability based on a typical
Gaussian distribution (Figure 20.7).
A total of 432 samples from four different centers
were analyzed (n ⫽ 211 embryo culture media;
n ⫽ 133 seminal plasma; n ⫽ 88 follicular fluid)
with assays that routinely achieved high sensitivity
and specificity of ⬎80%. The results of these
studies demonstrate a clear relationship between the
reproductive potential of human embryos and their
modification of the media in which they have been
cultured.These subtle modifications could be detected
through BSM profiling of OS biomarkers that appear
in the media during embryo culture. Specifically, there
were detectable differences in the metabolomic pro-
files found in culture media obtained from embryos
that resulted in pregnancy compared with those that
did not.
Likewise, significant differences were seen in the
biomarker profiles of normal, healthy semen donors
compared with those with various forms of male
infertility, including idiopathic male factor infertility,
0
500
1000
1500 2000 2500 3000 3500
–1
–0.5
0
0.5
A
B
1
–1
–0.5
0
0.5 1
1.5
2
0
0.5
1
1.5
2
Implantation index
Wavelength (nm)
Number of samples Relative signal
NH
C=C, SH groups
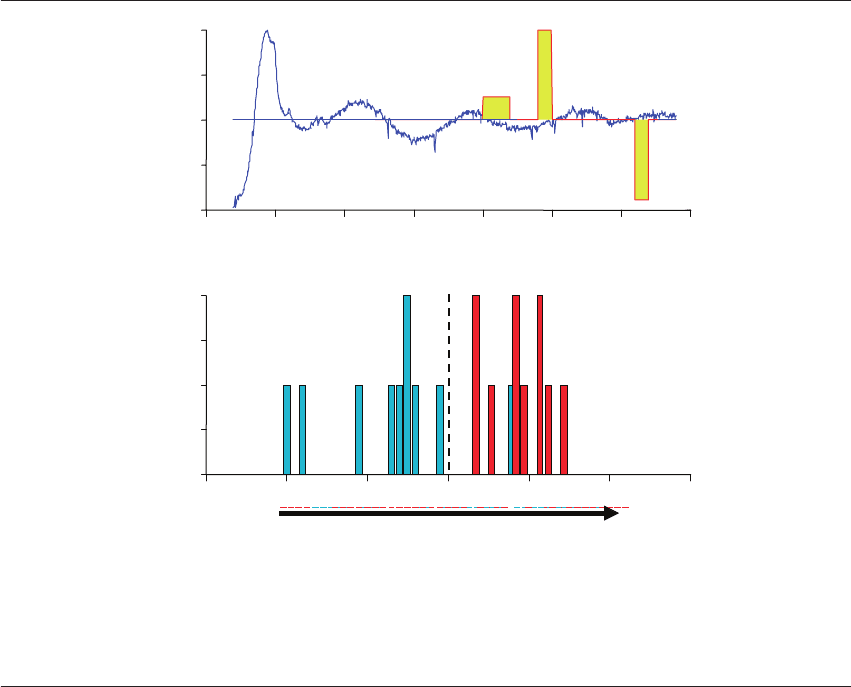
Figure 20.5 Raman spectral signature of three biomarkers in culture media. (A) A spectrum produced by Raman spectroscopy from
a single embryo media specimen. Three distinct biomarker regions are identified in yellow depicting C苷C, -SH, and -NH bonding at
specific wavelengths. The photon energy detected in these regions was captured and subjected to bioinformatic analysis. (B) The
implantation index score is calculated by the bioinformatic algorithms for each specimen. The distribution of metabolomic data is
shown for each of 20 media specimens: non-viable embryo scores are in blue vs viable embryo scores in red. The sensitivity and
specificity for this group of specimens were 100% and 90%, respectively.
HPE_Chapter20.qxp 7/17/2007 3:38 PM Page 256
METABOLOMIC PROFILING OF OXIDATIVE STRESS BIOMARKERS
varicocele, and vasectomy reversal (Figure 20.8).
Profiles characteristic of oocyte quality were also
detectable in follicular fluid and correlated to preg-
nancy outcomes of their respective transferred
embryos (Figure 20.9).
CONCLUSION
This technology offers significant potential as a
tool to allow rapid non-invasive assessment of
embryonic reproductive potential prior to transfer.
–0.6 –0.4 –0.2 0 0.2 0.4 0.6
0
50
100
Implantation index
% Implantation
Day 3
C
–0.6 –0.4 –0.2 0 0.2 0.4 0.6 0.8 1.0 1.2
3
5
Implantation index
Day
A
B
0.5 0.6 0.7 0.8 0.9 1.0 1.1 1.2 1.3
0
50
100
Implantation index
% Implantation
Day 5
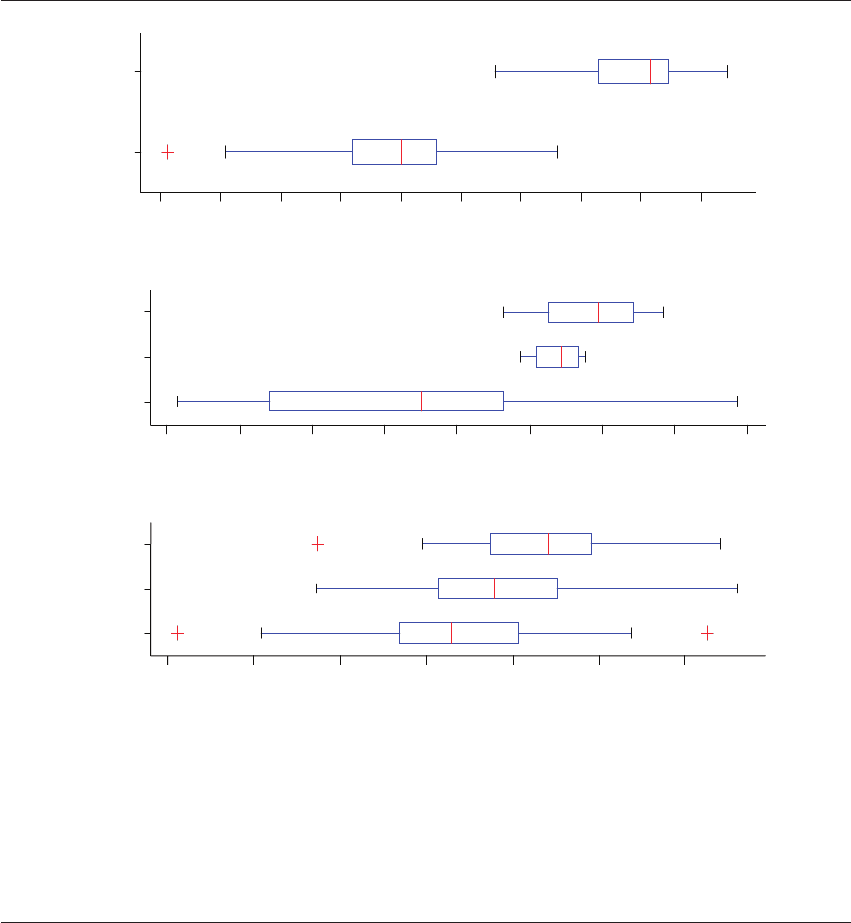
Figure 20.6 Metabolomic results by NIR of 0, 50%, and 100% implantation rate embryos. Media from embryos that produced 0,
50%, and 100% pregnancies were analyzed in a blinded prospective study; n ⫽ 65 with an equal number of specimens per group.
(A) A statistically significant difference was observed in the metabolomic profiles of day 3 embryos that produced a pregnancy
compared with day 5 embryos. Day 5 embryos showed a greater implantations index score than day 3 embryos. t Statistic ⫺7.86;
two-tailed p ⬍0.0001. (B) A comparison of implantation index scores of three groups of day 5 embryos: 0%, 50%, and 100% pregnancy
outcomes. The 0% and 100% groups are statistically different from each other (t Statistic 3.85; p ⫽ 0.0085).The implantations index
score of the 50% group falls between the 0 and 100% group scores. (C) A comparison of implantation index scores of three groups
of day 3 embryos: 0%, 50%, and 100% pregnancy outcomes. The 0% and 100% groups are statistically different from each other
(t Statistic 3.23; p ⫽ 0.003), while the implantation index score of the 50% group fell between the 0 and 100% group scores.
HPE_Chapter20.qxp 7/17/2007 3:38 PM Page 257
HUMAN PREIMPLANTATION EMBRYO SELECTION
Enhanced selection parameters based on metabolomic
profiling might allow a reduction in transfer order,
eventually to single embryo transfer, with the desir-
able effect of lowering multiple gestation rates while
maintaining or raising pregnancy rates.
The application of metabolomic profiling to
assess gamete viability appears to be equally prom-
ising. For example, this technique could be adapted
as a rapid diagnostic screening test for male factor
infertility. The ability to determine oocyte viability
prior to fertilization is expected to increase the num-
ber of viable embryos available for selection, transfer,
and cryopreservation. Conceivably, metabolomics
could work in concert with, or in place of, PGD as
a form of functional genomics testing. In certain
European countries where limitations on embryo
transfer are tightly regulated, viable oocyte selection
could provide a relevant means of sustaining IVF
success rates through improved oocyte fertilization
and cryopreservation techniques. Viability testing
of gametes and embryos used in cryopreservation
cycles is also a logical application of metabolomics
in IVF.
0.1
0.1
0.1
0.1
0.2
0.2
0.2
0.2
0.3
0.3
0.3
0.3
0.4
0.4
0.4
0.4
0.5
0.5
0.5
0.6
0.6
0.6
0.7
0.7
0.7
0.8
0.8
0.8
0.9
0.9
0.9
Integrated mean centered CH
Integrated mean centered SH
–2500 –2000 –1500 –1000 –500 0
–100
0
100
200
300
400
+Pregnancy +Non-pregnancy
Probability
of
viability
contours
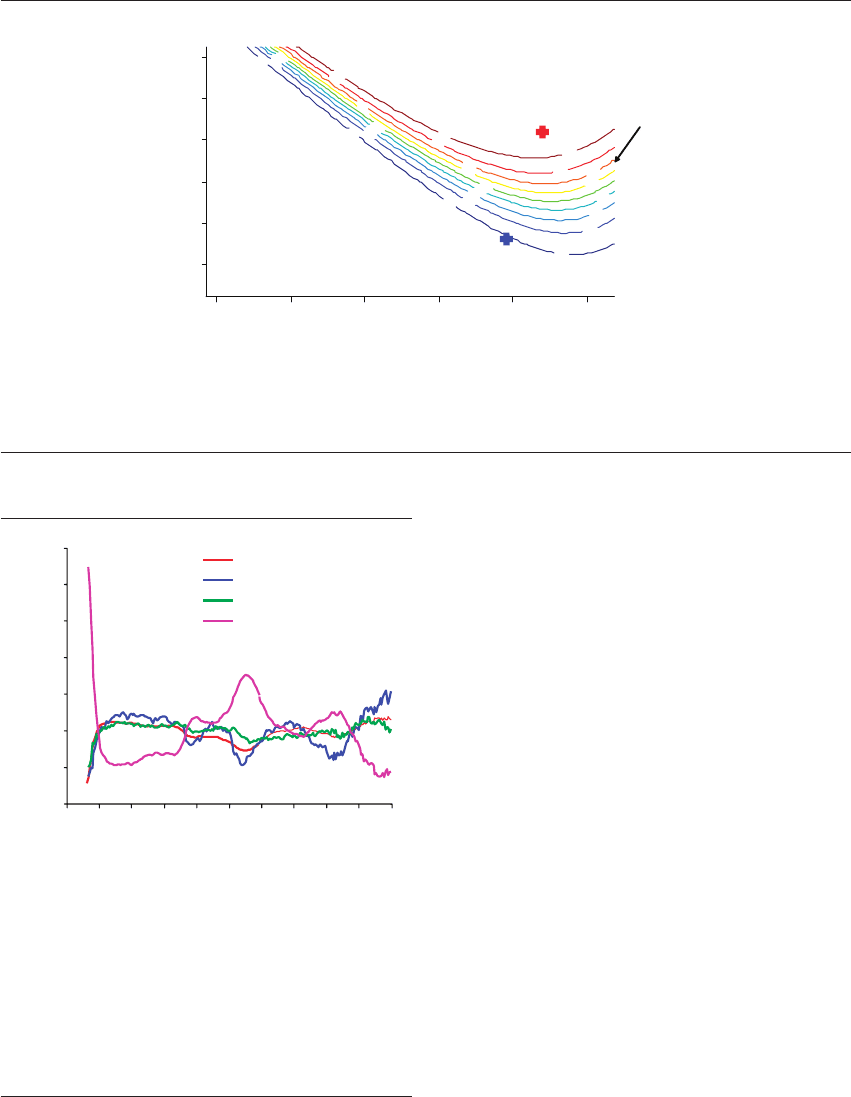
Figure 20.7 Bayesian probability statistics. A distribution of all results from embryos (pregnancy and non-pregnancy) was used to
create this Bayesian probability model. Only results from -SH and -CH biomarker populations were included to construct this curve.
The Bayesian database was then used to assess a single embryo’s metabolomic score, based on comparison with a representative
population of embryo scores between 0 and 100% implantation, to predict that embryo’s relative probability of implantation.
550 600 650 700 750 800
Wavelength (nm)
Relative absorbance
850 900 9501000 1050
–0.02
–0.01
0
0.01
0.02
0.03
0.04
0.05
Idiopathic
Varicocele
Vasectomy reversal
Normal donors
Figure 20.8 Near infrared spectra from seminal plasma. Seminal
plasma from four groups of men with infertility were studied by
metabolomic profiling: normal healthy donors, patients with
idiopathic male factor infertility, patients with varicocele, and
patients who underwent vasectomy reversal. A qualitative and
quantitative difference in metabolomic profiles could be discerned
between all groups of male infertility compared with normal
donors. The most pronounced difference was observed between
donors and patients with idiopathic male factor infertility. The
groups were statistically different from each other with assay
sensitivity and specificity of ⬎95% (data not shown).
HPE_Chapter20.qxp 7/17/2007 3:38 PM Page 258
METABOLOMIC PROFILING OF OXIDATIVE STRESS BIOMARKERS
ADDITIONAL ART STUDIES
Confirmation of these observations is planned through
a large prospective study involving several ART cen-
ters. Currently, a multicenter, multinational study is
underway in nine countries using a common Insti-
tutional Review Board (IRB), approved protocol and
informed consent. IVF programs in the USA, Canada,
Japan, Belgium, Italy, The Netherlands, Finland, Spain,
and Sweden are expected to add up to 500 additional
specimens to the current database. In addition, more
than half of the IVF cycles will be single embryo
transfer cycles. This refined study design will provide
unequivocal data demonstrating the relationship bet-
ween an embryo’s metabolomic profile and its implan-
tation potential.A Metabolomic Study Group for ART
is being formed in Europe to facilitate this and further
investigations in the field using metabolomic profiling.
ADDITIONAL INDICATIONS IN REPRODUCTIVE HEALTH
Beyond the immediate applications of embryo and
gamete viability testing, the Study Group will pursue
the use of metabolomics for functional genomics
testing and to assess endometrial receptivity for
implantation. In the former scenario, metabolomics
is viewed as a possible adjunct or replacement tech-
nology for aneuploidy screening, and perhaps other
genetic testing. With regard to implantation, study
designs employing metabolomics are being reviewed
that allow non-invasive examination of the endome-
trial lining of the uterus just prior to embryo transfer.
0
2
4
6
Implantation index
Number of samples
Pregnancy
No pregnancy
B
Relative absorbance
Wavelength (nm)
650
A
700 750 800 850 900 950
–1
0
1
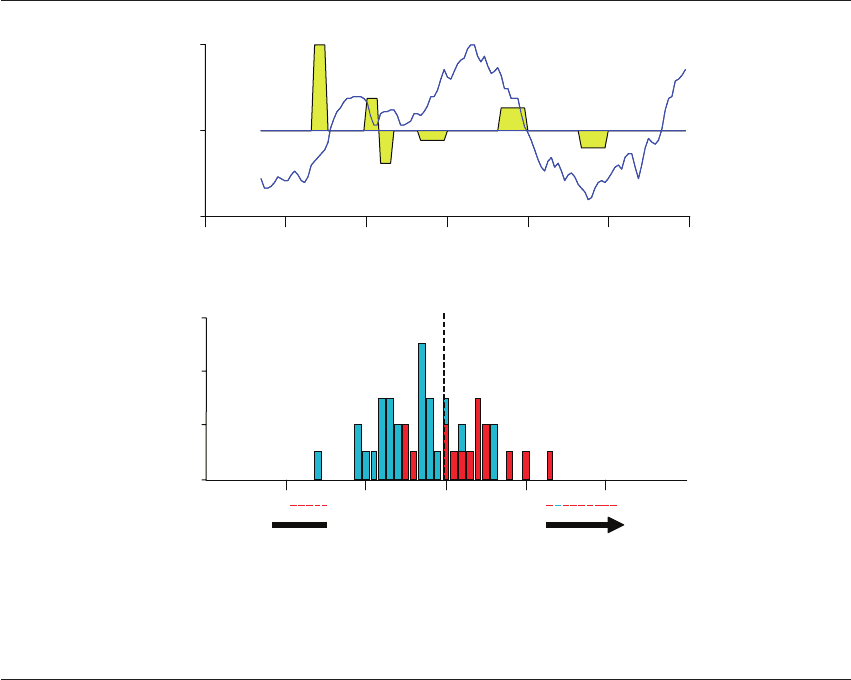
Figure 20.9 Near infrared spectra of follicular fluid. (A) A spectrum produced by NIR spectroscopy from a single follicular fluid
specimen. Six distinct biomarker regions are identified in yellow. The photon energy detected in these regions was captured and
subjected to bioinformatic analysis. (B) The implantation index score was calculated by the bioinformatic algorithms for each
specimen. The distribution of metabolomics data of each of 88 follicular fluid specimens showing the pregnancy outcome data of
oocytes from their respective follicular fluid that did not result in a pregnancy (blue) vs oocytes that produced viable embryos and a
pregnancy (red). The sensitivity and specificity for this group of specimens were 81% and 77%, respectively.
HPE_Chapter20.qxp 7/17/2007 3:38 PM Page 259
HUMAN PREIMPLANTATION EMBRYO SELECTION
In the field of maternal fetal medicine, Molecular
Biometrics (Chester, New Jersey, USA), has applied
metabolomics to assess fetal development by exam-
ining biomarkers in amniotic fluid. In several hun-
dred patients studied to date, final fetal birth weight,
intrauterine growth retardation/small for gestational
age, gestational diabetes mellitus, and preterm labor
have been characterized by metabolomic profiling
(Molecular Biometrics, data on file).
INDICATIONS FOR USE IN OTHER FIELDS
This same metabolomics platform has also been
applied to the assessment of neurodegenerative dis-
ease, using biomarker profiles to distinguish between
patients with Alzheimer’s disease, Parkinson’s dis-
ease, mild cognitive impairment, and age-matched
controls. Similarly, pulmonary edema and lactate lev-
els have been accurately diagnosed and monitored
non-invasively using this same metabolomics tech-
nique (Molecular Biometrics, data on file.) These
indications are also entering clinical development
programs for development of their respective med-
ical applications.
ACKNOWLEDGMENTS
The Metabolomics Study Group for ART is compro-
mised of the following investigators: Ashok Agarwal,
The Cleveland Clinic Foundation, Cleveland, Ohio;
Barry Behr, Stanford University, Palo Alto, California;
David Burns, McGill University, Montreal, Quebec,
Canada; Joe B Massey, Reproductive Biology Asso-
ciates, Atlanta, Georgia; Peter Nagy, Reproductive
Biology Associates, Atlanta, Georgia; Denny Sakkas,
Yale University, New Haven, Connecticut; Richard
J Scott, Reproductive Medicine Associates of New
Jersey, Morristown, New Jersey; and Emre Seli, Yale
University, New Haven, Connecticut.
REFERENCES
1. Sakkas D, Gardner DK. Noninvasive methods to assess embryo quality.
Curr Opin Obstet Gynecol 2005; 17: 283–8.
2. Patrizio P, Kovalevsky G. High rates of embryo wastage with use of
assisted reproductive technology: a look at the trends between 1995
and 2001 in the United States. Fertil Steril 2005; 84: 325–30.
3. Hollywood K, Brison D, Goodacre R. Metabolomics: current tech-
nologies and future trends. Proteomics 2006; 6: 4716–23.
4. Harington GG. Metabolomics Metabolomics: a “systems” contribu-
tion to pharmaceutical discovery and drug development. Drug Discov
World 2005; 39–46.
5. Lindon JC, Nicholson JK, Holmes E et al. Metabonomics: metabolic
processes studied by NMR spectroscopy of biofluids. Concepts Magn
Reson 2000; 12: 289–320.
6. Dunn WB, Bailey NJC, Johnson HE. Measuring the metabolome:
current analytical technologies. Analyst 2005; 130; 606–25.
7. Nicholson JK, Lindon JC, Holmes E. Metabolomics: understanding the
metabolic responses of living systems to pathophysiological stimuli via
multivariate statistical analysis of biological NMR data. Xenobiotica
1999; 29: 1181–9.
8. Ter Kuile BH,Westerhoff HV. Transcriptome meets metabolome: hier-
archical and metabolic regulation of the glycolytic pathway. FEBS Letts
2001; 500: 169–71.
9. Raamsdonk LM, Teusink B, Broadhurst D et al.A functional genomics
strategy that uses metabolome data to reveal the phenotype of silent
mutations. Nat Biotechnol 2001; 19: 45–50.
10. Pauling LC, Robinson AB, Teranishi R et al. Quantitative analysis of
urine vapor and breath by gas-liquid partition chromatography. Proc
Natl Acad Sci 1971; 68: 2374–6.
11. Oliver SG,Winson MK, Kell DB et al. Systematic functional analysis of
the yeast genome. Trends Biotechnol 1998; 16: 373–8.
12. Snoep JI, Westerhoff HV. From isolation to integration, a systems biol-
ogy approach for building the silicon cell. In: Alberghina L, Westerhoff
HV, eds. Systems Biology: Definitions and Perspectives. Springer-
Verlag, 2005: 7.
13. Kholodenko BN, Bruggeman FJ, Sauro HM. Mechanistic and modular
approaches to modeling and inference of cellular regulatory networks.
In Alberghina L, Westerhoff HV, eds. Systems Biology: Definitions and
Perspectives. Springer-Verlag, 2005: 143.
14. Burrill GS. Biotech 2005, Life Sciences. San Francisco: Burrill &
Company, LLC, 2005: 198.
15. Delneri D, Brancia FL, Oliver SG. Towards a truly integrative biology
through the functional genomics of yeast. Curr Opin Biotechnol 2001;
12: 87–91.
16. Jenkins H, Hardy N, Beckmann M et al.A proposed framework for the
description of plant metabolomics experiments and their results.
Nat Biotechnol 2004; 22: 1601–6.
17. Underwood BR, Broadhurst D, Dunn WB et al. Huntington’s disease
patients and transgenic mice have similar procatabolic serum metabo-
lite profiles. Brain 2006; 129: 877–8.
18. Clish CB. Integrative biological analysis of the APOE*3-leiden trans-
genic mouse. Omics 2004; 8: 3–13.
19. Forst CV. Host-pathogen systems biology. Drug Discov Today 2006;
11: 220–7.
20. Allwood JW, Ellis DI, Heald JK et al. Metabolomic approaches reveal
that phosphatidic and phosphatidyl glycerol phospholipids are major
discriminatory metabolites in responses by Brachypodium distachyon
to challenge by Magnaporthe grisea. Plant J 2006; 46: 351–68.
21. Goodacre R, Timmins EM, Burton R et al. Rapid identification of
urinary tract infection bacteria using hyperspectral, whole organism
fingerprinting and artificial neural networks. Microbiology 1998; 144:
1157–70.
22. Thomas N, Goodacre R, Timmins EM et al. Fourier transform infrared
spectroscopy of follicular fluids from large and small antral follicles.
Hum Reprod 2000; 15: 1667–71.
23. Jackson M, Sowa MG, Mantsch HH. Infrared spectroscopy: a new
frontier in medicine. Biophys Chem 1997; 68: 109–25.
HPE_Chapter20.qxp 7/17/2007 3:38 PM Page 260
METABOLOMIC PROFILING OF OXIDATIVE STRESS BIOMARKERS
24. Timmins EM, Howell SA, Alsberg BK. Rapid differentiation of closely
related candida species and strains by pyrolysis-mass spectroscopy
and Fourier transform-infrared spectroscopy. J Clin Microbiol 1998;
36: 367–74.
25. Houghton FD, Hawkhead JA, Humpherson PG et al. Non-invasive
amino acid turnover predicts human embryo developmental capacity.
Hum Reprod 2002; 17: 999–1005.
26. Houghton FD, Leese, HJ. Metabolism and developmental competence
of the preimplantation embryo.Eur J Obstet Gynecol Reprod Bio 2004;
115 (Suppl 1): S92–6.
27. Brison DR, Houghton FD, Falconer D et al. Identification of viable
embryos in IVF by non-invasive measurement of amino acid turnover.
Hum Reprod 2004; 19: 2319–24.
28. Lopes AS, Greve T, Callesen. Quantification of embryo quality by
respirometry. Theriogenology 2007; 67: 21–31.
29. Lopes AS, Madsen SE, Ramsing NB et al. Investigation of respiration
of individual bovine embryos produced in vivo and in vitro and corre-
lation with viability following transfer. Hum Reprod 2007; 22: 558–66.
30. Kenny LC, Dunn WB, Ellis DI et al. Novel biomarkers for pre-eclamp-
sia detected using metabolomics and machine learning. Metabolomics
2005; 1: 227–34.
31. Seli E, Sakkas D, Behr B et al. Non-invasive metabolomic profiling of
human embryo culture media correlates with pregnancy outcome.
Initial results of the Metabolomics Study Group for ART. Fertil Steril
2006; 86 (Suppl); 117.
32. Scott, RT, Miller K, Picnic S. A prospective blinded evaluation of the
relationship between metabolomic profiling of spent embryo culture
media and human embryonic reproductive potential. Fertil Steril
2006; 86 (Suppl); 235.
33. Halliwell B, Gutteridge JMC, eds. Free Radicals in Biology and
Medicine. Oxford: Oxford University Press, 1999.
34. Singh KK, ed. Oxidative Stress, Disease and Cancer. World Scientific
Publishing Company, 2006.
35. Agarwal A, Gupta S, Sharma R. Oxidative stress and its implications in
female infertility – a clinician’s perspective. Reprod BioMed Online
2005; 11: 641–50.
HPE_Chapter20.qxp 7/17/2007 3:38 PM Page 261
HPE_Chapter20.qxp 7/17/2007 3:38 PM Page 262

INTRODUCTION
Large-scale analyses of gene expression during
human oogenesis and embryogenesis has been ham-
pered by a scarcity of material for analysis coupled
with a lack of highly sensitive investigational tools.
Historically, a variety of classical molecular tech-
niques have been employed to examine cellular
mRNA levels.
1–3
However, these methods were gen-
erally crude in nature and thus lacked the precision
to accurately and reproducibly detect or quantify
transcripts in single cells. Reverse transcription-
polymerase chain reaction (RT-PCR) provided a
highly sensitive means to distinguish rare mRNA
species in individual cells.
4
Together with fluores-
cent dyes, RT-PCR can now be used to reliably
quantify gene expression in individual oocytes.
5,6
Subsequently, PCR-based methods, and, more
recently, analysis tools that provide genome-wide
perspectives such as microarray techniques have
been used to glean a wealth of knowledge regarding
the global expression profile of the oocyte and
embryo.
7–9
However, a great deal of work remains to
be done in order to fully dissect the complex regula-
tory pathways that direct early human development.
This chapter reviews our current understanding of
these developmental programs.
GENE EXPRESSION DURING
FOLLICULOGENESIS
The human follicular cycle culminates with the
expulsion of a single mature oocyte at ovulation,
which is arrested in metaphase II. However, the
process of follicular maturation begins in utero many
years, and even decades earlier. Primordial germ cells
(PGCs) migrate to the genital ridge in the develop-
ing embryo, where they actively engage in mitosis.
The oogonia subsequently initiate meiosis, and the
resulting primary oocytes arrest at the diplotene
phase of prophase I, invest themselves with granu-
losa cells, and thus give rise to primordial follicles.
Concomitantly, those oogonia that do not enter
meiosis I undergo atresia, resulting in the eradica-
tion of all remaining oogonia by the time of birth.
Shortly thereafter, primordial follicle formation
ceases, but the process of primary follicle develop-
ment has already begun. During this process, pri-
mordial follicles develop an extracellular matrix, the
zona pellucida, and a differentiated granulosa layer.
Further proliferation of the granulosa layer gives
rise to a secondary follicle, which then also acquires
a thecal layer.
10
Although the origin of PGCs and formation of
primordial follicles is not yet well understood at a
molecular level, several factors that are expressed
in adjoining somatic tissues have been identified
which may affect their development. Tumor necro-
sis factor-␣ (TNF␣) expression has been confirmed
in human primordial follicles.
11
TNF␣ appears to be
involved in the apoptosis of random oocytes, and
this is believed to facilitate primordial follicle
assembly.
12
Leukemia inhibitory factor (LIF) has
been shown to induce proliferation and differentia-
tion.
13
Expression profiles of Kit ligand (KL) and its
tyrosine kinase receptor (c-Kit) in the human ovary
suggest that they are involved in human fertility.
14
While their precise roles in the human are not known,
studies from mouse mutants have revealed that KL
and c-Kit are important in the establishment of the
PGCs within the ovary, oocyte survival and growth,
granulosa cell proliferation, theca cell recruitment,
and the maintenance of meiotic arrest.
14
21. Gene expression analysis in the human
oocyte and embryo
Nury M Steuerwald
HPE_Chapter21.qxp 7/13/2007 5:31 PM Page 263
HUMAN PREIMPLANTATION EMBRYO SELECTION
Genes expressed by the PGCs and oocytes them-
selves are believed to play a role in the develop-
ment of the oocyte and follicle. Human Factor in the
germline alpha (FIGLA)
15
appears to be associated
with cell–cell interactions and survival during
the process of primordial follicle formation.
16
The
murine counterpart of FIGLA also regulates the
expression of genes encoding zona pellucida pro-
teins. This role may possibly be conserved among
mammals.
17
It has been reported that there are four
human zona pellucida genes (ZP1, ZP2, ZP3, and
ZPB), not three as previously predicted by the
murine model.
18
The zona pellucida serves as the
first point of contact for sperm–egg interactions,
and it is also involved in establishing a block to
polyspermy and providing protection for the pre-
implantation embryo. The human Dazla (deleted in
azoospermia like autosome) gene is the autosomal
homolog of the Daz (deleted in azoospermia) gene
which maps to the long arm of the Y chromosome.
19
It encodes a cytoplasmic protein with RNA binding
domains and is believed to be involved with trans-
lational regulation. Dazla is expressed in oogonia as
well as in oocytes and granulosa cells of primordial
follicles. Its expression pattern in humans is similar
to that in mouse, where Dazla mutations cause male
and female sterility, suggesting that there may also
be functional parallels during gametogenesis.
19
The transition from primordial to mature follicle
is further mediated by the expression of several key
genes in the oocyte. Growth differentiation factor
(Gdf-9) and bone morphogenic protein (Bmp-15)
are oocyte-expressed genes that are both members
of the transforming growth factor  superfamily.
In the human, transcripts of these genes are present
in oocytes of primary follicles and persist follow-
ing ovulation.
20,21
GDF-9 is believed to promote
granulosa cell proliferation, as well as induce
cumulus cell differentiation and expansion.
22
GDF-9
modulates the expression of cumulus granulosa
genes such as hyaluronic acid synthase 2 (Has2),
prostaglandin-endoperoxide synthase 2 (Ptgs2), and
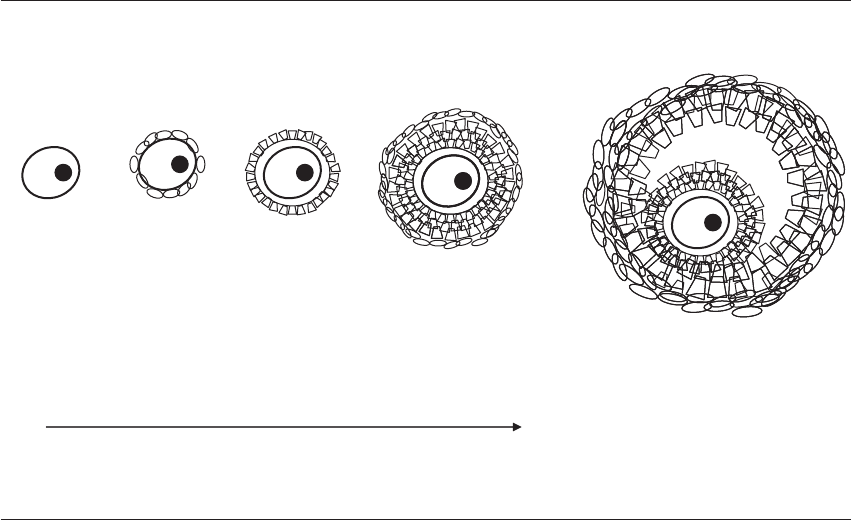
Primordial
germ cell
Primordial
follicle
Primary
follicle
Secondary
follicle
Antral
follicle
Ovarian somatic cells:
Tnf
a
, Lif, KL,
c-Kit
PGCs: Figla,
ZP genes, Dazla
Oocyte: Gdf9, Bmp15,
bFgf, Nobox
Cumulus cells: Has2,
Ptgs2, Grem1
Figure 21.1 Genes implicated in follicle development. Follicles at various stages of development are depicted. Ovarian somatic
tissues, primordial germ cells (PGC), oocytes, and cumulus cells express various factors throughout folliculogenesis. Genes expressed
by specific cell types are listed.
HPE_Chapter21.qxp 7/13/2007 5:31 PM Page 264

GENE EXPRESSION ANALYSIS IN THE HUMAN OOCYTE AND EMBRYO
gremlin (Grem1). The expression of these GDF-9
regulated genes has been shown to correlate with
morphological and physiological criteria consistent
with higher grade embryos, and this may serve as
a biochemical marker for embryo development
during in vitro fertilization procedures.
23
Bmp-15
shares a high degree of homology with Gdf-9 and is
also thought to regulate critical somatic cell activi-
ties during folliculogenesis, as evidenced by a recent
study implicating Bmp-15 mutations in the patho-
genesis of premature ovarian failure in a large
cohort of women.
24
Basic fibroblast growth factor
(BFGF) localizes to oocytes, granulosa, and theca
cells in the human ovary.
25
Human bFgf temporal
and spatial expression patterns hint at a potential
role in primordial follicle activation, as predicted by
studies in rodents.
26
Likewise, studies in the mouse
have identified another gene, Nobox (newborn
ovary homeobox), expressed in primordial and
growing oocytes, which appears to be essential for
oocyte survival and follicular development.
27
The
human gene encoding NOBOX has been identified,
but its precise function remains to be elucidated.
28
Figure 21.1 presents a schematic representation of
selected genes that have been implicated in oogene-
sis and folliculogenesis.
MESSENGER RNA EXPRESSION,
LOCALIZATION, STABILITY,
AND REGULATION DURING
OOGENESIS
Following many years of arrest at meiotic prophase,
the luteinizing hormone (LH) surge attending
ovulation induces the resumption of meiosis by
modulating cAMP levels, which is believed to bring
about the activation of maturation promoting fac-
tor (MPF).
29
MPF is a highly conserved complex
of proteins consisting of two subunits, one which
serves a regulatory role (cyclin) and another which
possesses kinase activity.
30,31
Activated MPF medi-
ates germinal vesicle breakdown by phosphorylat-
ing several proteins: histone protein phosphorylation
results in chromosomal condensation, and phos-
phorylation of nuclear envelope lamin proteins
results in its depolymerization and breakdown.
32
However, meiosis is again arrested at metaphase II,
presumably due to the action of a cytostatic factor
(CSF) which contains the protein products of the
c-mos and Cdk-2 genes.
33,34
CSF prevents the degra-
dation of cyclin until fertilization, when the increase
in calcium, probably initiated by the inositol
phosphate pathway,
35
activates a CSF protease which
allows meiosis to proceed.
36
It is undeniable that oocytes of several organ-
isms, including certain insects, nematode worms,
and amphibians are polarized with respect to the
distribution of maternal transcripts and proteins,
and this ultimately defines the axes of the embryo.
37
However, the details regarding the polarized nature
of mammalian oocytes have only recently begun to
emerge. Polarization in mammals is not as dramatic
morphologically as in low-order animals. Never-
theless, it has been described at the molecular level
in human oocytes.
38–40
Non-uniform allocation of
protein products encoded by the leptin and Stat3
genes could play a critical role in axis determination
in the oocyte as well as in the differentiation of the
embryo.
38
Their ultimate localization to trophecto-
dermal cells has prompted suggestions for their use
as trophectodermal markers. Members of several
classes of proteins serving a variety of functions
have also been found in polarized domains in
human oocytes and embryos.
39
These include
growth factors (TGF-2 and VEGF), growth factor
receptors (C-ERBB and C-KIT), and apopotosis
proteins (BCL-X and BAX). Asymmetric transcript
distributions of -human chorionic gonadotropin
and -LH amongst blastomeres in cleaving embryos
have also been reported.
36,40
Perturbations of the
spatial aspects and/or concentration gradients in
the oocyte could adversely affect its quality and
developmental potential.
41
Several structural features of eukaryotic mRNA
can affect the stability and may be responsible for
early embryonic message degradation and turnover.
These include polyadenylation and the formation of
secondary structures. The association of mRNAs
with cytoplasmic factors may also result in the
formation of complexes which may confer stability
to the message, or may promote its degradation.
HPE_Chapter21.qxp 7/13/2007 5:31 PM Page 265
