Elder K. Human preimplantation embryo selection
Подождите немного. Документ загружается.

HUMAN PREIMPLANTATION EMBRYO SELECTION
DEFINITIONS
Metabolomics is an emerging science that represents
the continuation of the ever-expanding disciplines
we call the ‘omics’.
3–6
Metabolomics represents the
study of cellular processes that are downstream
events of those studied in the fields of genomics
and proteomics. As a new scientific discipline, the
nomenclature of metabolomics is rapidly evolving
in parallel with the science itself, and is only now
being defined. As a consequence, there is currently
no universal definition of metabolomics, or its com-
panion techniques; some of the more commonly
used definitions that are used to frame the concept
of metabolomics include: (1) the study of global
metabolite profiles in a system (cell, tissue, or
organism) under a given set of conditions; (2) the
‘systematic study of the unique chemical finger-
prints that specific cellular processes leave behind’,
specifically, the study of their small molecule
metabolite profiles; and (3) the systematic analysis
of the inventory of metabolites, as small molecule
biomarkers, that represent the functional pheno-
type at the cellular level (Figure 20.1.)
The metabolome
3
refers to the complete inventory
of small-molecule, non-proteinaceous compounds,
such as metabolic intermediates, ATP, fatty acids,
glucose, cholesterol, hormones, and other signaling
molecules, and secondary metabolites that are
found within a biological sample. These molecules
are the ultimate products of cellular metabolism.
The metabolome is dynamic, changing from second
to second, based upon the activation and interaction
of different metabolic pathways within the cell; thus,
it is of particular importance to the rapidly changing
development of the embryo in nature and its interac-
tion with the changing conditions in the laboratory.
Ultimately, the metabolome represents the global
population of biomarkers that can be assessed in the
study of metabolomics. Mining of the metabolome
provides information that best describes the pheno-
type, and this can also be used to infer gene function.
The slightly different term metabonomics is
commonly used in the context of drug toxicity
assessment. There is some discussion over the exact
differences between ‘metabolomics’ and ‘metabo-
nomics’, but in general, the term metabolomics is
more commonly used in the context of emphasis on
comprehensive metabolic profiling, while metabo-
nomics attempts to quantify the multiple metabolic
changes caused by a biological perturbation. In
practice, there is still a large degree of overlap in the
way the terms are used, and they are often used
interchangeably.
THE CASE FOR METABOLOMICS
The dogma of molecular biology suggests that DNA is
transcribed into RNA which is then translated into
proteins. Protein and post-translational modifica-
tions of proteins ultimately give rise to small mole-
cules within the cell which become the biomarkers
that can be studied by metabolomics. It is generally
agreed that there are more than 25 000 genes, 100 000–
200 000 transcripts, and up to 1 000 000 proteins,
whereas there may be as few as 2500–3000 small
molecule metabolites/biomarkers that make up the
human metabolome. By systematically measuring this
population of small molecule biomarkers or metabo-
lites, scientists can establish ‘profiles’ or ‘signatures’ of
healthy individuals versus those with specific illnesses.
The realization that knowing the DNA sequence
of the human genome, or that of other organisms,
does not in itself explain the fundamental nature of
many normal biological processes and disease states
has triggered a marked increase in seeking paradigms
that can relate gene expression to the phenotype.
7
Metabolomics
Metabolomics can be thought of as the interface
between genomics and a functional phenotype:
Genomics → Transcriptomics → Proteomics
→ Metabolomics → Functional Phenotype
Definition: systematic analysis of the inventory of
metabolites – as small molecule biomarkers –
that represent the functional phenotype at the
cellular level.
Figure 20.1 Definitions of ‘metabolomics’. Metabolomics is a
natural offshoot of the fields of genomics and proteomics, and
represents the downstream events of those cellular processes.
HPE_Chapter20.qxp 7/17/2007 3:38 PM Page 246

METABOLOMIC PROFILING OF OXIDATIVE STRESS BIOMARKERS
Even with the sequencing of the total human
genome, it is clear that our knowledge of gene func-
tion is limited; for example, as many as 35–40%
of open reading frames have no known function.
3
Consequently, there is a need to better understand
the downstream effects of gene expression, which are
translated via the transcriptome or proteome, and
amplified at the level of the metabolome. This allows
for a greater degree of sensitivity when mining the
biomarker population of the metabolome compared
with other levels of cellular regulation. More impor-
tantly, metabolic fluxes are not regulated by gene
expression alone (considering post-transcriptional
and post-translational events), so the metabolome is
considered to be the closest expression of pheno-
type.
8
Metabolomics can provide indications of a
metabolic problem or lesion with high throughput at
lower costs than genomics, transcriptomics, or pro-
teomics, and is therefore well suited for widespread
investigations in the life sciences.
In summary, metabolomics is an extension of the
growing understanding of the body’s many different
genetic and molecular players, including genes and
proteins. We know that small changes in the flux
through an intracellular pathway can lead to large
changes in the concentration of metabolites.
9
These
changes can be detected by metabolomic profiling of
key biomarkers, thus signaling when crucial distur-
bances have occurred or are about to occur. In con-
trast, research focusing on genomics or proteomics
alone only examines a small portion of the essential
events that define normal biological function. While
genes can tell us what we might become or the poten-
tial for a biological error, and proteins tell us which
genes have been expressed, metabolomics provides us
with a real-time ‘snapshot’ of the downstream events
that characterize gene expression. For these reasons,
metabolomics is expected to be a relevant tool in the
management of various medical conditions, and, thus
an important method for studying basic biological
function in concert with other methods.
HISTORY
It is generally assumed that metabolomics was con-
ceived in 1970 by Arthur Robinson of the Department
of Chemistry at the University of California at San
Diego. Although it was not called metabolomics at
the time, Robinson and Pauling published their first
paper in 1971 in the Proceedings of the National
Academy of Sciences.
10
Robinson’s core concept was
that information-rich data that reflect the func-
tional status of a complex biological system reside in
the quantitative and qualitative pattern of metabo-
lites in body fluids. It was his expectation that body
fluid analysis could be optimized to create a low
cost, informative, and medically relevant means of
measuring metabolic changes even when standard
clinical chemistry markers were in the ‘normal
range’.
Later, the term ‘metabolomics’ was coined in
1998.
11
Six years later, in 2004, the Metabolomics
Society (http://www.metabolomicssociety.org) was
formed to promote the study of metabolomics. The
Human Metabolome Project (HMP) was launched in
Canada in 2005 and has identified approximately 2000
metabolites that make up the human metabolome.
The goal of HMP is to make this information
available worldwide through its database (www.
hmdb.ca). Many of the bioanalytical methods used
for metabolomics research have been adapted from
existing biochemical techniques and enhanced by
the advent of newer methodologies, such as various
forms of spectral analysis.
Three characteristics common to metabolomic
research are: (1) metabolite or small molecule bio-
markers are profiled without bias towards a specific
analyte or group of metabolites; (2) the biomarkers
or metabolites are identified and quantified by vari-
ous forms of analytical, biochemical and spectral
analysis; and (3) relationships between the metabo-
lites are characterized, primarily by multivariate
mathematical methods often referred to collectively
as bioinformatics or computational biology.
BIOINFORMATICS AND
COMPUTATIONAL BIOLOGY
In 1952, the British neurophysiologists and Nobel
Prize winners Hodgkin and Huxley constructed
a mathematical model of the nerve cell. In 1960,
HPE_Chapter20.qxp 7/17/2007 3:38 PM Page 247

HUMAN PREIMPLANTATION EMBRYO SELECTION
Denis Noble developed the first computer model of
a beating heart. Since then, mathematical computa-
tions of biological systems have rapidly escalated to
include analysis, quantification, and descriptions of
complex molecular level events. These progressively
important developments quickly gave rise to the
field of bioinformatics and computational biology,
which is defined as the use of techniques including
applied mathematics, informatics, statistics, com-
puter science, artificial intelligence, chemistry, and
biochemistry to solve biological problems, usually
at the molecular level. Research in computational
biology often overlaps with systems biology. Major
research efforts in the field include sequence align-
ment, gene finding, genome assembly, protein
structure alignment, protein structure prediction,
prediction of gene expression, and protein–protein
interactions, and the modeling of evolution.
The terms bioinformatics and computational
biology are often used interchangeably. However,
bioinformatics more properly refers to the creation
and advancement of algorithms, transformations,
discriminant analysis, computational, and statistical
techniques and theory to solve formal and practical
problems posed by or inspired from the management
and analysis of biological data. Computational biol-
ogy, on the other hand, refers to hypothesis-driven
investigation of a specific biological problem using
computers, carried out with experimental and sim-
ulated data, with the primary goal of discovery and
the advancement of biological knowledge. A typical
metabolomics experiment, for example, will gener-
ate volumes of data that need to be converted into
useful information and knowledge.
SYSTEMS BIOLOGY
Systems biology studies the interactions between
the components of a biological system, and how these
interactions give rise to the function and behavior
of that system; for example, the interactions between
enzymes and metabolites in a specific metabolic
pathway.
12
Typically, a cellular network is modeled
mathematically. Due to the large number of param-
eters, variables, and constraints in cellular networks,
numerical and computational techniques are used.
Other aspects of computer science are also used
in systems biology, including text mining to find
parameter data from literature, online databases,
and repositories for sharing data and models.
The systems biology approach is characterized
by a cycle of theory, computational modeling and
experiment to quantatively describe cells or cell
processes.
13
Since the objective is to model all the
interactions in a system, the experimental techniques
that most suit systems biology are those that are sys-
tem-wide and attempt to be as complete as possible.
Therefore, metabolomics, proteomics, and high-
throughput techniques are used to collect quantitative
data for the construction and validation of models.
ANALYTICAL TECHNIQUES USED
IN METABOLOMICS
Metabolite or biomarker analysis typically addresses
two issues: (1) separation of the biomarkers, usually
by a form of chromatography or electrophoresis,
particularly capillary electrophoresis; and (2) detec-
tion and quantification of the biomarkers, depend-
ing on the methodologies used. A summary of the
more commonly used separation and detection
techniques is provided below.
SEPARATION METHODS
GAS CHROMATOGRAPHY
Gas chromatography (GC) is often used in concert
with mass spectrometry (MS). This combination
method, often referred to as GCMS, is one of the
most widely used and powerful methods. It offers
very high chromatographic resolution, but requires
chemical derivatization for many biomolecules:
only volatile chemicals can be analyzed without
derivatization. Many large and polar metabolites
cannot be analyzed by GC.
HIGH PERFORMANCE LIQUID CHROMATOGRAPHY
Compared with GC, high performance liquid chro-
matography (HPLC) has lower chromatographic
HPE_Chapter20.qxp 7/17/2007 3:38 PM Page 248
METABOLOMIC PROFILING OF OXIDATIVE STRESS BIOMARKERS
resolution, but it does have the advantage that a much
wider range of analytes can potentially be measured.
CAPILLARY ELECTROPHORESIS
Capillary electrophoresis (CE) is the newest tool to be
added to the armamentarium of methods used for
metabolomic profiling. There are a number of advan-
tages of CE: it has a higher theoretical separation
efficiency of analytes than HPLC, and is suitable for
use with a wider range of metabolite classes than is
GC. As for all electrophoretic techniques, it is most
appropriate for charged analytes.
DETECTION METHODS
MASS SPECTROMETRY
Mass spectrometry is perhaps the most commonly
used technique to identify and, in some (but not all)
cases, to quantify metabolites after separation by
GC, HPLC, or CE. GCMS is the most ‘natural’ com-
bination of techniques, and was the first to be devel-
oped. Measuring time-of-flight of fragmentation
pattern constituents also provides useful informa-
tion regarding the makeup of the specimen being
analyzed by MS. In addition, mass spectral finger-
print libraries exist, or can be developed, allowing
identification of a metabolite according to its frag-
mentation pattern. MS is both sensitive and specific.
There are also a number of metabolomic studies
which use MS as a stand-alone technology: the sam-
ple is infused directly into the mass spectrometer
with no prior separation. In this scenario MS serves
to both separate and detect metabolites.A disadvan-
tage of mass spectrometry is that the specimen is
consumed in the analysis, and cannot be recovered.
MS instrumentation for metabolomic analysis is
generally very costly.
NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY
Nuclear magnetic resonance (NMR) has the added
advantage that the metabolites don’t always need to
be separated prior to analysis, and the sample can
thus be recovered for further analyses. Several small
molecule metabolites can be measured simultane-
ously. NMR is close to being a universal detector, as
it produces detailed structural information about
the compounds. However, it also possesses one major
disadvantage, which is that it is relatively insensitive
compared with mass spectrometry-based techniques.
NMR is not generally regarded as a reliable quanti-
tative method. It is also the most complex and
expensive instrumentation to acquire and maintain.
FOURIER TRANSFORM INFRARED SPECTROSCOPY
Fourier Transform Infrared spectroscopy (FTIR)
is a physicochemical method that measures vibra-
tions around chemical bonds within different
functional groups of molecules using light electro-
magnetic radiation at specific wavelengths. This
method gives quantitative information about the
total biochemical composition of a specimen without
consuming it. However, substantial sample prepara-
tion is required before analysis can be initiated by
FTIR, thus adding time and technical requirements
to the analysis.
RAMAN AND NEAR INFRARED SPECTROSCOPY
While MS and NMR are by far the two leading tech-
nologies used for metabolomics research, Raman
and near infrared (NIR) spectroscopy have also been
used successfully to identify and quantify biomark-
ers in metabolomics. These methods have similar
levels of analytical sensitivity to the previous meth-
ods but also have several added advantages: direct
sample measurement – no preparation is required;
little chemical bias; instrumentation costs are con-
siderably less; instrumentation size is much smaller
and the instruments are easier to run and maintain;
and rapid, simultaneous analysis of multiple bio-
markers at one time. These techniques are not always
suitable for all biomarker targets, but are becoming
more widely used for a number of metabolomic
applications. Our scientific team has successfully
utilized Raman and NIR spectroscopy as the plat-
form for its metabolomics technology and has coined
the phrase ‘biospectroscopy-based metabolomics’or
BSM for such applications.
HPE_Chapter20.qxp 7/17/2007 3:38 PM Page 249

HUMAN PREIMPLANTATION EMBRYO SELECTION
KEY APPLICATIONS
Metabolomics is an example of a true ‘disruptive
technology’ the availability of which is expected
to markedly impact discovery-driven as well as
hypothesis-driven research paradigms. As a result,
metabolomics is gaining interest across a wide
variety of disciplines, including functional genomics,
biomarker discovery, diagnostic and therapeutic
monitoring, drug discovery and development, phar-
macogenomics, and other life science applications.
As metabolic biomarkers are identified they can be
used to better understand specific disease processes
based on the pathways involved. This information
can give rise to therapeutic intervention through
new drug discovery. Metabolomic studies of cells in
culture has added immensely to the understanding
of biological diversity and complexity in both basic
science and medicine.
MOLECULAR DIAGNOSTICS
The sensitivity and specificity of metabolomic
profiling is expected to lead to the development
of a new class of molecular diagnostics that provides
rapid, cost-effective assessment of the functional phe-
notype in health and disease. Metabolomic analysis
can pinpoint changes in complex networks of meta-
bolic reactions, where outputs from one enzymatic
pathway are inputs to other chemical reactions,
thus identifying deviations in normal biology.
Metabolomics can be used to systematically distin-
guish between the often subtle differences that
separate normal physiology from the onset or pro-
gression of disease or an individual’s response to a
therapeutic compound, thus leading to (1) new
diagnostic and monitoring paradigms; (2) the for-
mation of new industry standards of specificity
and sensitivity; (3) the generation of reliable, new
measurements on which insurance providers can
establish more accurate and thus broader reimburse-
ment criteria; and (4) new designs for sensitive, high
throughput instruments that yield cost-effective,
user-friendly devices that produce rapid results
without the need for specimen preparation.
THERANOSTICS
Metabolomic technology will enable the healthcare
industry to rapidly move towards the practice of
personalized medicine, or ‘theranostics’, allowing the
pharmaceutical and biotechnology industries to shift
from products that only treat disease to new analytical
methods that can detect, monitor, and help stratify ill-
nesses. This trend in medical care looks at disease as a
process rather than a state, and allows physicians to
diagnose as well as track disease progression in order
to provide the most appropriate treatment interven-
tions on a patient-by-patient basis.
14
Pharmaceutical
and biotechnology companies are using metabolomics
to develop ‘companion diagnostics’ so that their new
therapeutic compounds can be used more effectively
and safely, following FDA approval.
DRUG DISCOVERY AND DEVELOPMENT:
PHARMACODIAGNOSTICS
Metabolomic profiling of biomarkers has the poten-
tial to revolutionize drug discovery and develop-
ment through novel diagnostic testing paradigms
referred to as pharmacodiagnostics. This approach
is expected to generate more efficacious clinical trial
designs that achieve the goals of cost-effective and
expeditious clinical research. Pharmacodiagnostic
strategies that are based on metabolomics and
pharmacokinetic monitoring have the potential
to reduce clinical trial failures and shorten drug
development timelines, as well as to create new
measurements of safety and efficacy that can be
used to predict and monitor adverse events. With
increased regulatory oversight, pharmacodiagnos-
tics are also expected to be used increasingly in the
design of phase IV or FDA-imposed postmarketing
studies.
BIOMARKER DISCOVERY
The identification of metabolites that could serve
as biomarkers of disease is a key application of
metabolomics. Over time, reliance on single ana-
lyte/biomarker efficiency has been replaced by the
HPE_Chapter20.qxp 7/17/2007 3:38 PM Page 250
METABOLOMIC PROFILING OF OXIDATIVE STRESS BIOMARKERS
concept of multivariate biomarker profiles that are
more sensitive and predictive. An initial approach in
biomarker development is to create databases from
metabolomic profiling of control populations, and
then test populations having a specific disease or
condition of interest. This approach statistically
compares the metabolomic profile of the diseased
population with the profile of healthy controls. The
data are mined by bioinformatics analysis in order
to identify the distribution of metabolites/biomarkers
that can discriminate the disease state and which
could then be used as predictors of that condition.
The challenge in this process is to utilize rigorous
statistical methodologies followed by validation of
the database. The biomarkers can be further vali-
dated for their predictive or monitoring value for
clinical applications, or can be adopted as surrogate
markers for pharmacokinetic and pharmaco-
diagnostic applications. Finally, once the biological
function of individual biomarkers is established,
they may become viable molecular targets for drug
development processes.
TOXICITY ASSESSMENT/TOXICOLOGY
Metabolomic profiling of biological fluids can be
used to detect the physiological changes caused by
toxic insult of a chemical or a mixture of chemicals.
These applications can address environmental,
agricultural, or medical issues. In many cases, the
observed changes can be related to specific syndromes,
e.g. a specific lesion in the central nervous system
(CNS), liver, or kidney. This is of particular rele-
vance to pharmaceutical companies that need to test
the efficacy as well as toxicity of potential drug can-
didates. Eliminating a compound on the grounds of
poor efficacy or adverse toxicity before it reaches
clinical trials could save enormous expense and
time in otherwise faulty trials.
FUNCTIONAL GENOMICS
Metabolomics is an excellent tool for determining
the phenotype created in nature or by genetic
manipulation such as gene deletion or insertion.
For example, metabolomics can be used to detect
any phenotypic changes in a genetically modified
organism or in a plant species intended for human
or animal consumption. More exciting is the prospect
of predicting the function of unknown genes by
studying the metabolomic profiles of biomarkers
that result from the deletion or insertion of known
genes. Such advances are likely to come from models
based on microorganisms,
15
plants,
16
and experimen-
tal animals. Transgenic mouse models of Huntington’s
disease, for example, have already demonstrated the
power of metabolomics to decipher the phenotypic
human clinical picture in an animal carrying the
transfected gene.
17
Likewise atherosclerosis in labo-
ratory mice has been diagnosed more efficiently and
accurately using metabolomics than by genomics or
proteomics alone.
18
HOST–PATHOGEN INTERACTIONS
Metabolomic investigations are also expected to
prove valuable in host–pathogen interactions.
19
Using plant models, MS was used to identify discrim-
inatory lipid metabolites and certain phospholipids
that are know to be important in protecting plants
from infectious diseases.
20
Parallel applications in
human infectious disease are equally plausible given
the cascade of metabolic events that are mounted by
a host’s defense mechanisms in response to infectious
agents.
21
NUTRIGENOMICS
This is a generalized term which links genomics,
transcriptomics, proteomics, and metabolomics to
human nutrition. In general the metabolome of
a given body fluid is influenced by endogenous
factors such as age, sex, body composition, and
genetics as well as underlying pathologies. Diet and
drugs are the main exogenous factors, and diet can
then be further broken down to nutrients and non-
nutrients. Metabolomics can be used to determine
the biological endpoints that reflect the balance
of these variables in an individual’s nutritional
makeup.
HPE_Chapter20.qxp 7/17/2007 3:38 PM Page 251

HUMAN PREIMPLANTATION EMBRYO SELECTION
APPLICATIONS OF METABOLOMICS IN THE
ASSISTED REPRODUCTIVE TECHNOLOGIES
METABOLOMICS-BASED EMBRYO SELECTION
A technology that is capable of identifying viable
gametes and embryos in an IVF program is expected
to lead to a reduction in the number of embryos
transferred and, eventually, the adoption of single
embryo transfer (SET). The availability of rapid,
non-invasive metabolomic profiling of embryos
could potentially empower practitioners to overcome
the IVF dilemma and move away from the current
paradigm of multiple embryo transfer. The practical
benefit of rapid, non-invasive metabolomic profil-
ing in IVF is the improvement of embryo and
gamete selection procedures thus allowing a reduc-
tion in the number of embryos transferred without
compromising pregnancy rates. The longer term
benefit of this technology to patients, physicians,
and insurance providers is the potential to limit the
incidence of multiple births and associated health-
care risks and expense.
CURRENT APPLICATIONS
The advent of metabolomics in the field of assisted
reproduction is very recent. Only a small number of
abstracts and papers have been published prior to
the work of the Metabolomics Study Group for
ART. The publications in the field so far are summa-
rized below.
(1) FTIR spectroscopy. Thomas et al
22
reported
substantial differences in the biochemical fin-
gerprints of follicular fluid from large follicles
(⬎17 mm) versus small (⬍15 mm) antral folli-
cles in patients undergoing IVF. The differences
in the distribution of steroid concentrations
between large and small follicles were signifi-
cant but did not follow protein bonding charac-
teristics as observed by FTIR. While not labeled
specifically as a metabolomics study, the authors
concluded that the differences detected in
metabolite profiles by FTIR reflect the develop-
mental stage of the follicle, and possibly oocyte
quality. FTIR has also been used successfully
to examine biological fluids for other medical
applications.
23,24
(2) HPLC. Houghton et al
25,26
and Brison et al
27
investigated amino acid turnover of embryo
culture media to establish metabolomic profiles,
or signatures, of developing human embryos.
Using HPLC as their analytical tool, the authors
observed differential amino acid compositions
in the culture media of viable versus non-viable
embryos and correlated their findings with preg-
nancy outcomes. The studies concluded that
amino acid analysis may be used as a non-invasive
test to predict embryonic viability.
(3) FTIR spectroscopy. Hollywood (in preparation)
used FTIR spectroscopy to examine the meta-
bolomic profile of biomarkers in embryo cul-
ture media. Cluster analysis of media specimen
FTIR spectra from embryos that did not pro-
duce a pregnancy differed significantly from that
of media from embryos that resulted in positive
biochemical and clinical (ultrasound) pregnan-
cies. The authors concluded that metabolomic
fingerprinting of media can be used to assess
the status of individual human embryos and
their potential for producing a pregnancy.
(4) Respirometry (oxygen consumption). Lopes
et al
28,29
studied oxidative pathways of metabo-
lism in individual bovine embryos by direct
quantification of oxygen consumption. Using a
non-invasive micro-electrode, these investigators
postulated that embryonic respiration rates, in
association with other embryo selection meth-
ods, allowed an accurate assessment of embryo
quality. The authors discussed the potential
use of this technology as a diagnostic tool for
improving embryo selection in IVF.
(5) GCMS. In a related, but non-ART study, inves-
tigators used GCMS in metabolomic studies to
assess biomarkers related to pre-eclampsia.
30
The authors identified metabolomic profiles in
plasma of pregnant women to discriminate those
with normal pregnancies from those at risk for
developing pre-eclampsia. The metabolomic
data showed high sensitivity and specificity,
targeting only three biomarkers in the analysis.
HPE_Chapter20.qxp 7/17/2007 3:38 PM Page 252
METABOLOMIC PROFILING OF OXIDATIVE STRESS BIOMARKERS
DATA FROM THE METABOLOMICS STUDY
GROUP FOR ART
The Metabolomics Study Group for ART has con-
ducted proof of principle studies of gamete and
embryo viability using a proprietary metabolomics
platform and bioinformatics developed by Molecular
Biometrics, LLC, Chester, NJ. Two retrospective
studies (Yale University, Stanford University and
Reproductive Biology Associates of Atlanta) and one
prospective blinded study (Reproductive Medicine
Associates of New Jersey) were performed.
31,32
STUDY AIMS
The Metabolomics Study Group for ART was organ-
ized to test the hypothesis that biomarkers of oxida-
tive stress (OS) could be targeted using metabolomics
technology based on various forms of biospec-
troscopy, coupled with specific bioinformatics, to
assess embryo and gamete viability in IVF; and that
the resulting metabolomic profiles could adequately
distinguish between viable and non-viable embryos
and gametes within a cohort group.
TECHNOLOGY PLATFORM
Free radicals such as reactive oxygen species (ROS)
exert their effect at the molecular level in all cell
types and play a role in both normal physiological
and pathological functions. Complex interactions
between the pro-oxidants and antioxidants are cru-
cial in the maintenance of intracellular homeostasis.
An imbalance in these reactions results in oxidative
stress, which has been implicated in a number of
medical conditions and diseases.
33,34
Moreover, bio-
markers of OS have been found in the male and
female reproductive tracts and are known to affect
the quality of gametes, early embryo development,
and implantation, which in turn affect pregnancy
outcomes. ROS have been shown to influence sperm,
oocytes and embryos and to modulate their interac-
tion in their respective microenvironments.
35
The
technology platform of Molecular Biometrics tar-
gets biomarkers of oxidative stress in culture media
of the developing embryo or in the respective
microenvironment of the gametes, i.e. in follicular
fluid (oocyte) and in seminal plasma (sperm).
Molecular Biometrics technology is based on the
confluence of two scientific disciplines: (1) biospec-
troscopy, the application of different forms of spectral
analysis in human biology that is used to identify,
quantify, and validate proteomic and small molecule
biomarkers; and (2) metabolomics, the science that
systematically examines and integrates the dynamic
interplay between multiple, small molecule biomark-
ers that are uniquely characteristic of complex biolog-
ical functions that define the normal phenotype. The
term ‘biospectroscopy-based metabolomics’ (BSM)
was coined to describe this technology platform.
These methodologies are used in concert to quantify a
sample’s molecular biomarker makeup, specifically,
biomarkers of oxidative stress, and then convert that
data into a novel ‘metabolomic profile’or ‘fingerprint’.
Each profile is analyzed using proprietary bioinfor-
matics and chemometrics to determine the probabil-
ity of normal, competent cellular function versus
the presence or predisposition to abnormal or even
pathological states of cellular activity. The technology
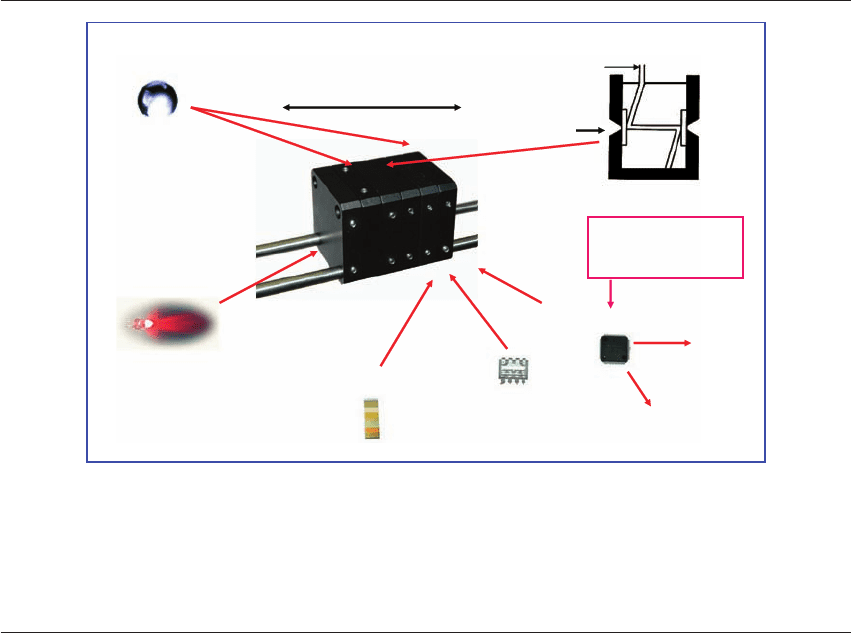
platform is illustrated in Figure 20.2.
Technology Platform
A methodology reduced to practice
Hypothesis driven
Discovery driven
Biomarker targets
(oxidative stress, proteomic, small molecules)
Spectral analysis
(Metabolomic profile by NMR, Raman, NIR)
Chemimetrics and bioinformatics
(Bayesian statistics, Haar transform, Matlab, Genetic algorithm)
Phenotypic function
Figure 20.2 Technology platform. Diagrammatic representation
of the pivotal steps that constitute the technology platform for
assessing biomarkers of oxidative stress using metabolomic
profiling. The platform can be driven from either a hypothesis or
discovery approach and includes biomarker targets, methods of
spectral analysis, chemometric and bioinformatic analysis of data,
leading to an assessment of the phenotypic function of the cell or
organism. NMR, nuclear magnetic resonance; NIR, near infrared.
Courtesy of Molecular Biometrics, LLC, Chester, NJ, USA.
HPE_Chapter20.qxp 7/17/2007 3:38 PM Page 253
HUMAN PREIMPLANTATION EMBRYO SELECTION
Metabolomic profiling was performed using
a small, prototype instrument (6 inches ⫻ 6 inches ⫻
4 inches) that was programmed with proprietary
spectroscopic and bioinformatics capabilities
(Figure 20.3.) Initially, sample cells were filled with
30 microliters of discarded culture media obtained
at the time of embryo assessment and selection. The
original prototype has been replaced by a commer-
cially manufactured instrument having a sample
size of 5–10 microliters. This high-speed procedure
takes about 1 minute to complete.
STUDY DESIGN
Infertility patients enrolling in an IVF program
were treated according to the standard IVF protocol
of the participating program. Informed consent was
Interference filter
Microprocessor
Array
detector
Ball lenses
LED light source
Disposable sample cell
Sample
Light
2.1”
USB To
computer
LED
display
electronic
‘telephone card’
OR
Figure 20.3 Instrument prototype design. The instrument design for obtaining spectral signatures of biomarkers utilizes a disposable
sample cell that contains 5–10 l of specimen. The sample cell is inserted into a fixed position in the light path within the instrument.
The stable light source (left) passes light (energy) into the sample, which then travels through a wavelength filter to select the specified
wavelength for the analysis. The photons of energy which emerge from the filter are captured by a light-detecting diode and delivered
to the bioinformatics microprocessor chip (far right) where the biomarker profiles are calculated. The total time of analysis is
approximately 1 minute. The instrument can be interfaced with any computer system to download data to patient files. This
prototype design has now been incorporated into a commercial product for large scale production.
INSTRUMENTATION
The metabolomics platform described here has
been reduced to practice within the least expensive
and most compact instrumentation available. The
goal was to engineer a generation of affordable,
point-of-care instruments for direct metabolomics
applications that are accessible to all IVF laborato-
ries. Unlike other metabolomics technologies that
rely on very large and costly NMR, FTIR, and MS
instruments, the current metabolomic applications
are based on Raman and NIR spectroscopy. These
instruments are robust, easy to use, do not require
specimen preparation and do not destroy or con-
sume the specimen. The device can interface with
any computer system and can download data
directly into patients’ medical records.
HPE_Chapter20.qxp 7/17/2007 3:38 PM Page 254
METABOLOMIC PROFILING OF OXIDATIVE STRESS BIOMARKERS
obtained from each patient enrolled in the study
and treatment information for that patient, includ-
ing pregnancy outcome data, was noted in the
patient’s medical record. Samples of normally dis-
carded embryo culture media were collected at the
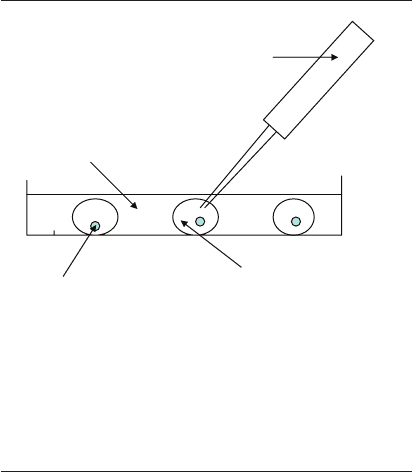
time of embryo selection/transfer (Figure 20.4). All
specimens were placed in Nunc vials and rapidly
frozen in liquid N
2
or dry ice and methanol and stored
at ⫺80°C or liquid N
2
. Appropriate media controls
were also prepared from media droplets that never
received an embryo but were handled in a parallel
manner with media droplets containing developing
embryos.
Pregnancy was confirmed via detection of fetal
heartbeat by ultrasound. The pregnancy outcome
groups were defined as follows: (1) no implantation or
0% pregnancy: regardless of the number of embryos
transferred, no pregnancies were recorded; 50%
pregnancy: four embryos were transferred and two
implanted producing twins while the two corre-
sponding embryos did not implant; and (2) 100%
pregnancy: regardless of the number of embryos
transferred, all embryos implanted and produced a
Sampling pipette
(5–10 µl volume)
Oil
Embryo
Embryo culture media
(35–65 µl microdrop)
Figure 20.4 Non-invasive study design. This illustration depicts
the non-invasive procedure for collecting culture media for
analysis of embryo viability. A small volume of media, usually
5–10 l, is collected directly from the media microdrop contain-
ing the developing embryo. Care is taken to avoid aspiration of
oil. The media is inserted directly into the disposable sample cell
(as described in Figure 20.3) for analysis.
pregnancy (e.g. two embryos transferred, twins;
three embryos transferred, triplets, etc.). All speci-
mens were shipped to McGill University, Montreal
Quebec for metabolomic measurements and bio-
informatic analysis.
METABOLOMIC PROFILING OF OS BIOMARKERS
Specimens were analyzed using both Raman and
near infrared spectroscopy, which measures the
bonds within functional groups of molecules at spe-
cific wavelengths. The spectra obtained from each
instrument were separately analyzed by proprietary
bioinformatics using a wavelength selective genetic
algorithm (GA) to determine regions predictive
of pregnancy outcome as determined by a logistic
regression of light attenuation at specified wave-
lengths. To avoid random correlations, a leave-one
out cross-validation was used. Sensitivity and speci-
ficity were calculated for each assay.
RESULTS
Two different forms of spectroscopic analysis were
used to report metabolomic parameters that were
established on the basis of observed OS biomarker
targets. Spectral profiles describing differences in
-CH, -NH, -SH, C⫽C, and -OH concentrations
showed distinct differences between culture media
of embryos that resulted in pregnancy compared
with those that did not. The ratio of C⫽C to -SH
content in the media that is reflective of oxidative
stress was also different between the two groups
(Figure 20.5). Using GA with Raman spectroscopy,
four spectral regions associated with these molecu-
lar species were reproducibly identified as predictive
of pregnancy outcome in two pilot studies. Compiled
outcomes from the leave-one-out cross-validation
of the logistic regression using Raman measure-
ments resulted in a specificity of 90% and a sensitiv-
ity of 100%.
In a separate blinded study, analysis of media
specimens by NIR required two wavelength regions
identified by GA. The specificity and sensitivity of
these assays was 83% and 73%,respectively. Significant
differences were observed between 0 and 100%
HPE_Chapter20.qxp 7/17/2007 3:38 PM Page 255
