Daniels M.J., Hogan J.W. Missing Data in Longitudinal Studies: Strategies for Bayesian Modeling and Sensitivity Analysis
Подождите немного. Документ загружается.


130 INFERENCE UNDER MAR
C(t
ij
,t
ik
; φ)=g(|t
ij
− t
ik
|; φ). An example is the exponential covariance
function (cf. Example 2.4) given by
C(t
ij
,t
ik
; σ
2
,φ)=σ
2
exp{−φ|t
ij
− t
ik
|}.
The corresponding correlation function is exp(−φ|t
ij
−t
ik
|), which for φ>0
decays exponentially with the lag between times. We use this covariance func-
tion to model the residual autocorrelation of CD4 trajectories in the HERS
data (described in Section 1.5) in Section 7.6.
An example of a nonstationary covariance function is the one based on an
integrated Ornstein-Uhlenbeck process (Taylor, Cumberland, and Sy, 1994),
which takes the form
C(t
ij
,t
ik
; σ
2
,α)=
σ
2
2α
3
{2α min(t
ij
,t
ik
)+exp(−αt
ij
)
+exp(−αt
ik
) − 1 − exp(−α|t
ij
− t
ik
|)} .
This is a structured nonstationary covariance function because the covariance
depends on the lag |t
ij
− t
ik
| between the observation times, and the times
themselves. These covariance functions have been used forlongitudinal CD4
counts (Taylor, Cumberland, and Sy, 1994).
6.5 Covariate-dependent covariance structures
The covariance structure can also depend on covariates. Recent work by Hea-
gerty and Kurland (2001) and Kurland and Heagerty (2004) has shown that
even for complete data, there can be bias in the mean regression coefficients in
generalized linear mixed models if the random effects variance depends on a
between-subject covariate and this dependence is not modeled. The problems
for incomplete data have already been documented.
6.5.1 Covariance/correlation matrices
Acomplication in allowing components of a covariance matrix to depend
on covariates is to ensure the resulting covariance matrices are positive def-
inite. Several parameterizations have been proposed that provide a new set
of parameters that are unconstrained, giving a natural parameterization on
which to introduce covariates. As we did to introduce structure, we use the
GARP/IV parameters of the modified Cholesky decomposition as a way to
introduce covariates. Other approaches are mentioned in Further Reading.
COVARIATE-DEPENDENT STRUCTURES 131
Modeling Σ as a function of covariates using the GARP/IV parameters
Recall that the GARP, {φ
ijk
},areunconstrained,asarethelogof the inno-
vation variances {log(σ
2
ij
)}.Covariatescan therefore be introduced as
φ
ijk
= v
ijk
γ, k =1,...,j− 1,j=2,...,J,
log(σ
2
ij
)=d
ij
λ, j =1,...,J,
where v
ijk
and d
ij
are design matrices for the GARP and log innovation
variances, respectively. These design vectorscontaincovariates of interest.
The form of these models is the same as the structured GARP/IV models,
but now the design vectors are also indexed by i and include covariates.
We again illustrate this approach using thedata from the schizophrenia
clinical trial (described in Section 1.2). The main covariate of interest in this
data was treatment, so we examine the GARP and IV parameters by treat-
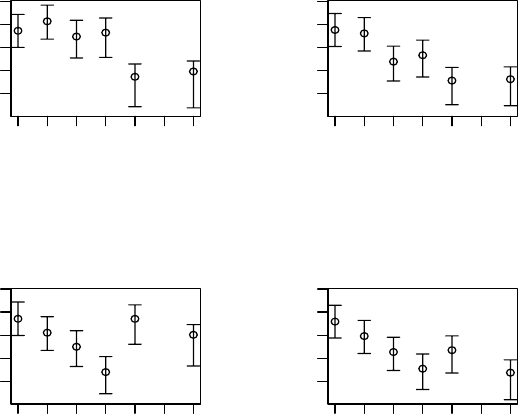
ment. Figure 6.4 shows the posterior means of the log innovation variances
for each treatment. Within each treatment, the innovation variances do not
show much structure as a function of time. However, there appear to be some
large differences across treatments. For example, the innovation variances for
the high dose at weeks4and6are considerably higher than for the other
three treatments. These plots suggest that the innovation variances at weeks
4and6couldbemodeled as a function of treatment.
Figure 6.5 shows the lag-1 GARP for each treatment, again plotted as a
function of time. Here, the lag-1 GARP at week 6 for the medium dose is
much smaller than for the other three treatments (with its credible interval
not overlapping with the standard dose treatment) and we could allow this
GARP to differ by treatment.
This exploratory analysis suggests the covariance matrix for the schizophre-
nia data does depend on treatment, and the GARP/IV parameterization pro-
vides a parsimonious way to model this as the individual parameters can de-
pend (or not) on (a subset of) treatment groups and the resulting covariance
matrices will be guaranteed to be positive definite.
These GARP/IV models will be explored more formally for the Growth
Hormone data in Chapter 7. For a detailed application of these models, see
Pourahmadi and Daniels (2002).
The modified Cholesky parameterization has also been used to introduce
covariates into the random effects covariance matrix in the normal random
effects model (Example 2.1) (Daniels and Zhao, 2003) and could also be used
for the random effects covariance matrixingeneralized linear mixed models
(Example 2.2); however, there should be some implicit or explicit ordering of
the random effects for this parameterization to be fully justified because the
parameterization is not invariant to the ordering of the components of b
i
.Ifthe
components of b
i
were the coefficients of orthogonal polynomials or regression
splines, there is an obvious ordering. For a detailed example of introducing
132 INFERENCE UNDER MAR
0123456
3.5 4.5 5.5
(a)
Weeks
Log(IV)
0123456
3.5 4.5 5.5
(b)
Weeks
Log(IV)
0123456
3.5 4.5 5.5
(c)
Weeks
Log(IV)
0123456
3.5 4.5 5.5
(d)
Weeks
Log(IV)
Figure 6.4 Schizophrenia trial: posterior means of log of the innovation variances
(as a function of week) with 95% credible intervals for each treatment: (a) low dose,
(b) medium dose, (c) high dose, (d) standard treatment.
covariates into the random effects matrix, we refer the reader to Daniels and
Zhao (2003). In Section 7.3, we allow the random effects covariance matrix in
the schizophrenia trial to differ by treatment.
There are other less computationally friendly ways to introduce covari-
ates into a covariance/correlation matrix. In the setting of the multivari-
ate normal model, there has been work on modeling the logarithm of the
marginal variances while keeping the correlation matrix constant across co-
variates (Manly and Rayner, 1987; Barnard, McCulloch, and Meng, 2000;
Pourahmadi, Daniels, and Park, 2007).
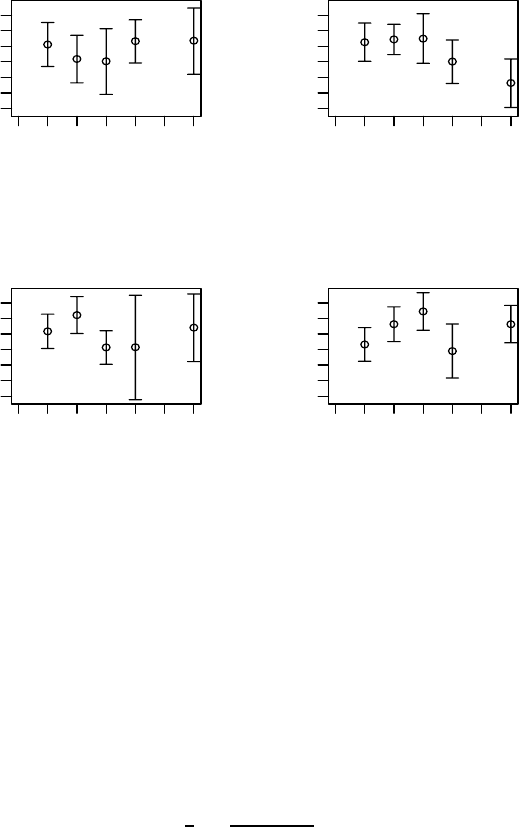
COVARIATE-DEPENDENT STRUCTURES 133
0123456
0.0 0.6 1.2
(a)
Weeks
GARP
0123456
0.0 0.6 1.2
(b)
Weeks
GARP
0123456
0.0 0.6 1.2
(c)
Weeks
GARP
0123456
0.0 0.6 1.2
(d)
Weeks
GARP
Figure 6.5 Schizophrenia trial: posterior means of lag-1 GARP (as a function of
week) with 95% credible intervals for each treatment: (a) low dose, (b) medium dose,
(c) high dose, (d) standard treatment.
Modeling the correlation matrix as a function of covariates in the
multivariate probit model
In the setting of multivariate probit models, we can model the individual
correlations as a function of covariates. The typical approach is to individually
transform the correlations to R using Fisher’s z-transform
z(ρ
ijk
)=
1
2
log
(1 − ρ
ijk
)
(1 + ρ
ijk
)
= v
ijk
γ
(Czado, 2000). This transformation of the individual correlations does not
guarantee the resulting correlation matrices are positive definite. Hence, the
vector of regression coefficients for the correlations γ will be constrained;
within a Gibbs sampling algorithm, values of γ sampled corresponding to a
134 INFERENCE UNDER MAR
non-positive definite correlation matrix will have prior probability zero and
will be rejected within a Metropolis-Hastings algorithm.
Modeling the covariance function in terms of covariates
For temporally misaligned data, similar approaches can be applied to co-
variance/correlation functions. Consider theexponential covariance function
introduced in Example 2.4,
C(t
ij
,t
ik
; σ
2
,φ)=σ
2
exp(−φ|t
ij
− t
ik
|).
Anatural way to introduce covariates here is through log(σ
2
)andlog(φ)
(assuming φ>0) as these transformations provide a set of unconstrained
parameters to maintain positive definiteness of C(·, ·; σ
2
,φ).
6.5.2 Dependence in longitudinal binary models
Forlongitudinal binary data models based on underlying normal latent vari-
ables (Example 2.6) or random effects (Example 2.2), covariates can be intro-
duced into their corresponding correlation/covariance matrix as discussed in
the previous section. For (marginalized)transition models, covariates can be
introduced through the (unconstrained) Markov dependence parameters.
Recall a marginalized transition model of order one (MTM(1)) has (Markov)
dependence parameters γ
ij
, j =2,...,J.Theγ
ij
are (unconstrained) log odds
ratios that can be modeled as a function of covariates via
γ
ij
= v
ij
α,
with few additional computations compared to models without covariates. For
higher-order MTMs, the dependence parameters corresponding to lags larger
than one also are unconstrained and can be similarly modeled.
These models are fit to data from the CTQ I trial in Section 7.4.
6.6 Joint models for multivariate processes
Multiple longitudinal processes are typically modeled separately, despite po-
tential gains in efficiency from modeling them jointly, unless there is inter-
est specifically in the relationship between the multiple processes (Liu and
Daniels, 2007). In the case of incomplete data, there are additional reasons
to build and base inference on joint models. Joint modeling allows the use of
all available data, on both the process of interest and the other processes, to
‘impute’ the missing values on the processofinterest. This can be especially
advantageous when (1) the processes are not all observed at the same times
and observed responses for other processes are ‘closer’ temporally than ob-
served responses from the same processand(2)the correlation between the
JOINT MODELS FOR MULTIVARIATE PROCESSES 135
processes is strong. By modeling the processes separately, we do not use the
information from the other processes to fill in the missing values.
Our primary interest here in developing joint models will be to incorporate
auxiliary variables under the auxiliary variable MAR (A-MAR) assumption
(Definition 5.11), where missingness in Y is MAR after conditioning on the
observed responses y
obs
,modelcovariates X, and auxiliary covariates V .Let
V
obs
be the observed values of the auxiliary covariate process. Then A-MAR
corresponds to the following form of the missing data mechanism,
p(r | y, x, v
obs
)=p(r | y
obs
, x, v
obs
).
In Bayesian inference, to allow for A-MAR mechanisms, we need to specify
the joint distribution p(y, v | x)andthenintegrate out v
p(y | x)=
p(y, v | x)dv (6.15)
to obtain the full data response model of interest; from a practical perspective,
joint models for which this integral can be obtained in closed form would be
preferred. When the auxiliary covariate(s) are time-varying, joint longitudinal
models can be constructed for the auxiliary process V and the process of
interest Y .
For the CTQ II example (Section 1.4), the A-MAR assumption implies
that probability of missingness of cessation status at time t is independent of
the unobserved cessation status at time t conditional on the previous weeks’
smoking responses and the previous weeks’ weight change responses (as op-
posed to conditional on just the previous weeks’ smoking responses). In a data
augmentation step, we draw Y
mis
from p(y
mis
| y
obs
, v
obs
), not p(y
mis
| y
obs
).
Hence, the A-MAR assumption is weaker than MAR to the extent that we can
correctly specify the distribution p(y, v | x). In the following, we review and
introduce some models for multivariate processes that are convenient when
one of the processes is of primary interest and the other process is an aux-
iliary covariate process (Carey and Rosner, 2001; Gueorgeiva and Agresti,
2001; O’Brien and Dunson, 2004; Liu and Daniels, 2007). In particular, the
models presented here lend themselves to the setting of auxiliary time-varying
covariates in that the full data responsemodelofinterestcan be obtained in
closed form (cf. (6.15)). We fit one of these models to the CTQ II smoking
cessation data in Section 7.5.
In the following examples, we denote Y
ij
as the primary response of interest
and V
ij
as the auxiliary covariate process at time t
ij
.
6.6.1 Continuous response and continuous auxiliary covariate
As a starting point, we assume both processes are (potentially) measured at
the same set of observation times (t
ij
= t
j
). For continuous data, the multi-
variate normal distribution is an obvious choice for the joint distribution of the
136 INFERENCE UNDER MAR
longitudinal processes. We model the joint distribution of the two processes,
(Y
i
, V
i
), as
/
Y
i
V
i
0
,
,
,
,
,
x
i
∼ N (x
i
β, Σ),
where
x
i
=
/
x
iy
x
iv
0
Σ =
/
Σ
yy
Σ
yv
Σ
T
yv
Σ
vv
0
.
The extension of this model to more than one auxiliary covariate process is
obvious. The dimension of Σ can be quite large, so structure is often imposed
on Σ using latent variables (Roy and Lin, 2000) or by inducing structure
directly on Σ (Carey and Rosner, 2001). To illustrate, consider models for
CD4 where the auxiliary covariate is viral load.
Consider the following (serial correlation) structure for Σ (Carey and Ros-
ner, 2001),
cov(Y
ij
,Y
ij
)=σ
2
y
γ
|t
j
−t
j
|
θ
y
y
cov(V
ij
,V
ij
)=σ
2
v
γ
|t
j
−t
j
|
θ
v
v
cov(Y
ij
,V
ij
)=σ
y
σ
v
γ
|t
j
−t
j
+1|
θ
yv
yv
.
The first two covariances correspond to within-process covariances for the
primary response (CD4) and the auxiliary covariate (viral load), and the third
corresponds to the between-process covariance. For each, it is assumed that
the correlation decreasesforresponses farther apart in time by restricting γ
y
,
γ
v
,andγ
yv
to be in [0, 1). The parameter γ
yv
determines the relationship
between CD4 and viral load; if γ
yv
=0,thenthey are independent processes.
Missing data is imputed in the data augmentation step by constructing
the appropriate conditional distributions from this multivariate normal model
with a structured covariance matrix. Clearly, this form of Σ implies that
observed responses closer in time tothemissing responses will carry more
weight in the imputation and the magnitude of γ
yv
will determine how much
information is used from the auxiliary covariates process to fill in values for
the primary process.
Multivariate normal models are natural for handling time-varying continu-
ous(auxiliary) covariates as the conditional distribution of one process given
the other can be written downinclosedform(asalinearregression)aswell
as the marginal distribution of the primary process of interest using standard
multivariate normal results.
JOINT MODELS FOR MULTIVARIATE PROCESSES 137
The covariance structures described here also can be used for misaligned
measurement times by inducing a structured covariance function within a
Gaussian process.
6.6.2 Binary response and binary auxiliary covariate
Models for a binary response and binary auxiliary covariate can be specified
similarly to those described in Section 6.6.1. Let Y
ij
= I{Z
Y
ij
> 0} and V
ij
=
I{Z
V
ij
> 0} where Z
Y
i
=(Z
Y
i1
,...,Z
Y
iJ
)
T
and Z
V
i
=(Z
V
i1
,...,Z
V
iJ
)
T
are latent
variables modeled as
/
Z
Y
i
Z
V
i
0
,
,
,
,
,
x
i
∼ N (x
i
β, Σ)
with
x
i
=
/
x
iy
x
iv
0
Σ =
/
Σ
yy
Σ
yv
Σ
T
yv
Σ
vv
0
.
The covariance matrix Σ is a correlation matrix for identifiability (cf. the
multivariate probit model in Example 2.6). Models using latent normal for-
mulations are convenient for specification of dependence and computations.
Recent work has also considered allowing the latent variables to follow a
multivariate t-distribution (O’Brien and Dunson, 2004). One reason for using
a t-distribution is that when the scale matrix and degrees of freedom are appro-
priately specified, the multivariate t-distribution approximates a multivariate
logistic model; that is, the marginal distribution of Z
Y
ij
follows (approximately)
alogistic distribution, which in turn implies P (Y
ijk
=1| x
i
)follows a logistic
regression model. Computations are as easy as the probit model given that
the multivariate t-distribution can be re-expressed as a gamma mixture of
multivariate normals (see Example 3.16).
Similar to the model described in Section 6.6.1, this class of models is well-
suited for handling time-varying auxiliary covariates as the conditional and
marginal distributions of the process of interest take a simple form due to the
underlying normal (or t-) latent structure.
We finish our discussion of joint modeling in the next section by introducing
amodel for a binary primary process of interest and a continuous auxiliary
covariate (or vice versa). This is motivated by the CTQ II smoking cessation
data (Section 1.4) where the primary process of interest was smoking cessation
(binary) and the auxiliary covariate was weight change (continuous).
138 INFERENCE UNDER MAR
6.6.3 Binary response and continuous auxiliary covariate
Astraightforward way to induce correlation between a continuous and a bi-
nary longitudinal process is to specify a joint multivariate normal distribution
for the continuous responses and the latent variables underlying the binary
process, with the restriction that the marginal covariance matrix correspond-
ing to the binary responses is a correlation matrix. This model has appeared
in various forms in the literature (Catalano and Ryan, 1992; Gueorgeiva and
Agresti, 2001; Liu and Daniels, 2007).
As in Section 6.6.2, Y
ij
= I{Z
ij
> 0} where Z is the vector of latent
variables underlying the vector of longitudinal binary responses Y ,and(Z, V )
are jointly multivariate normal as follows:
/
Z
i
V
i
0
,
,
,
,
,
x
i
∼ N (x
i
β, Σ),
where, as above,
x
i
=
/
x
iy
x
iv
0
Σ =
/
Σ
yy
Σ
yv
Σ
T
yv
Σ
vv
0
,
with Σ
yy
acorrelation matrix. Notice that the marginal distribution of Z
i
is
multivariate normal and the marginal distribution of Y
i
follows a multivari-
ate probit model.Thesubmatrix Σ
yv
characterizes the dependence between
the continuous and binary longitudinal processes. If Σ
yv
= 0,thenthe two
longitudinal processes are independent.
This model is used to address an auxiliary covariate (process) in the CTQ II
smoking cessation data in Section 7.5.
6.7 Model selection and model fit under ignorability
We now propose some modifications of the techniques for model comparison
and for assessing model fit, first introduced in Chapter 3, that can be extended
to incomplete data settings (under ignorability). Before we discuss these mod-
ifications, we remind the reader that we can only assess the fit of the full-data
model to the observed data;theadequacy of the model for the missing data
cannot be ascertained. Different full-data models that provide the same fit to
the observed data should be indistinguishable when using sensible model se-
lection criteria even though they may make very different assumptions about
the missing data.
Because the focus of this chapter has been on ignorable missingness, we
MODEL SELECTION AND MODEL FIT 139
assess the fit of the model using only the full-data response model p(y | θ),
and not the full data model p(y, r | ω)because it was unnecessary to specify
p(r | y
obs
, ψ). For nonignorable mechanisms, we do need to use p(y, r | ω);
this will be presented in detail in Chapter 8.
6.7.1 Deviance information criterion (DIC)
With complete data, the DIC is constructed using the likelihood based on the
full-data response model. The complication in using the DIC with incomplete
data under ignorability is that we do not observe the full data. The literature
is sparse on recommendations for constructing the DIC from incomplete data.
We consider two constructions based on those used in Pourahmadi and Daniels
(2002), Ilk and Daniels (2007) and Celeux et al. (2007). The recommendations
in Celeux et al., however, may not generalize well to our setting as they deal
with missing data that are latent (e.g., in the context of mixture and random
effects models). Latent data are of course fundamentally different than missing
response data. First, we can never observe the latent data, but we could have
observed the missing response data. Second, the observed vector of missingness
indicators has no analog in random effects and mixture models.
DIC based on the observed data likelihood
Afirst,andperhaps most obvious construction based on the development
in Chapter 5 and Section 6.1 would betoconstructtheDICbasedonthe
observed data likelihood L(θ | y
obs
),
DIC
O
=DIC
obs
(y
obs
)=−4E{ (θ | y
obs
)} +2(θ | y
obs
),
where
θ = E(θ | y
obs
). This approach has been applied by Pourahmadi and
Daniels (2002) and Ilk and Daniels (2007) and typically can be computed
directly in WinBUGS.
DIC based on the full-data likelihood
An alternative approach would be to constructtheDICbased on the full-data
response model likelihood,
DIC
full
(y
obs
, y
mis
)=−4E
θ
{(θ | y
obs
, y
mis
)} +2{θ(y
mis
) | y
obs
, y
mis
},
where E
θ
(·)istheexpectation with respect to p(θ | y)and
θ(y
mis
)=
E(θ | y
obs
, y
mis
). Since y
mis
is not observed, we take the expectation of
DIC
full
(y
obs
, y
mis
)withrespect to the posterior predictive distribution p(y
mis
|
