Castilho Leda R., Moraes Angela Maria (Ed.) Animal Cell Technology: From Biopharmaceuticals to Gene Therapy
Подождите немного. Документ загружается.


affinity for the operon. The benefit of this system is that the effector of the
induction is the culture temperature. A similar inducible system regulated
by tetracycline allows the expression of a gene by induction (Tet-On
1
)or
repression (Tet-Off
1
) in the presence of tetracycline or derivatives. This is
accomplished by modifications in the repressor protein, such as mutations
and/or fusions to repressors or transactivators. This system has the
advantage of low toxicity. Pleiotropic effects are its major drawback,
together with the loss of control in the repression (leakiness), evidenced
by a basal synthesis of protein in the absence of inducer (Blau and Rossi,
1999).
Other alternatives are based on mutated versions of endogenous ele-
ments that have lost the capacity to respond to the endogenous inducers or
to bind genomic sequences of animals, for example: (i) the systems
modulated by receptors of the steroid hormones that are induced with
mifepristone (RU486), an antagonist of progesterone; (ii) artificial chi-
meras that respond to ecdysone, a hormone that regulates the molting
process in insects; or (iii) human immunophilins (FK506 or FRAP), which
can be induced by rapamycin (Papadakis et al., 2004).
In some cases, it is preferable that the expression of the target gene
occurs in a predetermined phase of the cell cycle or growth and/or in a
specific cell type. The recent findings of promoters that fulfill these
requirements facilitated synchronizing the expression of recombinant
proteins with the cell cycle, and enlarged the list of suitable expression
hosts (Blau and Rossi, 1999; Nettelbeck et al., 1999).
Targeting signal sequences and tags. Transport signals guide readily
synthesized proteins through the different intracellular trafficking routes
and dictate their final location. The targeting signals appear in the form of
an N-terminal sequence (15–60 amino acids), which is generally removed
when the polypeptide reaches its destiny, and/or as a three-dimensional
peptide structure exposed on the surface of the mature protein. The
putative locations can be: the cytosol, peroxisomes, nucleus, mitochondria,
endoplasmic reticulum, Golgi apparatus, secretory lysosomes, vesicles, or
cellular surface. With regard to the production of recombinant proteins in
animal cells, it is preferred that the product is secreted to the culture
medium, thus simplifying its further isolation and purification. If the gene
to be expressed lacks a signal peptide for secretion, this can be either
introduced by genetic engineering or incorporated into the expression
vector. Some examples of secretion signals are found in the N-terminal
sequences of tissue plasminogen activator (tPA) and IgG1. The removal of
a targeting sequence from the gene of interest is only advisable: (i) if this
signal is not essential for the required post-translational processing of the
protein, since these modifications may affect the biological activity of the
product; (ii) if the accumulation of the recombinant protein in a given
cellular compartment may lead to toxicity or complicate its purification.
Tags comprise sequences that go from a few amino acids (His-, FLAG-
or c-myc-tags) to large polypeptides (serum albumin, protein A or G,
glutathione S-transferase, growth hormone, GFP, CAT, etc.). The purpose
of a tag is to confer a novel property such as an increased expression,
solubility, stability, and half-life (e.g. serum albumin, protein G, EBV-1
antigen), a specific affinity to simplify its isolation and purification (e.g.
52 Animal Cell Technology

GST, calmodulin-binding peptide, His-, FLAG-tag, protein A, glycopro-
tein D of HSV), or to enable its detection and/or selection, i.e. tags based
on protein-reporters (-galactosidase, GFP, CAT, hGH) (Makrides,
1999). Tags can be fused to N- or C-terminal ends of the protein and a site
for proteolytic cleavage is commonly included to eliminate the tag upon
exploitation of its functionality. The proteases most commonly used are
thrombin, enterokinase, factor Xa, and TEV (catalytic domain of Nia, the
nuclear inclusion protein from tobacco etch virus).
Positioning effect and site-directed chromosomal integration. The
term ‘‘positioning effect’’ refers to the factors associated with the struct-
ural organization of the chromatin that have an influence on the rate of
transcription of a heterologous gene (Zahn-Zabal et al., 2001). These
factors involve the condensation state of the chromatin, the direction and
location of the foreign gene with respect to other genes, and the structural
elements in the chromosomes of the host cell (Wurm, 2004). It must be
emphasized that nearly 95% of chromatin appears as heterochromatin,
which is transcriptionally inactive. In contrast, the euchromatin is less
condensed and transcriptionally active, wherein gene expression is influ-
enced by structural elements (i.e. LCRs, SARs, insulators, boundaries) that
determine the transcription of the locus in a cell-specific and/or cell cycle-
dependent manner (Bode et al., 2003). Given that chromosomal integration
of a heterologous gene is a random process, it becomes easy to understand
why the chances for integration into a transcriptionally active region of the
genome are low and, hence, why the expression pattern of the recombinant
product is heterogeneous and gives rise to clonal variation.
At present, there are three different alternatives to overcome the
positioning effect: (i) use of expression vectors containing elements such as
insulators, boundaries, SARs, and LCRs, and conserved anti-repressor
elements whose function is to generate a transcriptionally favorable
environment at the integration site (Zahn-Zabal et al., 2001); (ii) the
addition of butyrate or tricostatin to the culture, which block the deacety-
lation of histones and induce a structural relaxation of the chromatin,
making sites that were transcriptionally inactive accessible (Gorman et al.,
1983); and (iii) site-directed chromosomal integration of the expression
cassette by means of the CRE/LoxP of bacteriophage P1 or FLP/FRT
systems (for details see Section 3.6.2).
Expression of multiple genes. In animal cells most of the mRNA is
monocistronic and encodes a single protein. In some circumstances, a
coordinated expression of two or more heterologous genes may be
required. This is the case for the establishment of a stable cell line using a
selection marker, for metabolic engineering, or for the expression of
protein complexes (i.e. antibodies). Some of the strategies used with that
aim include: (i) the expression from a single vector of multiple genes, each
one controlled by its own promoter; (ii) the creation of fusion proteins,
where the genes are placed in tandem and separated by linkers, the expres-
sion being controlled by a single promoter; (iii) the transfection with
multiple expression vectors, each one carrying a single and distinct gene
and selection marker; (iv) the expression of large monocistronic transcripts
containing the different cDNAs of each gene linked by sequences encod-
ing protease cleavage sites, i.e. furine protease, which will subsequently
Cloning and expression of heterologous proteins in animal cells 53

allow each single polypeptide to be obtained; and (v) the use of IRES
(internal ribosomal entry site) elements located upstream of the gene
sequence, which allow binding of the ribosomal complex and, therefore,
the initiation of the translation of each individual open reading frame in a
cap-independent manner (Gurtu et al., 1996).
3.5 Cell lines and biotechnological processes
The choice of a suitable host cell line for recombinant protein production
depends on many considerations. On one hand, the host cell must show a
high transfectability, be able to transcribe, translate, fold, and process the
protein and, if possible, to secrete it to the culture medium. On the other
hand, it is advisable that the selected cell line grows in a serum-free
medium because recovery of the recombinant protein from culture media
with a low protein content is simpler (Mather, 1990). An intrinsic
resistance of the cells to shear forces and mechanical damage due to
agitation, aeration, and bubbling processes is also desirable. Other aspects
to be considered are the post-translational modifications that the protein
of interest requires and the optimal culture conditions of the selected cell
line (Andersen et al., 2000). In this regard, different studies have demon-
strated that changes in the carbohydrate content can alter the antigenicity,
stability, solubility, tertiary structure, biological activity, or in vivo half-
life of the recombinant protein (Delente, 1985). Nowadays, cell lines
displaying novel biochemical properties can be generated by means of
metabolic engineering. These modifications aim: (i) to allow robust cell
growth and/or inhibit apoptosis; (ii) to reduce the secretion of toxic
products and/or the consumption of nutrients; (iii) to express glycosyl-
transferases; and (iv) to control cell proliferation and direct energy
metabolism towards protein synthesis (Sanders, 1990). Relevant informa-
tion about metabolic engineering can be obtained from Kaufmann and
Fussenegger (2003).
Although numerous cell lines have been screened for their efficiency as
a host system for recombinant protein production, only a few have shown
favorable properties for the expression of biopharmaceuticals (Hauser,
1997). Regulatory and economic issues for large-scale production and the
intended application of the recombinant protein (diagnosis, therapy, etc.)
have to be carefully considered (Makrides and Prentice, 2003). Three
mammalian cell lines are now commonly used by the pharmaceutical
industry: Chinese hamster ovary (CHO) cells, the murine myeloma SP2/0
and the NS0 cell line (see Table 3.1). These cell lines have been used to
produce 11 of 21 therapeutic products approved from 1996 to 2000 (Chu
and Robinson, 2001).
3.6 Expression in animal cells
Once the DNA encoding the protein of interest is introduced into the
cells, expression may be transitory over a period of days or weeks until the
DNA is lost from the population. The ability to express the heterologous
DNA during a short period of time is called ‘‘transient expression’’ (Kauf-
54 Animal Cell Technology

man, 2000). A few transfected cells may incorporate the exogenous DNA
into their genome by a recombination mechanism leading to the ‘‘stable
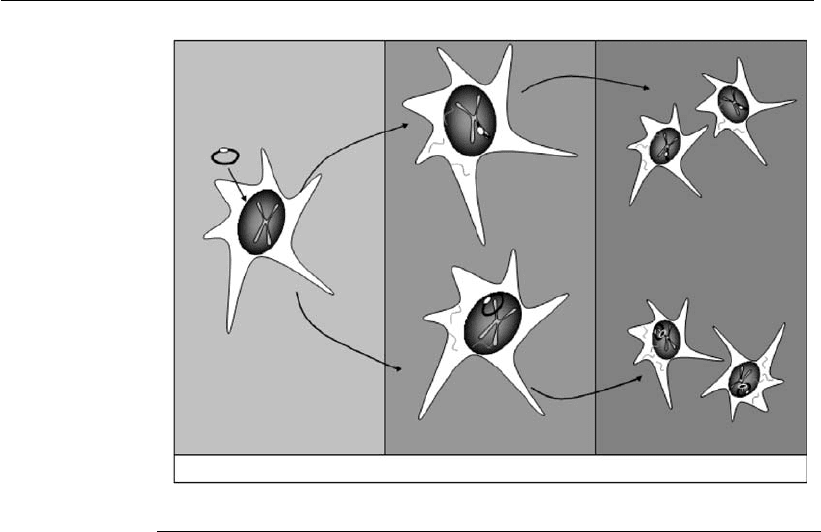
expression’’ of a gene (Kaufman, 1997). Figure 3.3 shows the different
phases involving the expression of a heterologous gene.
3.6.1 Transient expression
In general, transient expression allows fast production of a desired protein
to characterize it or to verify the integrity, functionality, and efficiency of
different recombinant vectors. This represents a convenient means of
comparing the performance of different vectors and ensuring their func-
tionality before the more laborious procedure of isolating and characteriz-
ing stably transfected cell clones (Kaufman, 1997; Makrides, 2003).
Transient expression obviates the clonal variation commonly observed in
stable transfections. The production of large amounts of recombinant
protein has recently been reported for transient expression systems on
large scales (Wurm and Bernard, 1999, 2001; Girard et al., 2002; Derouazi
et al., 2004).
Different cell lines can be used for transient expression. These include
COS, BHK, CHO, and two variants of genetically modified HEK-293
cells: HEK-293 T (Kim et al., 1997) and HEK-293 EBNA (Cachianes
et al., 1993). COS cells were generated by transfection of CV1 cells
(African green monkey kidney cells) with a mutant of the SV-40 virus that
has a defective replication origin but encodes the T antigen, necessary to
initiate viral replication. Hence, only vectors defective in the T antigen and
containing an SV-40 replication origin can multiply in these cells (Gluz-
man, 1981). This host/vector system allows the amplification of the copy
number of plasmid DNA (approximately 10 000 copies per cell), yielding
Table 3.1 Cell lines commonly used in the biopharmaceutical industry
Cell line Source Applications
COS Monkey Transient expression
Production of recombinant viruses
HEK-293 Human Transient and stable expression
Production of recombinant viruses
BHK-21 Hamster Transient and stable expression
Vaccine production
CHO.K1, CHO dhfr- Hamster Transient and stable expression
Hybridomas, NS0, SP2/0 Mouse Stable expression
Monoclonal antibodies production
MDCK Dog Stable expression
Vaccines production
Per.C6
TM
Human Stable expression
Production of recombinant viruses and
vaccines
Vero Monkey Vaccine production
Sf9, Sf21 Insect Production of recombinant proteins and
baculoviruses
Tn-368, High-Five
1
BTI-TN-5B1-4
Insect Recombinant protein production
Cloning and expression of heterologous proteins in animal cells 55
a high protein expression that is followed by cell lysis a few days after
transfection (Kaufman, 1997; Makrides, 2003). A novel cell line (named
HBK) showing high transient expression levels was recently generated
(Cho et al., 2002). This is a somatic hybrid of two human cell lines, HEK-
293 and Burkitt lymphoma cells, which combines high transfectability and
capacity for growth in suspension from each cell line, respectively.
3.6.2 Stable expression
In the case of stable transfections, the selection agent is added to the
cultures approximately 2 days post-transfection. The selection phase
normally lasts days to weeks. Those cells surviving the selection pressure
harbor the selection marker, grow as discrete colonies, and can either
express or not the protein of interest. These transfectants are soon isolated
by means of traditional techniques, which include single cell cloning by
cylinders, limiting dilution or soft agar. These methods have nowadays
been replaced at the industrial level by fully automated techniques that are
faster, less laborious, and enable high-throughput screening, for example,
Transfection
Transient expression
Selection Stable clones
Selection
medium
Integrated
stable
expression
Episomal stable
expression
Days:
0 1–3 3–14 7–14
Figure 3.3
Expression of a heterologous gene in mammalian cells. The transfection of host
cells with the recombinant vector can be performed by different methods (see
Section 3.7). During t he initial phase, first to third day upon t ransfection, the cells
express the gene transiently. In the absence of selection agent, the DNA is
eventually lost from the cell population. In a small proportion of transfected cells,
the exogenous DNA is incorporated, by non-homologous recombination, leading
to the generation of cell lines that stably express the gene of inter est. If the vector
contains episomal replication elements, then a stable episomal expression of the
exogenous gene takes place. If the vector integrates into the cell genome, then a
process known as integrated stable expression occurs.
56 Animal Cell Technology

by flow cytometry and using robots (Wurm, 2004). It is important to
stress that generally less than 1% of a cell population that transiently
expresses a gene will give rise to stable cell lines (Jordan and Wurm, 2003).
Table 3.2 compares the main advantages and disadvantages of transient
and stable expressions.
An alternative and novel strategy for the efficient generation of stable
cell lines uses the Cre/LoxP and the FLP/FRT recombination systems to
drive a site-specific integration of the heterologous gene. The Cre (cycliza-
tion recombination) recombinase from the P1 bacteriophage recombines
the DNA in a 34 bp site, known as LoxP (locus of crossover of P1),
whereas the FLP recombinase, isolated from the Saccharomyces cerevisiae
2 m plasmid, recognizes the FRT site (FLP recombination target), whose
size is 48 bp. The first phase of this method consists of selecting cell lines
that have randomly integrated a reporter gene flanked by LoxP or FLP
sites into a transcriptionally active chromosomal locus. In a second phase,
the reporter gene is replaced by the gene of interest through a recombina-
tion process mediated by Cre or FLP recombinases (Makrides, 2003).
Recently, Bode et al. (2003) have explored the potential of structural
elements located in the chromatin to direct a site-specific integration of
the gene, and were able to isolate high producer cell lines. A host/vector
system that makes use of FLP/FRT elements and different modified cell
lines, containing a single FRT site in a transcriptionally active locus, is
commercially available (Flp-In
TM
expression vectors, Invitrogen).
The success of transfection, either stable or transient, depends on several
factors that must be taken into account and can be summarized as follows:
(i) the transfectability and physiology of the cell line; (ii) the characteristic
of the genetic marker in the expression vector; (iii) the type of expression
desired; (iv) the size of the expression cassette and the quality of the DNA
to be introduced; (v) the compatibility of the transfection method and/or
reagents with the cell line; (vi) the type of assay to be used for detection of
Table 3.2 Comparis on of transient and stable expression
Transient expression Stable expression
Short time-frame for the generation of
the product
Long time-frame for the generation of the
product
Simple, does not require selection
marker
Selection and/or amplification markers
are required
No need to screen for recombinant
clones
Tedious and time-consuming screening
of the recombinant clones
Expression levels are not influenced by
the ‘‘positioning effect’’
The expression levels are influenced by
the ‘‘positioning effect’’ and the gene
copy number
Useful to confirm the integrity of
expression vectors, convenient for high-
throughput screening methods, e.g. to
study different mutant genes
Once a stable expressing clone is
identified, it constitutes an unlimited
source for protein production
Circular DNA is used The use of linearized or circular DNA
depends on the transfection method,
but, in general, linearized DNA is
advisable
Cloning and expression of heterologous proteins in animal cells 57
the recombinant product; and (vii) the presence of fetal bovine serum and/
or antibiotics in the culture medium. Additionally, the selection of an
appropriate vector is as important as the selection of the cell line and/or of
the transfection conditions.
3.7 Introduction of DNA into mammalian cells
A wide variety of methodologies and reagents are currently employed to
introduce different molecules into eukaryotic cells. The incorporation of
DNA can be achieved by two different mechanisms: infection or transfec-
tion. The first consists of a biological process mediated by a virus (the viral
infection of cells is mediated by receptors), while the second makes use of
physical or biochemical methods to incorporate the DNA into the cell.
Although the virus-mediated methods are more efficient, they are more
laborious and time-consuming compared with transfection. Additionally,
the nature of the infection process requires the presence of virus-specific
receptors in the host cell to allow viral penetration, which restricts the
spectrum of possible host cells. Another limitation of viral infection as a
method for DNA transfer is that, unlike plasmid transfection, it is not
possible to simultaneously transfer multiple recombinant viruses into the
cell (Wurm and Bernard, 1999).
In the next sections, the transfection methods most commonly used in
cell culture laboratories will be described. It is difficult to predict the best
method for transfection, so the expression vector, the cell type, and the
facilities of each laboratory should be taken into account before deciding
which technique to apply (Wurm, 2004). For infection techniques, an
updated compilation can be obtained from Heiser (2004).
3.7.1 Calcium phosphate co-precipitation method
Although this technique was originally used to increase the infectivity of
adenoviral DNA (Graham and Van der Eb, 1973), it became more popular
after extending its application to plasmid DNA (Maitland and McDougall,
1977; Wigler et al., 1977). Although the original method has been fre-
quently modified to increase the transfection efficiency, it consists basi-
cally of mixing purified DNA with calcium chloride and phosphate buffer
at neutral pH, which results in the formation of a fine, visible precipitate.
The precipitate (DNA–calcium phosphate complex), when added to the
cells, is incorporated by phagocytosis. The complexes fuse to the phago-
somes and are transported to different cellular organelles, including the
nucleus (Chen and Okayama, 1991). For many cell lines, the transfection
efficiency can be further increased by means of short exposures of the cells
to chemical agents like glycerol or dimethyl sulfoxide (DMSO) in concen-
trations between 10 and 20%. Variables such as pH, precipitate quality,
incubation time with the precipitate, DNA concentration, cellular state,
temperature, and concentrations of chemical agents, can influence the
transfection efficiency. This method is widely used because of its low cost,
the lack of requirements for sophisticated infrastructure, and its applic-
ability to a wide range of different adherent and suspension cell lines. On
58 Animal Cell Technology

the other hand, cytotoxicity, high mutation rates, and the need for
optimization and standardization of the transfection conditions are recog-
nized as the major disadvantages of this method. The critical parameters to
obtain an optimal precipitate were very well described by Chen and
Okayama (1991) and Jordan et al. (1996). Girard et al. (2002) reported the
establishment of a large-scale transfection method (100 L bioreactor) for
HEK-293 cells in suspension using calcium phosphate. More recently,
Lindell et al. (2004) developed a new technique named calfection, which
consists of the addition of a calcium chloride/DNA mixture to the culture
medium in agitation. Although this method presents good scale-up poten-
tial, the mechanism by which the DNA is incorporated remains unknown,
which complicates optimization of the process.
3.7.2 Cationic polymers
Several polycations with a good buffering capacity below physiological
pH, such as lipopolyamines and polyamidoamine polymers, have proved
to be efficient transfection agents (Boussif et al., 1995). Two of the most
popular methods based on polycations are discussed below: diethylami-
noethyl-dextran (DEAE-dextran) and polyethylenimine (PEI).
DEAE-dextran. Like the calcium phosphate co-precipitation method,
the DEAE-dextran technique was originally developed to increase the
viral infectivity of animal cells, and its application was later extended to
transfection processes. Although it is simple, efficient, and appropriate for
transient expression, its use for stable transfections has not given satisfac-
tory results. The transfection efficiency of this method can be increased by
treating cells with glycerol or DMSO. The DNA is incorporated by
endocytosis, and thus exposed to extreme pH levels and cellular nucleases,
which may explain, to a certain extent, the high frequency of mutations
observed when transfecting by this method (Calos et al., 1983). This
transfection technique can be applied to both adherent and suspension cell
lines. For detailed transfection protocols, the works by Keown et al.
(1990) and Kaufman (1997, 2000) are recommended.
PEI. Boussif et al. (1995) reported for the first time the use of PEI as a
vehicle for gene delivery into cells and, since then, there has been a
growing interest in the in vitro and in vivo applications of this method to
animal cells. PEI is an organic polymer with a high density of amino
groups that can be protonated. At physiological pH, the polycation
presents a high affinity for binding DNA and can mediate the transfection
of eukaryotic cells (Boussif et al., 1995). PEI presents two types of
structures, linear or branched, with variable molecular weights. Linear PEI
was reported to display a higher gene transfer efficiency under serum-free
conditions in CHO cells in comparison with branched PEI (Derouazi
et al., 2004). Moreover, the linear form with a molecular weight of 25 kDa
showed the highest level of recombinant protein expression while prevent-
ing aggregation and attachment of cells grown in suspension. Although the
precise mechanism mediating gene transfer by PEI remains unknown
(Bertschinger et al., 2004), it has been postulated that the buffer capacity
of the polycation at the lysosomal level would protect the DNA:PEI
particles from nuclease degradation (Boussif et al., 1995). The critical
Cloning and expression of heterologous proteins in animal cells 59

parameters for PEI-mediated transfections are the amount of DNA, the
PEI/DNA ratio, the cell density at the time of transfection, and the order
of addition of reagents. In fact, Boussif et al. (1995) described that the
transfection efficiency is one order of magnitude higher when the cationic
polymer is added to the plasmid solution rather than vice versa. This
method has been demonstrated to be effective for primary cultures and cell
lines, for adherent and suspension cells, and for serum-containing and
serum-free media. In addition, the cost of the PEI-based method enables
its use in transient transfections on a large scale (Boussif
et al., 1995; Derouazi et al., 2004).
3.7.3 Lipid-mediated gene transfer (lipofection)
Since the first work of Felgner et al. (1987), describing the use of a
synthetic cationic lipid for transfection, more than a dozen cationic
liposomes have been developed (Gao and Huang, 1995). Under optimal
conditions, the cationic lipids form small unilamellar liposomes when
formulated in water. The surface of these liposomes is positively charged
and interacts with the phosphate backbone of the DNA, forming com-
plexes that present high affinity for the negatively charged surface of the
cell membrane. The uptake and delivery of lipid–DNA complexes into
the intracellular compartment is mediated by endocytosis. Normally,
cationic liposomes contain an amphiphilic cationic lipid (DOSPA,
DOTMA, etc.) and a neutral helper lipid, generally DOPE. DOPE is
needed in the case of cationic lipids that do not form bilayers in order to
stabilize the formation of the cationic liposome (Gao and Huang, 1995).
This innovative technology stands out as being easy to accomplish, and its
diverse applications offer reproducible results associated with very good
yields. The large variety of formulations available has conferred to
lipofection a great versatility of applications: stable cell lines and primary
cultures, transient and stable transfections, adherent or suspension cells.
Different formulations presenting high transfection efficiencies are com-
mercialized by several companies. Unfortunately, the high cost of these
products precludes their use in large-scale processes. Multivalent cationic
lipids present better transfection efficiencies than those composed of
monovalent cationic lipids (Behr et al., 1989). For example,
Lipofectamine
TM
(composed of DOSPA:DOPE, i.e. a multivalent cationic
lipid associated with a helper lipid) turned out to be more effective than
Lipofectin (DOTMA:DOPE, a monovalent cationic lipid associated with
a helper lipid). Both these formulations are commercialized by Invitrogen.
Alternatively, the biopharmaceutical company Genentech reported the
routine use of a ‘‘home-made’’ cationic lipid for transient transfection of
CHO cells in a 5–10 L scale (Wurm and Bernard, 2001).
3.7.4 Electroporation
In this method, the delivery of DNA molecules to the cells is mediated by
short strong electrical pulses (Wong and Neumann, 1982). The exposure
of the cells to an electrical field induces a potential difference across the
cellular membranes that generates temporary pores through which the
60 Animal Cell Technology

DNA reaches the cytoplasm. Compared with other methods, such as
calcium phosphate co-precipitation and DEAE-dextran, electroporation
presents lower mutation frequencies (Bertling et al., 1987), perhaps
because the DNA remains free in the cytoplasm and nucleoplasm. Several
cell types resistant to transfection by other procedures, including lympho-
cytes (Potter et al., 1984), hematopoietic stem cells (Toneguzzo and
Keating, 1986) and murine embryonic stem cells (Torres and Ku
¨
hn, 1997),
have been successfully electroporated. Furthermore, this technique renders
higher efficiencies when linearized DNA is used (Sanders, 1990). The
advantages of this technique include its simplicity, reproducibility, applic-
ability for transient and stable transfections, for adherent and suspension
cells, and the possibility to control gene copy number in transfectants. On
the other hand, the pulse time and the intensity of the electrical field raise
concerns and must be determined empirically for each cell type, since
succesful transfection can be achieved in a very limited range of voltage. A
number of electroporation devices are commercially available, which are
safe, easy to handle, and allow the control of different electroporation
parameters. In addition to the physical properties of the electrical pulse,
the type of buffer solution, and the quality and concentration of cells
(exponentially growing cells, between 10
6
and 10
7
cells/ml) have to be
considered. In particular, cell concentration has proved to be critical, since
lower cell densities reduce the transfection efficiency and higher cell
densities favor cell fusion processes that have a detrimental effect on
transfection (Spencer, 1991).
3.8 Selection markers
Upon transfection, there are cells that incorporate the plasmid (trans-
fected) and others that do not (wild-type). A selection system allows the
separation of these two cell populations. In this regard, the detection of
morphologic changes or, more commonly, the use of some toxic metabo-
lite for which the vector confers resistance (biochemical markers) have
become the methods of choice to select positive transfectants.
3.8.1 Morphological changes
In general, cells transfected with DNA that contains transforming genes,
derived from oncogenic viruses, lose the growth feature called ‘‘contact
inhibition,’’ which limits their growth and proliferation. These transfec-
tants can be visualized under the microscope and isolated as colonies.
3.8.2 Biochemical markers and gene amplification
Many genes that encode proteins causing biochemical tranformations in
the cells have been isolated, characterized, and employed in eukaryotic
expression systems. Among these biochemical markers, those conferring
resistance to cytotoxic drugs are among the most commonly used. They
allow the selection of stably transfected cells from a heterogeneous popu-
lation cultured in the presence of toxic metabolites (Sanders, 1990). As
Cloning and expression of heterologous proteins in animal cells 61
