Voet D., Voet Ju.G. Biochemistry
Подождите немного. Документ загружается.

forward direction.Thus, the free energy of ATP hydrolysis,
a highly exergonic process, is harnessed to drive many oth-
erwise endergonic biological processes to completion (Sec-
tion 16-4C).
Appendix 61
To establish that the free energy of a substance is a func-
tion of its concentration, consider the free energy change
of an ideal gas during a reversible pressure change at con-
stant temperature (w¿ ⫽ 0, since an ideal gas is incapable of
doing non-P–V work). Substituting Eqs. [3.1] and [3.2] into
Eq. [3.9] and differentiating the result yields
[3.A1]
On substitution of the differentiated forms of Eqs. [3.3] and
[3.8] into this expression, it reduces to
[3.A2]
The ideal gas equation is PV ⫽ nRT,where n is the number
of moles of gas. Therefore
[3.A3]
This gas phase result can be extended to the more bio-
chemically relevant area of solution chemistry by applica-
tion of Henry’s law for a solution containing the volatile
solute A in equilibrium with the gas phase:
[3.A4]
Here P
A
is the partial pressure of A when its mole fraction
in the solution is X
A
, and K
A
is the Henry’s law constant of
A in the solvent being used. It is generally more conven-
ient, however, to express the concentrations of the rela-
tively dilute solutions of chemical and biological systems in
terms of molarity rather than mole fractions. For a dilute
solution
[3.A5]
where the solvent concentration, [solvent], is approxi-
mately constant.Thus
[3.A6]
where . Substituting this expression
into Eq. [3.A3] yields
[3.A7]
Free energy, as are energy and enthalpy, is a relative
quantity that can only be defined with respect to some ar-
bitrary standard state. The standard state is customarily
taken to be 25°C, 1 atm pressure, and, for the sake of math-
ematical simplicity, [A] ⫽ 1. The integration of Eq. [3.A7]
from the standard state, [A] ⫽ 1, to the final state, [A] ⫽
[A], results in
[3.A8]G
A
⫺ G°
A
⫽ n
A
RT ln[A]
dG
A
⫽ n
A
RT d(ln K¿
A
⫹ ln[A]) ⫽ n
A
RT d ln[A]
K¿
A
⫽ K
A
>[solvent]
P
A
⬇ K¿
A
[A]
X
A
⬇
n
A
n
solvent
⫽
[A]
[solvent]
P
A
⫽ K
A
X
A
dG ⫽ nRT
dP
P
⫽ nRT d ln P
dG ⫽ V dP
dG ⫽ dq ⫹ dw ⫹ P dV ⫹ V dP ⫺ T dS
where is the free energy of A in the standard state and
[A] really represents the concentration ratio [A]/1. Since
Henry’s law is valid for real solutions only in the limit of in-
finite dilution, however, the standard state is defined as the
entirely hypothetical state of 1M solute with the properties
that it has at infinite dilution.
The free energy terms in Eq. [3.A8] may be converted
from extensive quantities (those dependent on the amount
of material) to intensive quantities (those independent of
the amount of material) by dividing both sides of the equa-
tion by n
A
.This yields
[3.A9]
Equation [3.A9] has the limitation that it refers to solu-
tions that exactly follow Henry’s law, although real solu-
tions only do so in the limit of infinite dilution if the solute
is, in fact, volatile.These difficulties can all be eliminated by
replacing [A] in Eq. [3.A9] by a quantity, a
A
, known as the
activity of A. This is defined
[3.A10]
where ␥
A
is the activity coefficient of A. Equation [3.A9]
thereby takes the form
[3.A11]
in which all departures from ideal behavior, including the
provision that the system may perform non-P–V work, are
incorporated into the activity coefficient, which is an ex-
perimentally measurable quantity. Ideal behavior is only
approached at infinite dilution; that is, ␥
A
n 1 as [A] n 0.
The standard state in Eq. [3.A11] is redefined as that of
unit activity.
The concentrations of reactants and products in most
laboratory biochemical reactions are usually so low (on the
order of millimolar or less) that the activity coefficients of
these various species are nearly unity. Consequently, the
activities of most biochemical species under laboratory
conditions can be satisfactorily approximated by their mo-
lar concentrations:
[3.13]
However, the activity coefficient of a particular species
varies with the total concentration of all other species pres-
ent as well as with its own concentration. Thus, despite the
low concentrations of most biochemical species in the cell,
their extraordinarily high combined concentrations (e.g.,
see Fig. 1-13) make the activity coefficients of the individ-
ual species deviate significantly from unity. Unfortunately,
it is difficult to determine the values of these quantities in a
cellular compartment (where it is likewise difficult to de-
termine the concentration of any given species).
G
A
⫺ G°
A
⫽ RT ln[A]
G
A
⫺ G°
A
⫽ RT ln a
A
a
A
⫽␥
A
[A]
G
A
⫺ G°
A
⫽ RT ln[A]
G°
A
APPENDIX Concentration Dependence of Free Energy
JWCL281_c03_052-064.qxd 5/31/10 1:26 PM Page 61
62 Chapter 3. Thermodynamic Principles: A Review
1 First Law of Thermodynamics: Energy Is Conserved
The first law of thermodynamics,
[3.1]
where q is heat and w is work, is a statement of the law of con-
servation of energy. Energy is a state function because the en-
ergy of a system depends only on the state of the system. En-
thalpy,
[3.2]
where P is pressure and V is volume, is a closely related state
function that represents the heat at constant pressure under
conditions where only pressure–volume work is possible.
2 Second Law of Thermodynamics: The Universe Tends
Toward Maximum Disorder
Entropy, which is also a state
function, is defined
[3.5]
where W, the disorder, is the number of equivalent ways the
system can be arranged under the conditions governing it and
k
B
is the Boltzmann constant. The second law of thermody-
namics states that the universe tends toward maximum disor-
der and hence ⌬S
universe
⬎ 0 for any real process.
3 Free Energy: The Indicator of Spontaneity The Gibbs
free energy of a system
[3.9]G ⫽ H ⫺ TS
S ⫽ k
B
ln W
H ⫽ U ⫹ PV
¢U ⫽ q ⫹ w
decreases in a spontaneous, constant pressure process. In a
process at equilibrium, the system suffers no net change, so
that ⌬G ⫽ 0.An ideal process, in which the system is always at
equilibrium, is said to be reversible.All real processes are irre-
versible since processes at equilibrium can only occur at an in-
finitesimal rate.
4 Chemical Equilibria For a chemical reaction
the change in the Gibbs free energy is expressed
[3.15]
where ⌬G°, the standard free energy change, is the free energy
change at 25°C, 1 atm pressure, and unit activities of reactants
and products. The biochemical standard state, ⌬G°¿, is simi-
larly defined but in dilute aqueous solution at pH 7 in which
the activities of water and H
⫹
are both defined as unity. At
equilibrium
where is the equilibrium constant under the biochemical
convention. An endergonic reaction (⌬G ⬎ 0) may be driven
by an exergonic reaction (⌬G ⬍ 0) if they are coupled and if
the overall reaction is exergonic.
K¿
eq
¢G°¿ ⫽⫺RT ln K¿
eq
⫽⫺RT lna
[C]
c
eq
[D]
d
eq
[A]
a
eq
[B]
b
eq
b
¢G ⫽ ¢G° ⫹ RT lna
[C]
c
[D]
d
[A]
a
[B]
b
b
aA ⫹ bB Δ cC ⫹ dD
CHAPTER SUMMARY
Allen, J.P., Biophysical Chemistry Chapters 1–5, Wiley-Blackwell
(2008).
Atkins, P.W. and de Paula, J., Physical Chemistry for the Life Sci-
ences, Chapters 1–5, Freeman (2006). [Most physical chemistry
texts treat thermodynamics in some detail.]
Edsall, J.T. and Gutfreund, H., Biothermodynamics,Wiley (1983).
Hammes, G.G., Physical Chemistry for the Biological Sciences,
Chapters 1 and 2,Wiley (2007).
Haynie, D.T., Biological Thermodynamics (2nd ed.), Cambridge
University Press (2008).
Tinoco, I., Jr., Sauer, K., Wang, J.C., and Puglisi, J.C., Physical
Chemistry. Principles and Applications in Biological Sciences
(4th ed.), Chapters 2–5, Prentice Hall (2002).
van Holde, K.E., Johnson, W.C., and Ho, P.S., Principles of Physi-
cal Biochemistry (2nd ed.), Chapter 2, Prentice Hall (2006).
[The equivalence of the Boltzmann and Clausius formulations
of the second law of thermodynamics is demonstrated in
Section 2.3.]
REFERENCES
1. A common funeral litany is the Biblical verse: “Ashes to
ashes, dust to dust.” Why might a bereaved family of thermody-
namicists be equally comforted by a recitation of the second law
of thermodynamics?
2. How many 4-m-high flights of stairs must an overweight
person weighing 75 kg climb to atone for the indiscretion of eating
a 500-Cal hamburger? Assume that there is a 20% efficiency in
converting nutritional energy to mechanical energy. The gravita-
tional force of an object of mass m kg is F ⫽ mg, where the gravi-
tational constant g is 9.8 m ⴢ s
⫺2
.
3. In terms of thermodynamic concepts, why is it more diffi-
cult to park a car in a small space than it is to drive it out from such
a space?
4. It has been said that an army of dedicated monkeys, typing
at random, would eventually produce all of Shakespeare’s works.
How long, on average, would it take 1 million monkeys, each typ-
ing on a 46-key keyboard (space included but no shift key) at the
rate of 1 keystroke per second, to type the phrase “to be or not to
be”? How long, on average, would it take one monkey to do so at
a computer if the computer would only accept the correct letter in
PROBLEMS
JWCL281_c03_052-064.qxd 5/31/10 1:26 PM Page 62

the phrase and then would shift to its next letter (i.e., the com-
puter knew what it wanted)? What do these results indicate about
the probability of order randomly arising from disorder versus or-
der arising through a process of evolution?
5. Show that the transfer of heat from an object of higher tem-
perature to one of lower temperature, but not the reverse process,
obeys the second law of thermodynamics.
6. Carbon monoxide crystallizes with its CO molecules
arranged in parallel rows. Since CO is a very nearly ellipsoidal
molecule, in the absence of polarity effects, adjacent CO mole-
cules could equally well line up in a head-to-tail or a head-to-head
fashion. In a crystal consisting of 10
23
CO molecules, what is the
entropy of all the CO molecules being aligned head to tail?
7. The U.S. Patent Office has received, and continues to re-
ceive, numerous applications for perpetual motion machines. Per-
petual motion machines have been classified as those of the first
kind, which violate the first law of thermodynamics, and those of
the second kind, which violate the second law of thermodynamics.
The fallacy in a perpetual motion machine of the first kind is gen-
erally easy to detect.An example would be a motor-driven electri-
cal generator that produces energy in excess of that input by the
motor. The fallacy in a perpetual motion machine of the second
type, however, is usually more subtle. Take, for example, a ship
that uses heat energy extracted from the sea by a heat pump to
boil water so as to power a steam engine that drives the ship as
well as the heat pump. Show, in general terms, that such a propul-
sion system would violate the second law of thermodynamics.
8. Using the data in Table 3-4, calculate the values of ⌬G°at
25°C for the following metabolic reactions:
(a)
Glucose
(b)
Glucose Ethanol
(c)
Glucose Lactate
[These reactions, respectively, constitute oxidative metabolism, al-
coholic fermentation in yeast deprived of oxygen, and homolactic
fermentation in skeletal muscle requiring energy faster than ox-
idative metabolism can supply it (Section 17-3B).]
*9. The native and denatured forms of a protein are generally
in equilibrium as follows:
Protein (denatured) Δ protein (native)
C
6
H
12
O
6
Δ 2 CH
3
CHOHCOO
⫺
⫹ 2H
⫹
C
6
H
12
O
6
Δ 2 CH
3
CH
2
OH ⫹ 2 CO
2
(aq)
C
6
H
12
O
6
⫹ 6 O
2
Δ 6 CO
2
(aq) ⫹ 6 H
2
O(/)
For a certain solution of the protein ribonuclease A, in which the
total protein concentration is 2.0 ⫻ 10
⫺3
M, the concentrations of
the denatured and native proteins at both 50 and 100°C are given
in the following table:
(a) Determine ⌬H° and ⌬S° for the folding reaction assuming that
these quantities are independent of temperature. (b) Calculate
⌬G° for ribonuclease A folding at 25°C. Is this process sponta-
neous under standard state conditions at this temperature?
(c) What is the denaturation temperature of ribonuclease A under
standard state conditions?
*10. Using the data in Table 3-4, calculate ⌬G°
f
¿ for the follow-
ing compounds at 25°C: (a) H
2
O(/); (b) sucrose (sucrose ⫹
H
2
O glucose ⫹ fructose: ⫺29.3 kJ ⴢ mol
⫺1
); and
(c) ethyl acetate (ethyl acetate ⫹ H
2
O ethanol ⫹ acetate
⫺
⫹
H
⫹
: ⫺19.7 kJ ⴢ mol
⫺1
; the pK of acetic acid is 4.76).
11. Calculate the equilibrium constants for the hydrolysis
of the following compounds at pH 7 and 25°C: (a) phospho-
enolpyruvate (⌬G°¿ ⫽⫺61.9 kJ ⴢ mol
⫺1
); (b) pyrophosphate
(⌬G°¿ ⫽⫺33.5 kJ ⴢ mol
⫺1
); and (c) glucose-1-phosphate (⌬G°¿ ⫽
⫺20.9 kJ ⴢ mol
⫺1
).
12. ⌬G°¿ for the isomerization reaction
is ⫺7.1 kJ ⴢ mol
⫺1
. Calculate the equilibrium ratio of [G1P] to
[G6P] at 25°C.
13. For the reaction A S B at 298 K, the change in enthalpy is
⫺7 kJ ⴢ mol
⫺1
and the change in entropy is ⫺25 J ⴢ K
⫺1
ⴢ mol
⫺1
.Is
the reaction spontaneous? If not, should the temperature be in-
creased or decreased to make the reaction spontaneous?
14. Two biochemical reactions have the same K
eq
⫽ 5 ⫻ 10
⫺8
at temperature T
1
⫽ 298 K. However, reaction 1 has ⌬H° ⫽
⫺28 kJ ⴢ mol
⫺1
and Reaction 2 has ⌬H° ⫽⫹28 kJ ⴢ mol
⫺1
.The two
reactions utilize the same reactants.Your lab partner has proposed
that you can get more of the reactants to proceed via Reaction 2
rather than Reaction 1 by lowering the temperature of the reac-
tion. Will this strategy work? Why or why not? How much would
the temperature have to be raised or lowered to change the value
of K
2
/K
1
from 1 to 10?
Glucose-1-phosphate(G1P) Δ glucose-6-phosphate(G6P)
¢G
o
¿ ⫽
Δ
¢G
o
¿ ⫽Δ
Problems 63
Temperature [Ribonuclease A [Ribonuclease A
(ºC) (denatured)] (M)(native)] (M)
50 5.1 ⫻ 10
⫺6
2.0 ⫻ 10
⫺3
100 2.8 ⫻ 10
⫺4
1.7 ⫻ 10
⫺3
JWCL281_c03_052-064.qxd 5/31/10 1:26 PM Page 63
This page intentionally left blank

PART
II
BIOMOLECULES
The digestive enzyme
bovine carboxypeptidase
A showing its central
 sheet.
JWCL281_c04_065-081.qxd 11/2/10 6:43 PM Page 65
This page intentionally left blank

6767
CHAPTER 4
Amino Acids
1 The Amino Acids of Proteins
A. General Properties
B. Peptide Bonds
C. Classification and Characteristics
D. Acid–Base Properties
E. A Few Words on Nomenclature
2 Optical Activity
A. An Operational Classification
B. The Fischer Convention
C. The Cahn–Ingold–Prelog System
D. Chirality and Biochemistry
3 “Nonstandard” Amino Acids
A. Amino Acid Derivatives in Proteins
B. Specialized Roles of Amino Acids
also energy metabolites and, in animals, many of them are
essential nutrients (Chapter 26). In addition, as we shall
see, many amino acids and their derivatives are of bio-
chemical importance in their own right (Section 4-3B).
1 THE AMINO ACIDS OF PROTEINS
The analyses of a vast number of proteins from almost
every conceivable source have shown that all proteins are
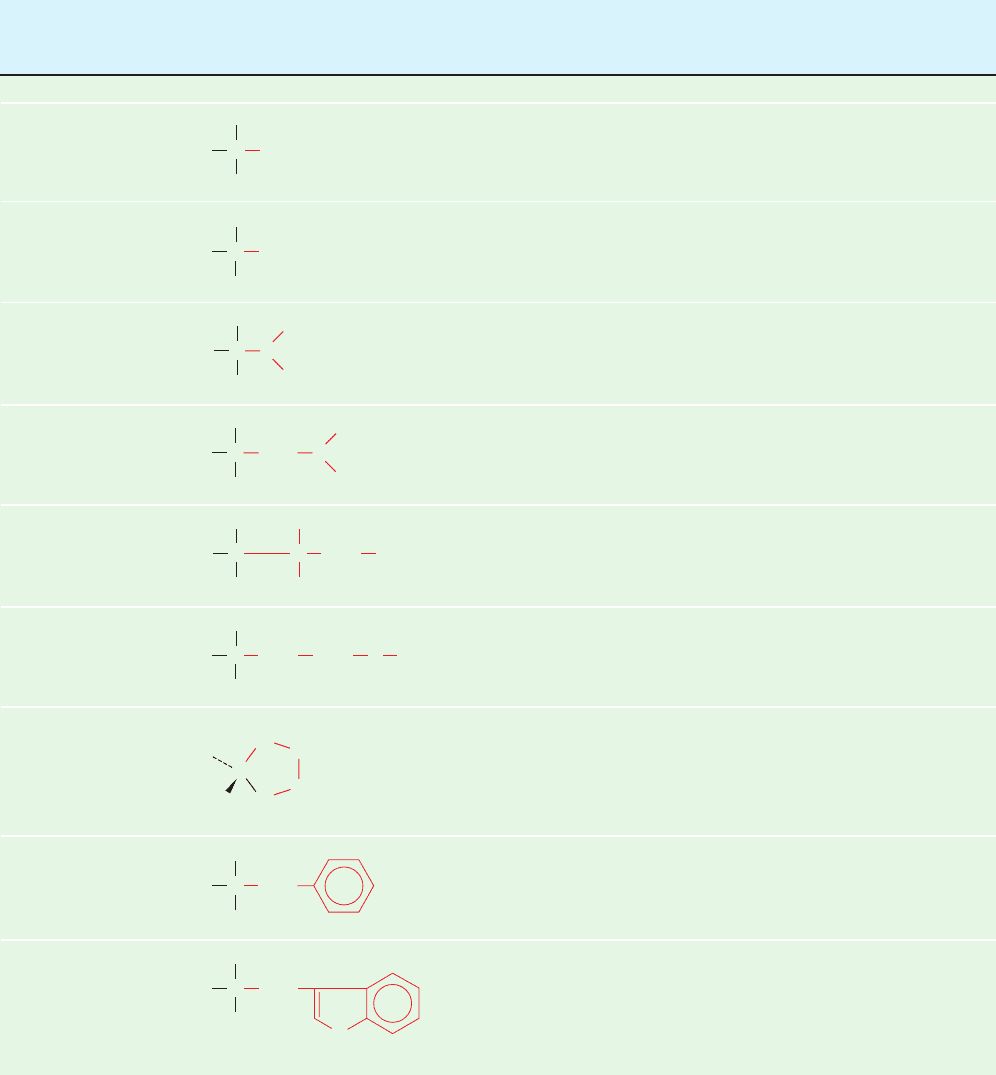
composed of the 20 “standard” amino acids listed in Table 4-1.
These substances are known as ␣-amino acids because,
with the exception of proline, they have a primary amino
group and a carboxylic acid group substituent on the same
carbon atom (Fig. 4-1; proline has a secondary amino
group).
A. General Properties
The pK values of the 20 “standard” ␣-amino acids of pro-
teins are tabulated in Table 4-1. Here pK
1
and pK
2
,re-
spectively, refer to the ␣-carboxylic acid and ␣-amino
groups, and pK
R
refers to the side groups with acid–base
properties. Table 4-1 indicates that the pK values of the
␣-carboxylic acid groups lie in a small range around 2.2
so that above pH 3.5 these groups are almost entirely in
their carboxylate forms. The ␣-amino groups all have pK
values near 9.4 and are therefore almost entirely in their
ammonium ion forms below pH 8.0.This leads to an impor-
tant structural point: In the physiological pH range,both the
carboxylic acid and the amino groups of -amino acids are
completely ionized (Fig. 4-2). An amino acid can therefore
act as either an acid or a base. Substances with this prop-
erty are said to be amphoteric and are referred to as am-
pholytes (amphoteric electrolytes). In Section 4-1D, we
shall delve a bit deeper into the acid–base properties of the
amino acids.
a
It is hardly surprising that much of the early biochemical
research was concerned with the study of proteins. Proteins
form the class of biological macromolecules that have the
most well-defined physicochemical properties, and conse-
quently they were generally easier to isolate and character-
ize than nucleic acids, polysaccharides, or lipids. Further-
more, proteins, particularly in the form of enzymes, have
obvious biochemical functions. The central role that pro-
teins play in biological processes has therefore been recog-
nized since the earliest days of biochemistry. In contrast,
the task of nucleic acids in the transmission and expression
of genetic information was not realized until the late 1940s
and their catalytic function only began to come to light in
the 1980s, the role of lipids in biological membranes was
not appreciated until the 1960s, and the biological func-
tions of polysaccharides are still somewhat mysterious.
In this chapter we study the structures and properties of
the monomeric units of proteins, the amino acids. It is from
these substances that proteins are synthesized through
processes that we discuss in Chapter 32. Amino acids are
Figure 4-1 General structural formula for ␣-amino acids.
There are 20 different R groups in the commonly occurring
amino acids (Table 4-1).
Figure 4-2 Zwitterionic form of the ␣-amino acids that occurs
at physiological pH values.
H
2
N
COOH
C
R
H
␣
COO
⫺
C
R
H
+
H
3
N
JWCL281_c04_065-081.qxd 5/31/10 1:37 PM Page 67
68 Chapter 4. Amino Acids
Table 4-1 Covalent Structures and Abbreviations of the “Standard” Amino Acids of Proteins, Their Occurrence, and the pK
Values of Their Ionizable Groups
Name, Residue Average
Three-Letter Symbol, Structural Mass Occurrence pK
1
pK
2
pK
R
and One-Letter Symbol Formula
a
(D)
b
in Proteins (%)
c
␣-COOH
d
␣- Side Chain
d
Amino acids with nonpolar side chains
Glycine 57.0 7.1 2.35 9.78
Gly
G
Alanine 71.1 8.3 2.35 9.87
Ala
A
Valine 99.1 6.9 2.29 9.74
Va l
V
Leucine 113.2 9.7 2.33 9.74
Leu
L
Isoleucine 113.2 6.0 2.32 9.76
Ile
I
Methionine 131.2 2.4 2.13 9.28
Met
M
Proline 97.1 4.7 1.95 10.64
Pro
P
Phenylalanine 147.2 3.9 2.20 9.31
Phe
F
Tryptophan 186.2 1.1 2.46 9.41
Tr p
W
NH
⫹d
3
C
COO
⫺
CH
2
H
NH
3
⫹
C
COO
⫺
NH
3
HH
⫹
C
COO
⫺
CH
3
H
NH
3
⫹
C
COO
⫺
CH
CH
3
CH
3
H
NH
3
⫹
C
COO
⫺
CHCH
2
CH
3
CH
3
H
NH
3
⫹
C
COO
⫺
CCH
2
CH
3
CH
3
H
H
*
NH
3
⫹
C
COO
⫺
SCH
2
CH
2
CH
3
H
NH
3
⫹
H
2
C
COO
⫺
N
H
2
CH
2
CH
2
5
1
⫹
4
3
2
H
C
C
COO
⫺
N
H
CH
2
H
1
2
3
4
5
6
7
NH
3
⫹
(continued)
a
The ionic forms shown are those predominating at pH 7.0 (except for that of histidine
e
), although residue mass is given for the neutral compound. The C
␣
atoms, as well as those atoms marked with an asterisk, are chiral centers with configurations as indicated according to Fischer projection formulas.The
standard organic numbering system is provided for heterocycles.
b
The residue masses are given for the neutral residues. For molecular masses of the parent amino acids, add 18.0 D, the molecular mass of H
2
O, to the
residue masses. For side chain masses, subtract 56.0 D, the formula mass of a peptide group, from the residue masses.
c
The average amino acid composition in the complete SWISS-PROT database (http://www.expasy.ch/sprot/relnotes/relstat.html), Release 55.11.
d
From Dawson, R.M.C., Elliott, D.C., Elliott, W.H., and Jones, K.M., Data for Biochemical Research (3rd ed.), pp. 1–31, Oxford Science Publications (1986).
e
Both the neutral and protonated forms of histidine are present at pH 7.0 because its pK
R
is close to 7.0.The imidazole ring of histidine is numbered here
according to the biochemistry convention. In the IUPAC convention, N3 of the biochemistry convention is designated N1 and the numbering increases
clockwise around the ring.
f
The three- and one-letter symbols for asparagine or aspartic acid are Asx and B, whereas for glutamine or glutamic acid they are Glx and Z. The one-
letter symbol for an undetermined or “nonstandard” amino acid is X.
JWCL281_c04_065-081.qxd 5/31/10 1:37 PM Page 68
Section 4-1. The Amino Acids of Proteins 69
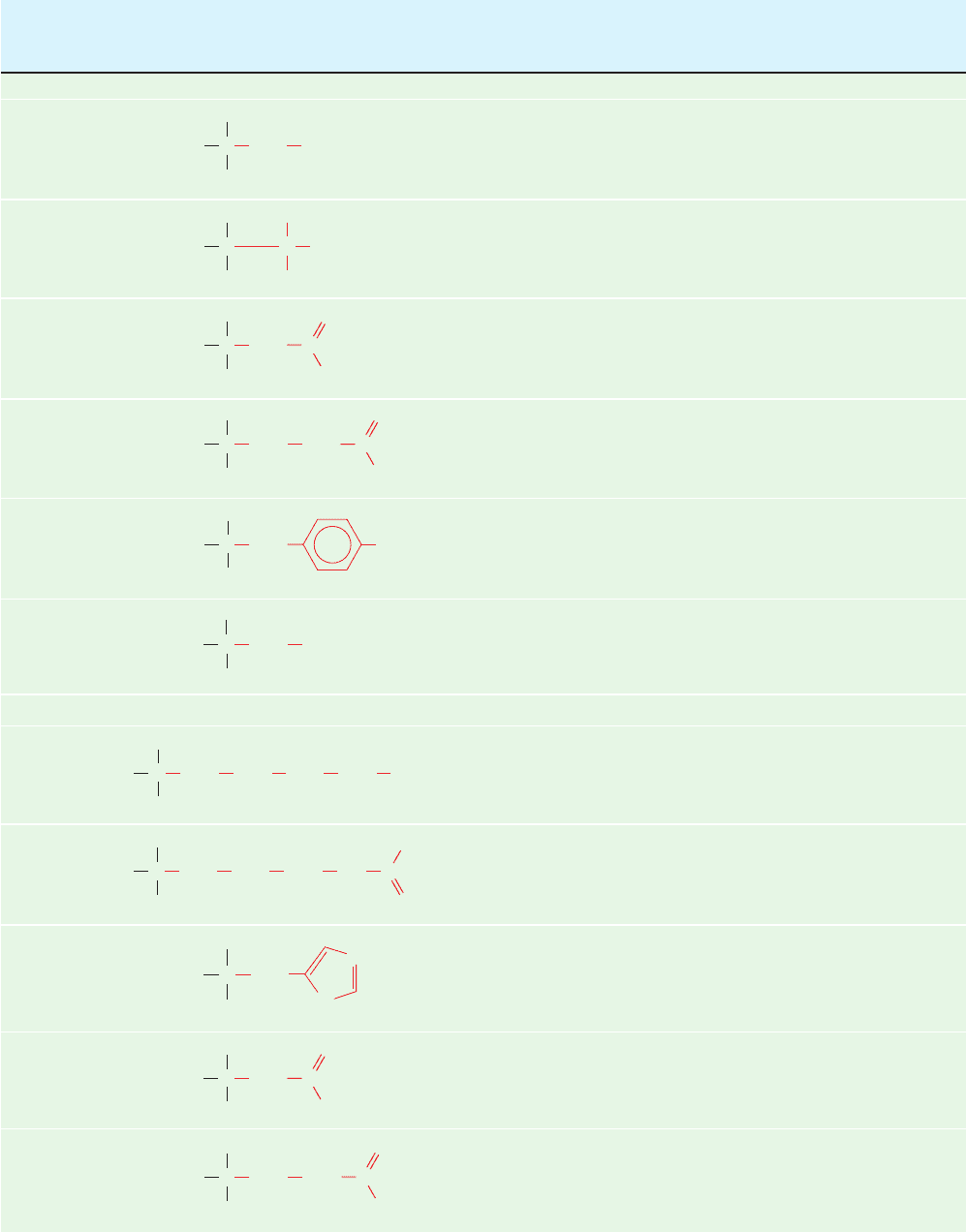
Table 4-1 (Continued)
Name Residue Average
Three-Letter Symbol, Structural Mass Occurrence pK
1
pK
2
pK
R
and One-Letter Symbol Formula
a
(D)
b
in Proteins (%)
c
␣-COOH
d
␣-
d
Side Chain
d
Amino acids with uncharged polar side chains
Serine 87.1 6.5 2.19 9.21
Ser
S
Threonine 101.1 5.3 2.09 9.10
Thr
T
Asparagine
f
114.1 4.0 2.14 8.72
Asn
N
Glutamine
f
128.1 3.9 2.17 9.13
Gln
Q
Tyrosine 163.2 2.9 2.20 9.21 10.46 (phenol)
Ty r
Y
Cysteine 103.1 1.4 1.92 10.70 8.37 (sulfhydryl)
Cys
C
Amino acids with charged polar side chains
Lysine 128.2 5.9 2.16 9.06 10.54 (ε-NH
⫹
3
)
Lys
K
Arginine 156.2 5.5 1.82 8.99 12.48 (guanidino)
Arg
R
Histidine
e
137.1 2.3 1.80 9.33 6.04 (imidazole)
His
H
Aspartic acid
f
115.1 5.4 1.99 9.90 3.90 (-COOH)
Asp
D
Glutamic acid
f
129.1 6.8 2.10 9.47 4.07 (␥-COOH)
Glu
E
NH
⫹d
3
C
COO
⫺
O
⫺
C
O
CH
2
CH
2
H
NH
3
⫹
C
COO
⫺
C
CH
2
CH
2
CH
2
NHH
NH
2
NH
3
⫹
NH
2
⫹
N
H
1
2
3
4
5
C
COO
⫺
NH
⫹
CH
2
H
NH
3
⫹
C
COO
⫺
O
⫺
C
O
CH
2
H
NH
3
⫹
C
COO
⫺
CH
2
OH
H
NH
3
⫹
C
COO
⫺
CH
2
SHH
NH
3
⫹
C
COO
⫺
CH
2
CH
2
CH
2
CH
2
H
NH
3
⫹
NH
3
⫹
C
COO
⫺
NH
2
C
O
CH
2
CH
2
H
NH
3
⫹
C
COO
⫺
NH
2
C
O
CH
2
H
NH
3
⫹
C
COO
⫺
OHCH
2
H
NH
3
⫹
C
COO
⫺
CCH
3
H
OH
H
*
NH
3
⫹
JWCL281_c04_065-081.qxd 5/31/10 1:37 PM Page 69
70 Chapter 4. Amino Acids
Molecules that bear charged groups of opposite polarity
are known as zwitterions (German: zwitter, hybrid) or
dipolar ions. The zwitterionic character of the ␣-amino
acids has been established by several methods including
spectroscopic measurements and X-ray crystal structure
determinations (in the solid state the a-amino acids are
zwitterionic because the basic amine group abstracts a pro-
ton from the nearby acidic carboxylic acid group). Because
amino acids are zwitterions, their physical properties are
characteristic of ionic compounds. For instance, most
␣-amino acids have melting points near 300⬚C, whereas their
nonionic derivatives usually melt around 100⬚C. Further-
more, amino acids, like other ionic compounds, are more
soluble in polar solvents than in nonpolar solvents. Indeed,
most ␣-amino acids are very soluble in water but are
largely insoluble in most organic solvents.
B. Peptide Bonds
The ␣-amino acids polymerize, at least conceptually,
through the elimination of a water molecule as is indicated
in Fig. 4-3. The resulting linkage, which was inde-
pendently characterized in 1902 by Emil Fischer and Franz
Hofmeister, is known as a peptide bond. Polymers com-
posed of two, three, a few (3–10), and many amino acid
residues (alternatively called peptide units) are known, re-
spectively, as dipeptides, tripeptides, oligopeptides, and
polypeptides. These substances, however, are often re-
ferred to simply as “peptides.” Proteins are molecules that
consist of one or more polypeptide chains. These polypep-
tides range in length from ⬃40 to ⬃34,000 amino acid
residues (although few have more than 1500 residues)
and, since the average mass of an amino acid residue is
⬃110 D, have molecular masses that range from ⬃40 to
over ⬃3700 kD.
Polypeptides are linear polymers; that is, each amino
acid residue is linked to its neighbors in a head-to-tail fash-
ion rather than forming branched chains. This observation
reflects the underlying elegant simplicity of the way living
systems construct these macromolecules for, as we shall
see, the nucleic acids that encode the amino acid sequences
CO¬NH
of polypeptides are also linear polymers. This permits the
direct correspondence between the monomer (nucleotide)
sequence of a nucleic acid and the monomer (amino acid)
sequence of the corresponding polypeptide without the
added complication of specifying the positions and se-
quences of any branching chains.
With 20 different choices available for each amino acid
residue in a polypeptide chain, it is easy to see that a huge
number of different protein molecules can exist. For exam-
ple, for dipeptides, each of the 20 different choices for the
first amino acid residue can have 20 different choices for
the second amino acid residue, for a total of 20
2
⫽ 400 dis-
tinct dipeptides. Similarly, for tripeptides, there are 20 pos-
sibilities for each of the 400 choices of dipeptides to yield a
total of 20
3
⫽ 8000 different tripeptides. A relatively small
protein molecule consists of a single polypeptide chain of
100 residues.There are 20
100
⫽ 1.27 ⫻ 10
130
possible unique
polypeptide chains of this length, a quantity vastly greater
than the estimated number of atoms in the universe (9 ⫻
10
78
). Clearly, nature can have made only a tiny fraction of
the possible different protein molecules. Nevertheless, the
various organisms on Earth collectively synthesize an enor-
mous number of different protein molecules whose great
range of physicochemical characteristics stem largely from
the varied properties of the 20 “standard” amino acids.
C. Classification and Characteristics
The most common and perhaps the most useful way of
classifying the 20 “standard” amino acids is according to
the polarities of their side chains (R groups). This is be-
cause proteins fold to their native conformations largely in
response to the tendency to remove their hydrophobic side
chains from contact with water and to solvate their hy-
drophilic side chains (Chapters 8 and 9). According to this
classification scheme, there are three major types of amino
acids: (1) those with nonpolar R groups, (2) those with un-
charged polar R groups, and (3) those with charged polar R
groups.
a. The Nonpolar Amino Acid Side Chains Have a
Variety of Shapes and Sizes
Nine amino acids are classified as having nonpolar side
chains. Glycine (which, when it was found to be a compo-
nent of gelatin in 1820, was the first amino acid to be iden-
tified in protein hydrolyzates) has the smallest possible
side chain, an H atom. Alanine (Fig. 4-4), valine, leucine,
and isoleucine have aliphatic hydrocarbon side chains
ranging in size from a methyl group for alanine to isomeric
butyl groups for leucine and isoleucine. Methionine has a
thiol ether side chain that resembles an n-butyl group in
many of its physical properties (C and S have nearly equal
electronegativities and S is about the size of a methylene
group). Proline, a cyclic secondary amino acid, has confor-
mational constraints imposed by the cyclic nature of its
pyrrolidine side chain, which is unique among the “stan-
dard” 20 amino acids. Phenylalanine, with its phenyl moi-
ety (Fig. 4-4), and tryptophan, with its indole group,contain
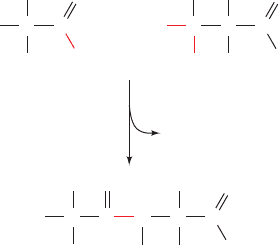
Figure 4-3 Condensation of two ␣-amino acids to form a
dipeptide. The peptide bond is shown in red.
H
3
N
C
C
R
1
H
O
–
O
+
+
H
2
O
H
N
H
H
+
C
C
R
2
O
–
O
H
3
N
C
C
R
1
H
H
O
+
C
C
R
2
H
O
–
O
N
H
JWCL281_c04_065-081.qxd 5/31/10 1:37 PM Page 70
