Vidakovic B. Statistics for Bioengineering Sciences: With Matlab and WinBugs Support
Подождите немного. Документ загружается.


17.2 Logistic Regression 661
data in this example comes from Munich’s Klinikum Großharden (Fahrmeir
and Tutz, 1996) and concerns infections in births by C-section. The response
variable of interest is the occurrence or nonoccurrence of infection. Three co-
variates, each at two levels, were considered as important for the occurrence
of infection:
noplan – The C-section delivery was planned (0) or not planned (1);
riskfac – Risk factors for the mother, such as diabetes, overweight, previ-
ous C-section birth, etc., are present (1) or not present (0); and
antibio – Antibiotics as a prophylaxis are given (1) or not given (0).
Table 17.1 provides the results.
Table 17.1 Caesarean-section delivery data.
Planned No Plan
Infection Infection
yes no total yes no total
Antibiotics
Risk factor yes 1 17 18 11 87 98
Risk factor no 0 2 2 0 0 0
No Antibiotics
Risk factor yes 28 30 58 23 3 26
Risk factor no 8 32 40 0 9 9
Here is the MATLAB code that uses built-in functions glmfit and glmval
to fit and present the model.
infection = [ 1 11 0 0 28 23 8 0];
total = [18 98 2 0 58 26 40 9];
proportion = infection./total;
noplan = [ 0 1 0 1 0 1 0 1];
riskfac = [ 1 1 0 0 1 1 0 0];
antibio = [ 1 1 1 1 0 0 0 0];
[b,dev,stats] = glmfit([noplan’ riskfac’ antibio’],...
[infection’ total’],’binomial’,’logit’);
logitFit = ...
glmval(b,[noplan’ riskfac’ antibio’],’logit’);
The resulting additive model (no interactions) is
log
P(infection)
P(no infection)
=β
0
+β
1
·noplan +β
2
·riskfac +β
3
·antibio
with estimators of
βs as
b
0
b
1
b
2
b
3
–1.8926 1.0720 2.0299 –3.2544
The interpretation of the estimators for β coefficients is illuminating if we
look at the odds ratio
P(infection)
P(no infection)
:
662 17 Regression for Binary and Count Data
P(infection)
P(no infection)
=exp(β
0
) ·exp(β
1
noplan) ·exp(β
2
riskfac) ·exp(β
3
antibio).
For example, when
antibio=1, i.e., when antibiotics are given, the estimated
odds of infection
P(infection)/P(no infection) increase by the factor exp(−3.25) =
0.0388, that is, the odds decrease 25.79 times. Of course, these statements are
valid only if the model is accurate. Other competing models (such as probit or
clog-log) may result in different changes in risk ratios.
1 2 3 4 5 6 7 8
0
0.1
0.2
0.3
0.4
0.5
0.6
0.7
0.8
0.9
Code for (noplan, riskfact, antibio)
Probability of Infection
Observed Proportions
Logistic Fit
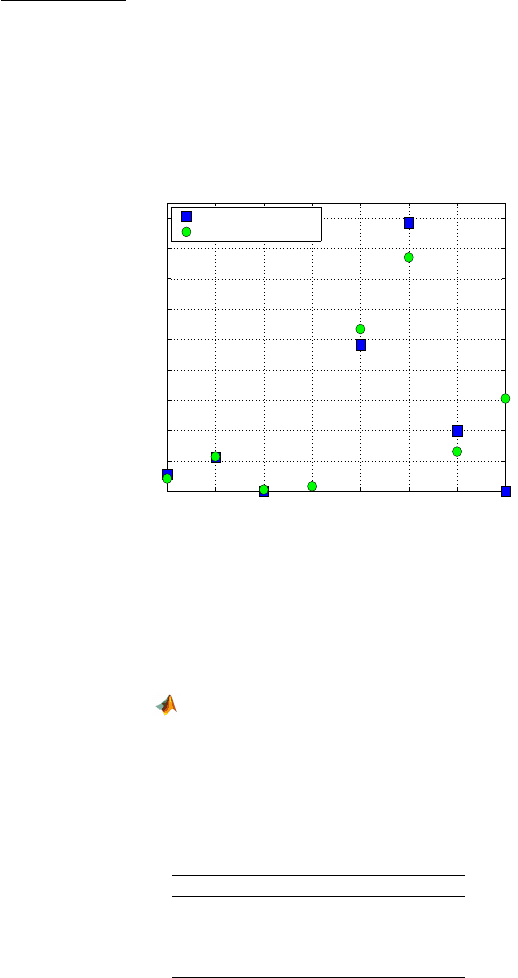
Fig. 17.1 Caesarean delivery infection predictions. For a triple (noplan, riskfac, antibio), the
numbers on the x-axis code as follows: 1=(0,1,1), 2=(1,1,1), 3=(0,0,1), 4=(1,0,1), 5=(0,1,0),
6=(1,1,0), 7=(0,0,0), and 8=(1,0,0). Blue squares are the observed relative frequencies and
green circles are the model-predicted probabilities of infection. Note that point 4 does not
have an observed proportion.
The m-function logisticmle.m also gives standard errors for estimators
of
βs. Table provides t-values, that is, ratios of coefficients and their stan-
dard deviations, for testing if the coefficients are significantly different from
0. These are known as Wald’s Z statistics, since they are approximately nor-
mal.
Table 17.2 t-ratios (Wald’s Z statistic) for the estimators b =
ˆ
β.
b s
b
t
Intercept –1.8926 0.4124 –4.5893
noplan 1.0720 0.4253 2.5203
riskfac 2.0299 0.4553 4.4588
antibio –3.2544 0.4812 –6.7624
The deviance (p. 665) of this model as a measure of goodness of fit is dis-
tributed as
χ
2
with 3 degrees of freedom. The number of degrees of freedom is

17.2 Logistic Regression 663
calculated as 7 (number of groups with observations) minus 4 (four estimated
parameters
β
0
−β
3
). Since it is found to be significant,
dev = 10.9967;
pval = 1 - chi2cdf(dev, 7-4) % 0.0117
the fit of this model is not good. To improve the fit, one may include the inter-
actions.
One may ask why the regression model was needed in the first place.
The probabilities of interest could be predicted by relative frequencies. For
example, in the case (
noplan = 0, riskfac = 1, antibio = 1), the relative fre-
quency of infection was 1/18
= 0.0556, just slightly larger than the model-
predicted
ˆ
p
=0.0424. There are two benefits to this approach. First, the model
is able to predict probabilities in the cases where no patients are present, such
as for (
noplan = 1, riskfac = 0, antibio = 1). Second, the predictions for the
cases where y
= 1 is not observed are “borrowing strength” from the model
and are not modeled individually. For example, zero as an estimator in the
case (
noplan = 1, riskfac = 0, antibio = 0) is not reasonable; the model-based
estimator
ˆ
p
= 0.3056 is more realistic. Figure 17.1 compares observed and
model-predicted infection rates. For a triple of covariates (noplan, riskfac, an-
tibio), the numbers on the x-axis code as follows: 1=(0,1,1), 2=(1,1,1), 3=(0,0,1),
4=(1,0,1), 5=(0,1,0), 6=(1,1,0), 7=(0,0,0), and 8=(1,0,0). Note that point 4 does
not have an observed proportion, however the model-predicted proportion can
be found. For computational aspects refer to file
caesarean.m.
Next, we provide a Bayesian solution to this example and compare the
model fit with the classical fit above. The comparisons are summarized in Ta-
ble 17.1.
C-SECTION INFECTIONS
model{
for(i in 1:N){
inf[i] ~ dbin(p[i],total[i])
logit(p[i]) <- beta0 + beta1
*
noplan[i] +
beta2
*
riskfac[i] + beta3
*
antibio[i]
}
beta0 ~dnorm(0, 0.00001)
beta1 ~dnorm(0, 0.00001)
beta2 ~dnorm(0, 0.00001)
beta3 ~dnorm(0, 0.00001)
}
DATA
list(inf=c(1, 11, 0, 0, 28, 23, 8, 0),
total = c(18, 98, 2, 0, 58, 26, 40, 9),
noplan = c(0,1,0,1,0,1,0,1),
riskfac = c(1,1, 0, 0, 1,1, 0, 0),
antibio =c(1,1,1,1,0,0,0,0), N=8)
664 17 Regression for Binary and Count Data
INITS
list(beta0 =0, beta1=0, beta2=0, beta3=0)
mean sd MC error val2.5pc median val97.5pc start sample
beta0 –1.964 0.4258 0.001468 –2.853 –1.945 –1.183 1001 1000000
beta1 1.111 0.4339 8.857E-4 0.2851 1.102 1.986 1001 1000000
beta2 2.104 0.4681 0.00159 1.226 2.09 3.066 1001 1000000
beta3 –3.335 0.4915 9.756E-4 –4.337 –3.322 –2.411 1001 1000000
deviance 32.24 2.873 0.00566 28.67 31.59 39.49 1001 1000000
Table 17.3 Comparison of classical and noninformative Bayes estimators b =
ˆ
β, with esti-
mators of standard deviations.
b s
b
ˆ
β
B
ˆ
σ
B
Intercept −1.8926 0.4124 −1.964 0.4258
noplan 1.0720 0.4253
1.111 0.4339
riskfac 2.0299 0.4553
2.104 0.4681
antibio
−3.2544 0.4812 −3.335 0.4915
17.2.2 Assessing the Logistic Regression Fit
The measures for assessing the goodness of linear regression fit that we cov-
ered in Chap. 16, R
2
, F, MSE, etc., are not appropriate for logistic regression.
As in the case of linear regression, there is a range of measures for assessing
the performance of logistic regression and we will briefly overview a few.
The significance of model parameters
β
0
,β
1
,... is tested by the so-called
Wald’s test. One finds the statistic Z
i
=
b
i
s(b
i
)
that has an approximate normal
distribution if the coefficient
β
i
is 0. Equivalently, W
i
=
b
2
i
s
2
(b
i
)
with an approx-
imate
χ
2
-distribution with 1 degree of freedom can be used. Large values of
|Z
i
| or W
i
are critical for H
0
: β
i
=0.
The sample variances s
2
(b
i
) are diagonal elements of (X
0
V X )
−1
, where
X
=
1 x
11
x
12
... x
1,p−1
1 x
21
x
22
... x
2,p−1
...
1 x
n1
x
n2
... x
n,p−1
is the design matrix and

17.2 Logistic Regression 665
V =
ˆ
p
1
(1 −
ˆ
p
1
) 0 ... 0
0
ˆ
p
2
(1 −
ˆ
p
2
) ... 0
...
0 0 ...
ˆ
p
n
(1 −
ˆ
p
n
)
.
The customary measure for goodness of fit is deviance, defined as
D
=−2log
likelihood of the fitted model
likelihood of the saturated model
.
For the logistic regression in (17.2), where y
i
is the number of 1s and n
i
−y
i
is
the number of 0s in class i, the likelihood is L
=
Q
k
i
=1
p
y
i
i
(1 − p
i
)
n
i
−y
i
and the
deviance is
D
=−2
k
X
1=1
½
y
i
log
µ
ˆ
y
i
y
i
¶
+
(n
i
− y
i
)log
µ
n
i
−
ˆ
y
i
n
i
− y
i
¶¾
,
where
ˆ
y
i
= n
i
ˆ
p
i
is the model fit for y
i
. The saturated model estimates p
i
as
ˆ
p
i
= y
i
/n
i
and
ˆ
y
i
= y
i
providing the fit that matches the observations.
The deviance statistic in this case has a
χ
2
-distribution with k − p degrees
of freedom, where k is the number of classes/groups and p is the number of
parameters in the model. Recall that in the previous example the deviance of
the model was distributed as
χ
2
with k − p =7 −4 =3 degrees of freedom.
For both Bernoulli and binomial observations, the mean and variance de-
pend on a single parameter, p. When the mean is well fitted, the variance
could be underfitted (overdispersion in data) or overfitted (underdispersion in
data). The ratio D/df is often used to indicate over- or underdispersion in the
data.
The traditional
χ
2
-statistic for the goodness of fit in model (17.2) is defined
as
χ
2
=
k
X
i=1
·
(y
i
−
ˆ
y
i
)
2
ˆ
y
i
+
(y
i
−
ˆ
y
i
)
2
n
i
−
ˆ
y
i
¸
,
where n
i
is the number of observations in class i, i =1, . .., k. This statistic has
an approx.
χ
2
-distribution with k − p degrees of freedom.
Goodness of fit measure G is defined as the difference of deviance between
the null model (intercept-only model) and the model under consideration. G
has a
χ
2
-distribution with p −1 degrees of freedom, and small values of G are
critical, suggesting that the deviance did not improve significantly by adding
covariates.
The logistic model can always be expressed in terms of Bernoulli outcomes,
where y
i
is 0 or 1, as in (17.1). Then k = n, n
i
= 1, the likelihood for the sat-
urated model, is
Q
n
i
=1
y
y
i
i
(1 − y
i
)
1−y
i
= 1 (we assume that 0
0
= 1), and the de-
viance for the Bernoulli representation becomes

666 17 Regression for Binary and Count Data
D =−2
n
X
i=1
[
y
i
log
ˆ
p
i
+(1 − y
i
)log(1 −
ˆ
p
i
)
]
.
Statistic D does not follow any specific distribution, irrespective of sample
size. Likewise, the Pearson
χ
2
becomes
χ
2
=
n
X
i=1
(y
i
−
ˆ
p
i
)
2
ˆ
p
i
(1 −
ˆ
p
i
)
(17.3)
in model (17.1) and does not follow any specific distribution, either.
To further evaluate the model, several types of residuals are available.
Deviance residuals are defined as
r
D
i
=sign(y
i
−
ˆ
y
i
)
s
2
½
y
i
log
µ
y
i
ˆ
y
i
¶
+
(n
i
− y
i
)log
µ
n
i
− y
i
n
i
−
ˆ
y
i
¶¾
, i =1, . .., k,
for model (17.2) and
r
D
i
=sign(y
i
−
ˆ
p
i
)
p
2y
i
log
ˆ
p
i
+(1 − y
i
)log(1 −
ˆ
p
i
), i =1, . .., n,
for model (17.1). The deviance D is decomposed to the sum of squares of de-
viance residuals in an ANOVA-like fashion as D
=
P
(r
D
i
)
2
. The squared resid-
ual (r
D
i
)
2
measures the contribution of the ith case to the deviance.
Deviance residuals can be plotted against the order of sampling to explore
for possible trends and outliers. Also useful for checking the model are half-
normal plots where the ordered absolute values r
D
i
are plotted against the
normal quantiles
Φ
−1
³
i+n−1/8
2n+1/2
´
. These kinds of plots are an extension of Atkin-
son’s (1985) half normal plots in regular linear regression models. Deviation
from a straight line in a half-normal plot indicates model inadequacy.
For the model in (17.1), the Pearson residual is defined as
r
pea
i
=
y
i
−
ˆ
p
i
p
ˆ
p
i
(1 −
ˆ
p
i
)
,
and the sum of squares of r
pea
i
constitutes Pearson’s χ
2
statistic,
n
X
i=1
(r
pea
i
)
2
=
n
X
i=1
(y
i
−
ˆ
p
i
)
2
ˆ
p
i
(1 −
ˆ
p
i
)
,
as in (17.3). This statistic represents a discrepancy measure; however, as we
mentioned, it does not follow the
χ
2
-distribution, even asymptotically.
In the case of continuous covariates, large n and small n
i
, Hosmer and
Lemeshow proposed a
χ
2
-statistic based on the grouping of predicted values
ˆ
p
i
. All
ˆ
p
i
are ordered and divided into g approximately equal groups, usually
ten. For ten groups, sample deciles of ordered
ˆ
p
i
could be used.
The Hosmer–Lemeshow statistic is
17.2 Logistic Regression 667
χ
2
HL
=
g
X
i=1
(n
i
−n p
i
)
2
np
i
,
where g is the number of groups, n
i
is the number of cases in the ith group,
and
p
i
is the average of model (predicted) probabilities for the cases in the
ith group. The
χ
2
hl
-statistic is compared to χ
2
g
−2
quantiles, and small p-values
indicate that the fit is poor. In the case of ties, that is, when there are blocks
of items with the same predicted probability
ˆ
p, the blocks are not split but
assigned to one of the two groups that share the block. The details of the algo-
rithm can be found in Hosmer and Lemeshow (1989).
In the case of linear regression, R
2
, as a proportion of model-explained
variability in observations, had a strong intuitive appeal in assessing the re-
gression fit. In the case of logistic regression, there is no such intuitive R
2
.
However, there are several proposals of R
2
-like measures, called pseudo-R
2
.
Most of them are defined in terms of model likelihood or log-likelihood. The
model likelihood and log-likelihood are calculated using model logit,
`
i
= b
0
+b
1
x
i1
+···+b
p−1
x
i,p−1
,
LL
p
= LL(b
0
,... , b
p−1
) =
n
X
i=1
(
y
i
×`
i
−log(1 +exp{`
i
})
)
,
and the model likelihood is L
p
= exp{LL
p
}. The null model is fitted without
covariates, and
`
0
= b
0
LL
null
= LL(b
0
) =
n
X
i=1
(
y
i
×`
0
−log(1 +exp{`
0
})
)
.
The null model likelihood is L
null
=exp{LL
null
}.
By analogy to linear regression, R
2
=
SSR
SST
=
SST−SSE
SST
,
R
2
m f
=
LL
null
−LL
p
LL
null
=1 −
LL
p
LL
null
,
defines McFadden’s pseudo-R
2
. Some other counterparts of R
2
are
Cox–Snell: R
2
cs
= 1 −
·
L
null
L
p
¸
2/n
;
Nagelkerke: R
2
n
=
1 −
h
L
null
L
p
i
2/n
1 −
(
L
null
)
2/n
;
Effron: R
2
e
= 1 −
P
n
i
=1
(y
i
−
ˆ
p
i
)
2
P
n
i
=1
(y
i
− y)
2
, y =
P
n
i
=1
y
i
n
.
668 17 Regression for Binary and Count Data
Example 17.2. Arrhythmia. Patients who undergo coronary artery bypass
graft surgery (CABG) have an approximately 19 to 40% chance of developing
atrial fibrillation (AF). AF is a quivering, chaotic motion in the upper cham-
bers of the heart, known as the atria. AF can lead to the formation of blood
clots, causing greater in-hospital mortality, strokes, and longer hospital stays.
While this can be prevented with drugs, it is very expensive and sometimes
dangerous if not warranted. Ideally, several risk factors that would indicate an
increased risk of developing AF in this population could save lives and money
by indicating which patients need pharmacological intervention. Researchers
began collecting data from CABG patients during their hospital stay such as
demographics like age and sex, as well as heart rate, cholesterol, operation
time, etc. Then the researchers recorded which patients developed AF during
their hospital stay. The goal was to evaluate the probability of AF given the
measured demographic and risk factors.
The data set
arrhythmia.dat, courtesy of Dr. Matthew C. Wiggins, con-
tains the following variables:
Y Fibrillation
X
1
Age
X
2
Aortic Cross Clamp Time
X
3
Cardiopulmonary Bypass Time
X
4
Intensive Care Unit (ICU) Time
X
5
Average Heart Rate
X
6
Left Ventricle Ejection Fraction
X
7
Anamnesis of Hypertension
X
8
Gender [1 - Female; 0 - Male]
X
9
Anamnesis of Diabetes
X
10
Previous MI
The MATLAB script arrhythmia.m provided a logistic regression fit. The
script calculates deviance and several goodness-of-fit measures.
load ’Arrhythmia.mat’
Y = Arrhythmia(:,1);
X = Arrhythmia(:,2:11); %Design matrix n x (p-1) without
%vector 1 (intercept)
Xdes =[ones(size(Y)) X]; %with the intercept: n x p
n = length(Y); %number of subjects
alpha = 0.05; %alpha for CIs
[b, dev, stats]=glmfit(X,Y, ’binomial’,’link’,’logit’)
lin = Xdes
*
b %linear predictor, n x 1 vector
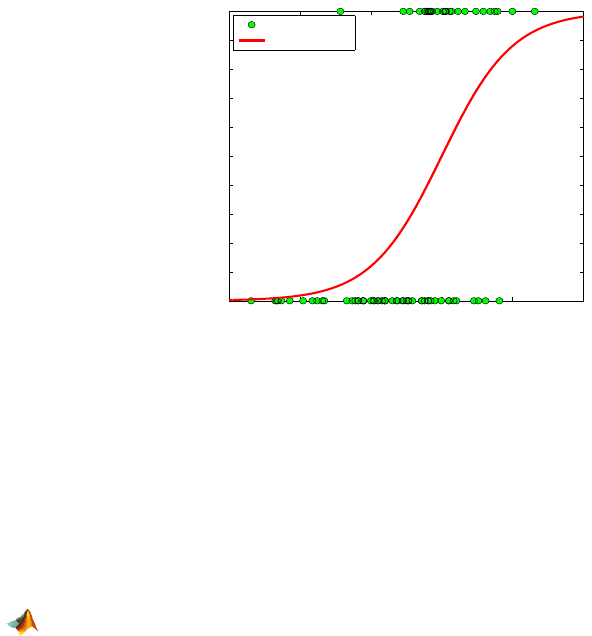
Figure 17.2 shows observed arrhythmia responses (0 or 1) with their logis-
tic fit.
With the linear predictor, fitted probabilities for
{Y
i
= 1} are given as
ˆ
p
i
.
Also, the estimators of the
βs and their standard deviations and p-values for
17.2 Logistic Regression 669
−6 −4 −2 0 2 4
0
0.1
0.2
0.3
0.4
0.5
0.6
0.7
0.8
0.9
1
Linear Predictor: b
0
+ b
1
X
1
+ ... + b
10
X
10
Probability of Arrhythmia
Observations
Logistic Fit
Fig. 17.2 Arrhythmia responses 0 or 1 with their logistic fit. The abscise axis is the linear
predictor
lin.
the Wald test are given next. The intercept is significantly nonzero (0.0158),
and the variable
X1 (age) is strongly significant (0.0005). This agrees with the
inference based on confidence intervals, only the intervals for
β
0
and β
1
do not
contain 0, or, equivalently, the intervals for the odds ratio, exp
{β
0
} and exp{β
1
},
do not contain 1.
phat = exp(lin)./(1 + exp(lin));
V = diag( phat .
*
(1 - phat) );
sqrtV = diag( sqrt(phat .
*
(1 - phat) ))
sb = sqrt( diag( inv( Xdes’
*
V
*
Xdes ) ) )
% inv( Xdes’
*
V
*
Xdes ) is stats.covb
% Wald tests for parameters beta
z = b./sb %tests for beta
_
i = 0, i=0,...,p-1
pvals = 2
*
normcdf(-abs(z)) %p-values
%[0.0158; 0.0005; 0.3007; 0.2803; 0.1347; 0.8061
% 0.4217; 0.3810; 0.6762; 0.0842; 0.5942]
%(1-alpha)
*
100% CI for betas
CIs = [b - norminv(1 - alpha/2)
*
sb , b + norminv(1-alpha/2)
*
sb]
%(1-alpha)
*
100% CIs for odds ratios
CIsOR = exp([b-norminv(1-alpha/2)
*
sb , b+norminv(1-alpha/2)
*
sb])
% 0.0000 0.1281
% 1.0697 1.2711
% 0.9781 1.0744
% ...
% 0.8628 10.3273
% 0.4004 4.9453
Figure 17.3 shows estimators of β
0
−β
10
(as green circles) and 95% con-
fidence bounds. Since the intervals for
β
0
and β
1
do not contain 0, both the
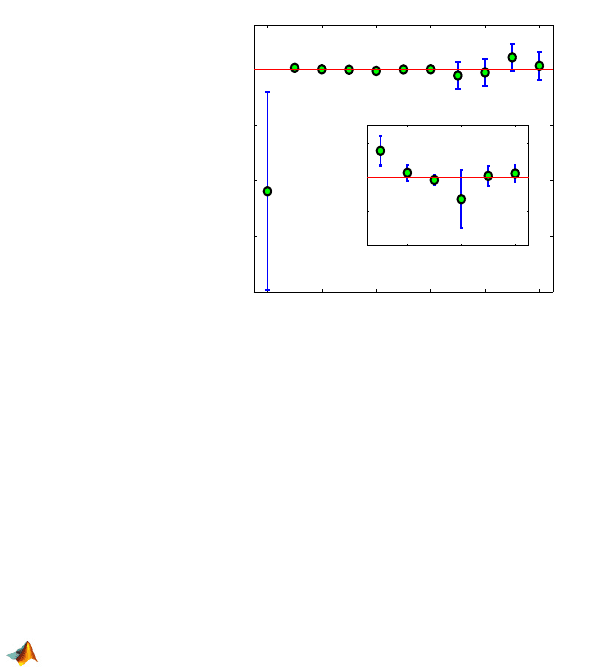
670 17 Regression for Binary and Count Data
intercept and covariate age are important in the model. It is tempting to do
variable/model selection based on outcomes of Wald’s test – but this is not
advisable. Exclusion of a parameter/variable from the model will necessarily
change the estimators and confidence intervals for the remaining parameters
and previously insignificant parameters may become significant. As in linear
regression, best subset, forward, and backward variable selection procedures
exist and may be implemented.
0 2 4 6 8 10
−20
−15
−10
−5
0
index
b ±1.96s
b
2 4 6
−0.4
−0.2
0
0.2
index
b ±1.96s
b
Fig. 17.3 Estimators of β
0
−β
10
are shown as green circles, and 95% confidence intervals
are given. For comparison, the intervals for
β
1
−β
6
are shown separately on a different scale.
Next, we find the log-likelihoods for the model and null model. The model
deviance is 78.2515, while the difference of deviances between the models is
26.1949. This would be a basis for a likelihood ratio test if the response were
grouped. Since in a Bernoulli setup the distributions of deviance and G are
not
χ
2
, the testing needs to be done by one of the response-grouping methods,
such as the Hosmer–Lemeshaw method.
%Log-likelihood
loglik = sum( Y .
*
lin - log( 1 + exp(lin) )) %-39.1258
%fitting null model.
[b0, dev0, stats0] = glmfit(zeros(size(Y)),Y,’binomial’,’link’,’logit’)
%b0=-0.6381, dev0=104.4464, stats=... (structure)
loglik0 = sum( Y .
*
b0(1) - log(1 + exp(b0(1))) ) %-52.2232
%
G = -2
*
(loglik0 - loglik) % 26.1949
dev0 - dev %26.1949, the same as G, difference of deviances
%
%model deviance
devi = -2
*
sum( Y .
*
log( phat + eps) + (1-Y ).
*
log(1 - phat + eps) )
