Ojima I. (ed.) Fluorine in Medicinal Chemistry and Chemical Biology
Подождите немного. Документ загружается.


16
Fluorinated Amino Acids and
Biomolecules in Protein Design and
Chemical Biology
He Meng , Ginevra A. Clark , and Krishna Kumar
16.1 Introduction
While materials scientists have long appreciated the usefulness of fl uorinated compounds,
chemical biologists and protein scientists have just begun to uncover their value. Fluorine
has unique and fascinating properties. The judicious replacement of hydrogen with fl uorine
can provide tools for unraveling the workings of biological systems. Rational design using
fl uorinated amino acids has been successful in deciphering collagen stability, in mapping
ligand – receptor interactions, in unveiling protein folding and dynamics, and in creating
extra - biological structures. In addition, the third - phase properties of perfl uorocarbons have
been utilized in biomolecule enrichment, artifi cial membranes, and small molecule micro-
arrays. Unusual properties of fl uorinated molecules will continue to proffer novel ways to
perturb and observe biological systems.
16.1.1 Fluorine in the Context of Biological Systems
Noncovalent interactions are central to many key biological functions. The self - assembly
of the plasma and organelle membranes, association between receptors and ligands, and
folding of RNA and protein molecules are all controlled by noncovalent interactions.
Noncovalent forces in the design of proteins have proved useful in gaining insight into
Fluorine in Medicinal Chemistry and Chemical Biology Edited by Iwao Ojima
© 2009 Blackwell Publishing, Ltd. ISBN: 978-1-405-16720-8
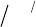
412 Fluorine in Medicinal Chemistry and Chemical Biology
sequence/structure and function relationships. We and others have employed fl uorocarbons
as building blocks to construct biomolecule analogues and exploited their unique charac-
teristics in biological systems. Organic fl uorine exhibits unusual properties. At the level
of substitution of hydrogen by a single fl uorine atom or a few fl uorine atoms (e.g., H to
F, CH
3
to CF
3
), the effects are mainly manifested in electronic, steric, and hydrophobic
properties of the resulting compounds. On the other hand, for perfl uorocarbon chains,
third - phase properties, as distinct from water and hydrocarbons, become more relevant.
We fi rst review here the enrichment and self - assembly of biomolecules based on fl uorous
phase separation. We then highlight some recent uses of fl uorinated amino acids in protein
design and chemical biology.
16.1.2 Unique Properties of Fluorine
Fluorine is the most electronegative element. While fl uorine forms the strongest hydrogen
bonds in the ionic state, it does not readily participate in hydrogen bonding once covalently
bound to carbon [1] . This observation was at fi rst surprising, but careful scrutiny of crystal
structures has revealed few organic molecules displaying F
…
H intermolecular distances
required for hydrogen bonding. These interactions only occur in the absence of better
hydrogen - bond acceptors. This property illustrates an essential feature of fl uorine: it holds
its unshared electrons tightly, evident in its low polarizability ( α = 0.557 × 10
− 24
c m
− 3
,
σ
α
= +0.13 kcal/mol) [2, 3] . The C – F bond (485 kJ/mol) [4] is the strongest formed by
carbon bonded singly to any element. Perfl uorocarbons are chemically inert and thermally
stable. The C – F bond has a reversed and large dipole moment ( µ = 1.85 D) [5] compared
with that of a C – H bond ( µ = 0.4 D) [2] . Despite electron localization on fl uorine, perfl uo-
rocarbons are minimally polarizable, resulting in weak intermolecular interactions and
high vapor pressures.
16.1.3 Phase Separation Properties of Fluorocarbons
The most striking feature of perfl uorocarbons is their insolubility. They phase separate
from nonpolar organic solvents and water at room temperature. This property was exploited
by Horv á th in 1994 with the introduction of fl uorous biphasic catalysis (FBC) [6] . This
new paradigm for separation and purifi cation of catalysts and organic molecules has been
used widely [7 – 9] . Gladysz and Curran [10] have formalized the defi nition of the term
“ fl uorous ” in analogy to “ aqueous ” as “ of, relating to, or having characteristics of highly
fl uorinated saturated organic materials, molecules or molecular fragments. Or, more simply
(but less precisely), ‘ highly fl uorinated ’ or ‘ rich in fl uorines ’ and based upon sp
3
- hybridized
carbon. ”
The Hilderbrand – Scatchard solubility parameter δ (Equation 16.1 ) can be used to
estimate the miscibility of fl uorocarbons with organic solvents [11 – 13] .
δν=
()
∆E
v
12
(16.1)

Fluorinated Amino Acids and Biomolecules in Protein Design and Chemical Biology 413
∆ E
v
is the vaporization energy of the pure component (cal/mol) and v is the molar volume
(cm
3
/mol) at temperature T . Perfl uorocarbons have relatively small δ values due to large
molar volumes and exceedingly low propensities for intermolecular interactions [11] .
In contrast, water has a δ value [14] of
23 5
12 32
. cal cm
−
. The mutual solubility of two
nonpolar liquids (1 and 2) can be estimated primarily on the difference between δ values
| δ
1
− δ
2
|. Solubility also depends on the temperature ( T ) and molar volumes (v
1
and v
2
)
(Equation 16.2 ).
νν δδ
12 12
2
4+
()
⋅−
()
< RT
(16.2)
In general, when δ
1
= δ
2
, there is no heat of mixing and two liquids are miscible in all
proportions. When | δ
1
− δ
2
| is less than
35
12 32
. cal cm
−
for an average molar volume
of 100 mL, the liquids are still completely miscible at room temperature [12] . When the
differences in δ become substantial, phase separation occurs.
16.2 Fluorous - based Methods in Chemical Biology
16.2.1 Fluorous Enrichment of Peptides
Fluorous affi nity separation was originally used to remove catalysts from complex reaction
mixtures [6] . A perfl uoroalkyl moiety (generally no shorter than − C
6
F
13
) is appended to a
compound of interest. Tagged molecules are then rapidly separated from other components
in the mixture by either liquid – liquid extraction or liquid – solid - phase extraction. Fluorous
affi nity - based separation has recently been used in biomolecule purifi cation, proteomics,
and microarray experiments [15 – 20] .
Solid - phase peptide synthesis (SPPS) [21] has greatly facilitated the preparation of
these important biomolecules. The fi nal products often reside in a mixture containing
similar compounds, due to incomplete coupling of certain residues. Purifi cation of the
desired product from the crude mixture is tedious, costly, and environmentally unfriendly.
Our laboratory has developed a new fl uorous capping reagent, a trivalent iodonium salt,
to simplify the purifi cation of polypeptides synthesized on solid support (Figure 16.1 a)
[15, 16] . In analogy to routine capping steps using acetic anhydride (Ac
2
O)/diisopropyl
ethylamine (DIEA), this reagent reacts aggressively with free amines of α - amino acids to
deliver secondary amines with fl uoroalkyl chains. The newly formed bonds between fl uo-
roalkyl tags and peptides are stable enough to survive subsequent coupling, deprotection,
and cleavage conditions in both t - Boc and Fmoc chemistry. The capped peptides can be
removed either by centrifugation or by fl uorous solid - phase extraction (FSPE). Removal
of deletion products greatly simplifi es the purifi cation of desired peptides on reversed -
phase high performance liquid chromatography (RP - HPLC).
Complementary to our capping approach, Boom and Overkleeft have pursued a dif-
ferent strategy using the tagging method (Figure 16.1 b) [22, 23] . This method is suitable
for SPPS using Fmoc chemistry. Peptides are assembled with routine capping steps
(Ac
2
O/DIEA). Upon the completion of the peptide chain, benzyloxycarbonyl - (or
414 Fluorine in Medicinal Chemistry and Chemical Biology
Figure 16.1 Fluorous purifi cation of peptides synthesized on solid phase. (a) Capping method and trivalent iodonium salt reagent
used in both t - Boc and Fmoc chemistry. (b) Tagging method and reagents for Fmoc chemistry.
Fluorinated Amino Acids and Biomolecules in Protein Design and Chemical Biology 415
methylsulfonylethoxycarbonyl - ) based fl uorous ponytails are appended to the full - length
products. The cleavage step frees all peptides from the resin. Tagged full - length peptides
are easily purifi ed on Fluophase ™ columns or by FSPE. An additional required detagging
step then delivers the fi nal products.
In addition, fl uorous tags have been developed for purifi cation of oligonucleotides
[18] and oilgosaccahrides [19] synthesized on solid support.
Peters and colleagues have implemented a similar strategy in proteomics [24] . Pro-
teins of interest were fi rst subjected to enzymatic digestion. Peptide fragments were then
selectively labeled with fl uorous tags via cysteine residues or by β - elimination/Michael
addition to phosphorylated peptides. Tagged peptides were signifi cantly enriched and
isolated from complex mixtures by FSPE. These samples can be directly analyzed by
MALDI - MS or ESI - MS. Fluoroalkyl tags do not undergo fragmentation, providing a
distinct advantage over biotin - based affi nity reagents. This fl uorous derivatization and
enrichment strategy has been extended to analyze small molecules and peptides using
desorption/ionization on silicon mass spectrometry (DIOS - MS) [25] .
16.2.2 Fluorous Small - Molecule Microarrays
Microarrays have tremendously accelerated gene sequencing and biological sample screen-
ing [26, 27] . The distinct advantages are the high - throughput nature and the minuscule
amounts of sample required. In contrast, small molecules of interest in pharmacology
and biology are discovered by tedious and expensive procedures. Small - molecule microar-
rays (SMMs) offer an attractive alternative for this discovery process. One challenge
confronting SMMs is the immobilization of small molecules onto glass slides. This step
conventionally relies on covalent modifi cations of small molecules, often requiring
multiple chemical steps. Fluorous SMMs appear to be promising for alleviating this
diffi culty.
Pohl and co - workers have developed fl uorous - based carbohydrate microarrays [28] .
The noncovalent interactions between fl uorous tagged carbohydrates and fl uorous func-
tionalized slides are the focus here in the array fabrication. Sugars (mannose, galactose,
N - acetylglucosamine, and fucose) were selected as initial targets. The C
8
F
17
tails were
appended to protected trichloroacetimides of these monosaccharides to produce fl uorous
tagged sugars. Next, fl uorous sugars were spotted onto a commercially available glass
slide coated with a Tefl on/epoxy mixture. The fl uorous - based microarrays were then inter-
rogated using fl uorescein isothiocyanate - labeled jack bean lectin concanavalin A (FITC -
ConA). FITC - ConA bound exclusively to mannose as expected. This experiment
demonstrated that the C
8
F
17
groups anchored fl uorous sugars onto the glass slide, and
that this type of microarray could be used for investigating carbohydrate – protein
interactions.
Spring and co - workers have applied the same principle for fabricating fl uorous - based
SMMs to illustrate the recognition of small - molecule ligands by proteins [29] . The
prototype binding pair of biotin – avidin was employed, in which biotin was conjugated
to fl uorous tags and “ printed ” onto fl uoroalkyl - coated glass slides. Avidin interacted with
biotin on the glass surface as judged by the fl uorescence emanating from dyes linked
to avidin.
416 Fluorine in Medicinal Chemistry and Chemical Biology
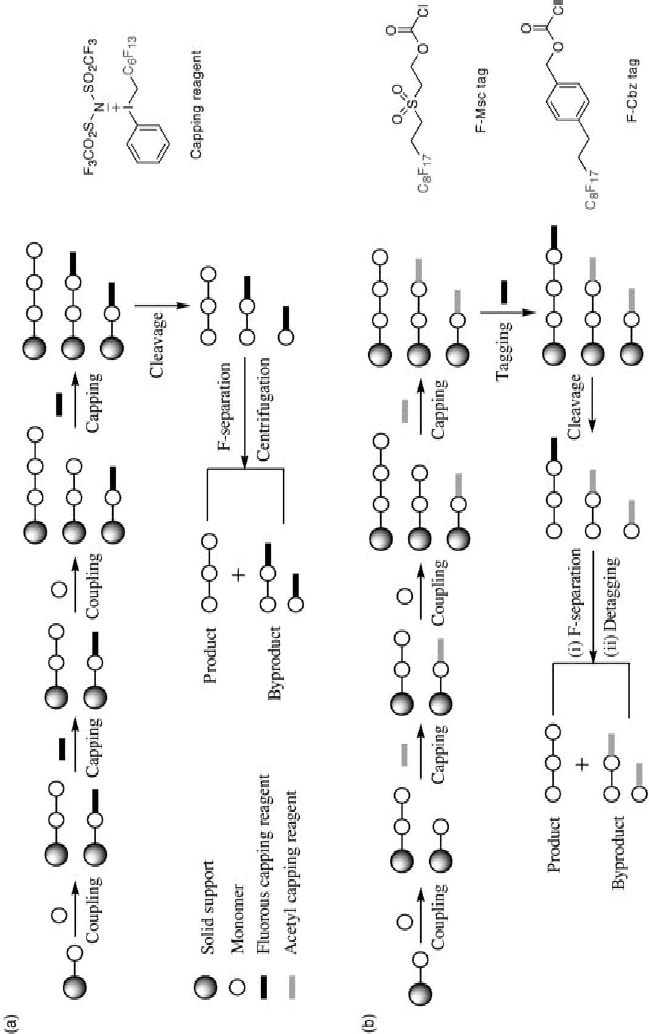
Schreiber and co - workers have also developed fl uorous - based SMMs (Figure 16.2 ) for
the discovery of histone deacetylase (HDAC) inhibitors [30] . Twenty small molecules com-
prised of putative active and inactive HDAC inhibitors were appended to C
8
F
17
- based
anchors. Among them was included suberoylanilide hydroxamic acid (SAHA), a known
inhibitor of multiple members of the HDAC family. Once the array was produced as above,
purifi ed His - tag fused HDAC2, HDAC3/NCoR2, and HDAC8 were allowed to interact
with the array. Detection by Alexa - 647 - labeled anti - His antibody revealed several binders
for each enzyme. The nonfl uorous tagged compounds were examined to check their inhibi-
tory capacity against their targets in a fl uorescence - based biochemical activity assay.
Further, these compounds were also studied by surface plasmon resonance (SPR) for
binding to HDAC3/NCoR2. It is important to note that the results obtained from these three
different techniques are in good agreement, validating the fl uorous - based SMM method.
Figure 16.2 Fluorous small - molecule microarrays. Small - molecule histone deacetylase
(HDAC) binders are noncovalently immobilized onto a glass slide coated with fl uorocarbon
compounds. An antibody labeled with a fl uorescent dye recognizes HDAC proteins. See color
plate 16.2.
( Source: Vegas, A. J., Brander, J. E., Tang, W. et al. , Fluorous - based small - molecule microarrays
for the discovery of histone deacetylase inhibitors, Angew. Chem. Int. Ed. (2007), 46 , 7960 –
7964. Copyright Wiley-VCH Verlag GmbH & Co. KGaA. Reproduced with permission.)
Fluorinated Amino Acids and Biomolecules in Protein Design and Chemical Biology 417
16.2.3 Fluorinated Lipids
Phospholipids are ubiquitous components of biological membranes and play key roles in
confi ning cellular components, regulating cell – cell interactions, and supporting membrane
protein and lipid functions. They are therefore commonly used to fabricate model mem-
branes such as vesicles and supported lipid bilayers for studies in cell biology, drug
delivery, and materials science. Clustered display of ligands and receptors has been pos-
tulated to be mediated by phase - separated microdomains (or “ lipid rafts ” ). Using fl uoro-
carbons to control lipid self - assembly and present clustered biological ligands would be
very useful for probing such mechanisms.
Bendas, Schmidt, and Tanaka have designed fl uorinated glycolipids with a glycerol
diether core. The glycolipid sLexFL88 (sialyl Lewis
x
hexasaccharide with a n - perfl uorooc-
tanyl tail) self - associates into atypical clusters in 1,2 - distearoyl - sn - glycero - 3 - phosphocho-
line (DSPC) lipids [31] . With 1 mol% of sLexFL88 in DSPC, the domains average ∼ 300 nm
in diameter. The size of these clusters increases with increasing concentrations of sLexFL88.
CHO (Chinese hamster ovary) cells expressing E - selectins were used to explore leukocyte
rolling on these model membranes. The size of sLexFL88 clusters and the distance
between clusters dramatically infl uenced cell binding and rolling [32] . Clustering of
sLexFL88 enhanced the cell - binding behavior; however, cells migrated more slowly on
membranes composed of sLexFL88 and LacFL88 than on those made from sLexEO6 and
DSPC [32] .
Our group has fabricated supported lipid bilayers using 2 - dipalmitoyl - sn - glycero - 3 -
phosphocholine (DPPC) and its fl uorinated derivative [33] . The fl uorinated derivative was
synthesized by replacing the terminal n - hexyl groups of acyl chains with n - perfl uorohexyl
groups. Such membranes contain intricate composition - dependent structures, which are
stripes of ∼ 50 – 100 nm interspersed between ∼ 1 µ m sized domains. Furthermore, variable -
temperature atomic force microscopy (AFM) revealed that domains and stripes are intrin-
sic features of two gel phases of DPPC and its fl uorinated counterpart at low temperatures.
These results suggest that fl uorinated lipids self - assemble into segregated microdomains
in biologically relevant lipid environments. Clustering derived from fl uorination could be
used for investigating polyvalent interactions and producing biomaterials with specifi c
surface functionalities.
16.3 Fluorinated Amino Acids in Protein Design
16.3.1 Unique Properties of Fluorinated Amino Acids
There are myriad ways by which fl uorine can be exploited to perturb the properties of
biomolecules. Fluorine is scarce in living systems and the use of
19
F NMR for studying
protein structure and dynamics is extremely useful. Fluorine is massively electron with-
drawing and signifi cantly perturbs acidity constants of amino acids [34] . For example, the
p K
a
values for pentafl uorophenylalanine (2.2 and 8.3) [35] and hexafl uorovaline (3.17 and
6.30) [36] are greatly shifted from their hydrocarbon counterparts (2.16 and 9.1 for Phe;
2.61 and 9.71 for Val) [35] . It can also infl uence hydrogen - bonding properties of the
418 Fluorine in Medicinal Chemistry and Chemical Biology
backbone. The inductive properties reverse the quadrupole of aromatic rings, altering
magnitudes of π – π interactions and π – cation interactions [37] . Fluorine substitution also
infl uences the conformational preferences of proline, resulting in a change in the trans/cis
ratio of the peptide bond [38] . Finally, substitution of hydrogen by fl uorine infl uences the
α - helical propensities of amino acids [39] .
While a C – F substitution dramatically changes the electronic properties of a C – H
bond, it exerts only a minor steric infl uence [40] . The van der Waals radius of fl uorine
(1.47 Å ) is only slightly larger than that of hydrogen (1.2 Å ) [41] . In general, a C– F group
is nearly isosteric with a C – H group. A CH
3
to CF
3
substitution, on the other hand, signifi -
cantly increases the bulk. The volume of the van der Waals hemisphere changes from
16.8 Å
3
for CH
3
to 42.6 Å
3
for CF
3
[4] . The van der Waals volume of CF
3
is comparable
to an ethyl group and is only slightly smaller than that of an isopropyl group [42] .
Another important consideration is how substitution of CH
3
with CF
3
infl uences
hydrophobicity. The driving force for folding of globular proteins, and many ligand
binding events, is hydrophobic interactions [43, 44] . The strength of the hydrophobic effect
is related to the size and the molecular nature of the hydrophobic group. Whitesides and
co - workers have demonstrated that the interaction of a hydrophobic ligand with carbonic
anhydrase is directly proportional to the surface area of the hydrophobic group [45] .
Fluorocarbons were more hydrophobic than hydrocarbons as the surface area is larger for
fl uorocarbons. (Hansch parameters for CF
3
( Π = 1.07) and CH
3
( Π = 0.5) groups) [46] .
The Hansch parameters for side - chains of hexafl uoroleucine and Leu are 1.87 and 1.58
[47] , pointing to the superior hydrophobicity of the fl uorinated amino acid. As such, the
substitution of CH
3
with CF
3
should result in an increase in the conformational stability
of a protein if the CF
3
group is shielded from exposure to solvent in the folded
structure.
16.3.2 Incorporation of Fluorinated Amino Acids into Proteins
In order to use fl uorinated amino acids to study biological systems, they need to be syn-
thesized and incorporated. Despite the challenges in both steps, there are several methods
available. For instance, enantiomerically pure fl uorinated amino acids may be prepared by
asymmetric synthesis or by stereochemical resolution using enzymatic methods [48] .
Fluorinated amino acids can be introduced into proteins biosynthetically, or chemically
by SPPS. Several reviews that detail the synthesis of enantiomerically pure fl uorinated
amino acids and incorporation methods into proteins are available [48 – 51] .
16.3.3 Fluorine NMR for Structure Determination
Nuclear magnetic resonance (NMR) methods have emerged as an important complement
to X - ray crystallography for determining protein structure. Structural details can be
obtained for proteins that are not readily crystallized, such as membrane proteins or molten
globules. However, even with NMR methods, it is inherently diffi cult to obtain structural
information on dynamic protein states. One major problem is that
1
H NMR resonances
tend to overlap, making interpretation of spectra diffi cult. Because
19
F has a large magne-
Fluorinated Amino Acids and Biomolecules in Protein Design and Chemical Biology 419
togyric ratio, is present in 100% natural abundance, and displays a broad dispersion in
chemical shifts, it represents an invaluable probe for exploring protein dynamics and
protein – protein interactions [52] .
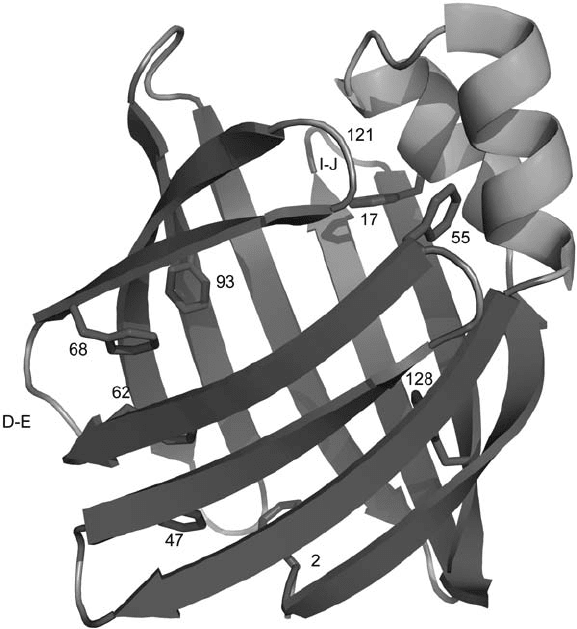
Rat intestinal fatty acid binding protein (IFABP) binds and transfers fatty acids to
their metabolic destination. Structures of apo - and holo - IFABP have been solved by NMR
and X - ray crystallography. IFABP consists of two short α - helices and 10 antiparallel β -
sheets arranged to form a ligand - binding cavity (Figure 16.3 ). The side - chains of several
aromatic amino acids in IFABP play an important role in both structure and ligand binding.
Knowledge of the details of the conformation and dynamics of these side - chains would
allow a deeper understanding of the protein – ligand interaction and protein folding. To
extract such details, Frieden and co - workers replaced eight phenylalanine (Phe) residues
with 4 - F - Phe [53] . One - dimensional (1D)
19
F resonance assignments were based on the
Figure 16.3 The structure of apo - IFABP (PDB code: 1IFB). Eight Phe residues (shown in
stick representation), the D
– E and I – J regions, and location of G121 are indicated by labels.
The structure was generated using MacPyMOL (DeLano Scientifi c LLC, Palo Alto, CA, U.S.A.).
See color plate 16.3.
420 Fluorine in Medicinal Chemistry and Chemical Biology
chemical shift of individual proteins containing one or two 4 - F - Phe residues. The ligand -
bound (oleic acid) and ligand - free forms were then analyzed by 2D
19
F –
19
F NOE (nuclear
Overhauser effect) spectra and linewidth measurements. Upon ligand binding, the NOE
spectrum revealed more exchange cross - peaks, and the 1D
19
F spectrum indicated that
most of the peaks were broadened. These results suggested that aromatic side - chains in
the binding cavity surprisingly become more fl exible upon ligand binding [54] , even
though the backbone is more rigid in the ligand - bound form [55] .
Fluorine labels have also been used to explore the folding process of IFABP. Protein
folding involves interactions among side - chains and backbone hydrogen bonds [56] . One
challenge in elucidating this process is the characterization of “ intermediates ” between
folded and unfolded states. The “ acid state ” of IFABP may well resemble such an inter-
mediate – also referred to as a “ molten globule. ” Pulsed - fi eld gradient (PFG) NMR indi-
cated that apparent hydrodynamic radii of the acid state IFABPs were larger than that of
the native state, but smaller than that of the fully denatured state. One - dimensional
19
F
spectra showed that IFABP was structured at pH 2.8, whereas it was mostly unfolded at
pH 2.3. It is of note that even at pH 2.3, there was still a small portion of the protein in
its native - like structure. More importantly, at pHs lower than 4.8, signifi cant changes in
chemical shift and linewidths were observed for Phe128, 17, 68, and 93, suggesting that
the D – E turn and I – J regions are involved in the early stages of unfolding. The overall
structure of the hydrophobic core remained intact at pH 2.8, as indicated by
19
F –
19
F NOE
between Phe68 and Phe93 as well as circular dichroism (CD) measurements. Collectively,
changes in side - chain orientations occur before structural changes in the backbone upon
lowering the pH, even though these side - chains are buried in the hydrophobic core. Upon
oleic acid binding, the spectrum at pH 2.3 resembles the spectrum at higher pH, suggesting
that the ligand binding may shift the unfolded protein to a native - like structure [57] . The
folding kinetics of IFABP were also monitored by
19
F NMR on a slow - folding mutant in
which Val replaced Gly121 [58] . This single - site replacement is postulated to disrupt a
normal nucleation site in the I – J region. The G121V mutant folds more slowly, is less
stable, and exhibits the same structure and dynamics as wild type, allowing for a closer
examination of its folding behavior by stop - fl ow
19
F NMR and CD. In the process of
refolding, the secondary structures formed twice as quickly as the stabilized conformation
of side - chains. A local, nonnative - like structure involving Phe62, Phe68, and Phe93
appeared within milliseconds, followed by arrangement of Phe2 and Phe17, and fi nally
Phe47 into their conformations. This is followed by an overall rearrangement. Without
19
F
NMR, it would have been a serious challenge to shed light on both side - chain dynamics
and intermediate states during folding of IFABP [58] .
Mehl and co - workers have incorporated 4 - CF
3
- Phe into proteins to elucidate struc-
tural changes in nitroreductase upon addition of the cofactor fl avin mononucleotide. When
4 - CF
3
- Phe was introduced in the active site (Phe124), the
19
F signal shifted upon ligand
addition. If it was introduced at a distant site (Phe36), the change in chemical shift was
quite subtle. In addition, substitution of 4 - CF
3
- Phe did not alter the activity of the enzyme
[59, 60] . These results suggest that 4 - CF
3
- Phe could be a sensitive probe for probing ligand
binding to proteins.
Membrane proteins are good pharmaceutical targets, since they are displayed on the
cell surface where drugs can bind without permeating lipid bilayers. Furthermore, they are
involved in cell - signaling interactions, viral entry, and a host of other processes important
