Moss Tom. DNA-protein interactions: principles and protocols
Подождите немного. Документ загружается.

Footprinting with Exonuclease III 45
nondenaturing acrylamide gel, as described in Subheading 3.1., provided the
complexes show different gel mobilities and have half-lives long enough to
survive the electrophoresis. Such a purification step requires the use of 10 times
more radioactively labeled DNA.
The procedure is as follows:
1. Form the complex.
2. Subject the complex to digestion with Exo III according to Subheading 3.3.,
step 2.
3. Dialyze the complex by drop dialysis against a low salt buffer (e.g., 10 mM
Tris-HCl, pH 7.9) in order to avoid salt effects during electrophoresis (Subhead-
ing 3.1., step 5).
4. Apply the complex to a nondenaturing gel as described in Subheading 3.1., steps
6 and 7. The half-life of the complex is in most cases not changed by the Exo III
digestion of the DNA.
5. Expose the gel at –70°C to X-ray film using an intensifying screen. A 1-h expo-
sure should be enough to recognize the complexed bands. If not, the recovered
DNA will be insufficient for the subsequent sequencing gel analysis.
6. Before removing the film for development mark exactly its position on the gel.
7. Excise the complex bands of interest with a spatula. The band representing the
free DNA will be visible as a smear after Exo III digestion.
8. To elute the complexed DNA, put the excised gel slice in 600 µL of bidistilled
water. Heat the complex to 90°C for 3 min and shake overnight at room tempera-
ture. The effectiveness of the elution can be easily monitored by comparing the
radioactivity of the eluate with the radioactivity of the gel slice.
9. Vacuum-dry the eluate in a SpeedVac concentrator.
10. Dissolve the pellet in 10 µL formamide buffer and spin down the gel residue.
11. Transfer the supernatant to a new Eppendorf tube and apply the sample to a
sequencing gel as described in Subheading 3.2., steps 7–9.
4. Notes
1. The gel concentration has to be adjusted according to the molecular weight of the
protein–DNA complex. Here we describe the conditions established for the study
of the E. coli RNA polymerase (MW 455,000) and a DNA fragment of 130 bp
carrying a promoter (9). For some applications, another widely used non-
denaturing gel system may be appropriate: 1X TBE buffer, 4% acrylamide, and
0.1% bis-acrylamide. Recirculation of the buffer is not necessary here.
2. It has been observed that many batches of commercially available Exo III contain an
activity that removes the 5' label. A 5'-phosphatase or a 5'–3' exonuclease activ-
ity could account for this phenomenon. Filling in the 5' protruding ends using α-
thio-dNTPs as described by some authors (10,11) may eliminate the problem.
Addition of E. coli tRNA can reduce the effect, but will not completely avoid it.
46 Metzger and Heumann
3. Investigation of the complexes of specific binding of proteins and DNA in crude
extracts using Exo III requires additional precautions in order to avoid problems
caused by endogenous nuclease activities during Exo III exposure. To avoid this
problem, sodium-phosphate, tRNA, deoxyoligonucleotides and fragmented
phage DNA (e.g., 2 mM sodium phosphate, 1 µg of FX 174 DNA cut with HaeIII,
10 µg of yeast tRNA, and 1 µg mixed p[dN]
5
) should be added to the assay (5).
We find this suppresses nuclease and possibly phosphatase activities contained
in the crude extracts (see also Note 2).
4. Testing different concentrations of Exo III and different incubation periods can pro-
vide additional information about the nature of the protein–DNA complex under
study. If Exo III is able to “nibble” into a protected area with increasing exposure
time, this indicates differences in the strength of protein–DNA interaction (10).
5. Different binding sites for one or more proteins may be detected as distinct stop
points for Exo III, as shown in refs. 12–14. This applies as much when working
with crude extracts as when using purified factors. It is necessary, however, that
the ratio of DNA to binding proteins be >1.
6. Heparin, which is often used as a DNA competitor for E. coli RNA–polymerase
and other DNA-binding proteins, also interacts with Exo III and reduces its activ-
ity markedly.
References
1. Rogers, S. G. and Weiss, B. (1980) Exonuclease III of Escherichia coli K-12, an
AP endonuclease. Methods Enzymol. 65, 201–211.
2. Shalloway, D., Kleinberger, T., and Livingston, D. M. (1980) Mapping of SV 40
DNA replication origin region binding sites for the SV 40 DNA replication anti-
gen by protection against Exonuclease III digestion, Cell 20, 411–422.
3. Metzger, W., Schickor, P., and Heumann, H. (1989) A cinematographic view of
Escherichia coli RNA polymerase translocation. EMBO J. 8, 2745–2754.
4. Pavco, P. A. and Steege, D. A. (1990) Elongation by Escherichia coli RNA poly-
merase is blocked in vitro by a site specific DNA binding protein. J. Biol. Chem.
265, 9960–9969.
5. Wu, C. (1985) An exonuclease protection assay reveals heat-shock element and
TATA box binding proteins in crude nuclear extracts. Nature 317, 84–87.
6. Loh, T. P., Sievert, L. L., and Scott, R. W. (1990) Evidence for a stem cell-spe-
cific repressor of Moloney murine leukemia virus expression in embryonic carci-
noma cells. Mol. Cell. Biol. 10, 4045–4057.
7. Carnevali, F., La Porta, C., Ilardi, V., and Beccari, E. (1989) Nuclear factors spe-
cifically bind to upstream sequences of a Xenopus laevis ribosomal protein gene
promoter. Nucleic Acids Res. 17, 8171–8184.
8. Fried, M. and Crothers, D. M. (1981) Equilibria and kinetics of lac repressor-
operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 9,
6505–6525.
9. Heumann, H., Metzger, W., and Niehörster, M. (1986) Visualization of interme-
diary transcription states in the complex between Escherichia coli DNA-depen-
Footprinting with Exonuclease III 47
dent RNA polymerases and a promoter-carrying DNA fragment using the gel
retardation method. Eur. J. Biochem. 158, 575–579.
10. Straney, D. C. and Crothers, D. M. (1987) A stressed intermediate in the formation
of stably initiated RNA chains at the Escherichia coli lac UV 5 promoter. J. Mol.
Biol. 193, 267–278.
11. Straney, D. C. and Crothers, D. M. (1987) Comparison of the open complexes
formed by RNA polymerase at the Escherichia coli lac UV 5 promoter. J. Mol.
Biol. 193, 279–292.
12. Gaur, N. K., Oppenheim, J., and Smith, I. (1991) The Bacillus subtilis sin gene, a
regulator of alternate developmental processes, codes for a DNA-binding protein.
J. Bact. 173, 678–686.
13. Owen, R. D., Bortner, D. M., and Ostrowski, M. C. (1990) ras Oncogene activa-
tion of a VL30 transcriptional element is linked to transformation. Mol. Cell. Biol.
10, 1–9.
14. Wilkison, W. O., Min, H. Y., Claffey, K. P., Satterberg, B. L., and Spiegelman, B.
M. (1990) Control of the adipisin gene in adipocyte differentiation. J. Biol. Chem.
265, 477–482.
Further Reading
Kow, Y. W. (1989) Mechanism of action of Escherichia coli Exonuclease III.
Biochemistry 28, 3280–3287.

Hydroxyl Radical Footprinting 49
49
From:
Methods in Molecular Biology, vol. 148: DNA–Protein Interactions: Principles and Protocols, 2nd ed.
Edited by: T. Moss © Humana Press Inc., Totowa, NJ
5
Hydroxyl Radical Footprinting
Evgeny Zaychikov, Peter Schickor, Ludmilla Denissova,
and Hermann Heumann
1. Introduction
The basic principle of the DNA footprinting technique is the measurement
of accessibility of the DNA using a probe. The probe can be any enzyme or a
chemical reagent that is able to cut the DNA backbone. When the target DNA
is a fragment containing a signal sequence for a sequence-specific binding
protein, sites on the DNA that interact with the protein are inaccessible to the
probe. After electrophoretic separation based on molecular weight, these inac-
cessible sites appear as blanks in an otherwise regular DNA cleavage pattern,
thus revealing the characteristic interaction footprint for the binding protein.
Although the footprinting pattern is a characteristic of the protein–DNA
interaction, it is also greatly affected by the type of probe used. Hydroxyl radi-
cals provide DNA footprinting probes, which are very convenient to handle
and are distinguished by a number of distinct advantages:
1. Hydroxyl radicals cut the DNA with almost no sequence dependence.
2. Because the probe is very small, the resolution of the footprint is very high (1 bp).
3. The cleavage reaction is effective over a wide range of buffer compositions, salt
concentrations, pHs, and temperatures. Only glycerol, a radical scavenger, inter-
feres with the cutting when present at concentrations higher than 0.5%.
4. All chemicals needed are easily available and uncomplicated in their handling.
1.1. Generation and Action of Hydroxyl Radicals
Hydroxyl radicals are generated according to the Fenton reaction by reduc-
tion of iron(II) with hydrogen peroxide as follows:
ascorbate or
diothiothreited
Fe
2+
(EDTA
4–
) + H
2
O
2
→
Fe
3+
(EDTA
4–
) + OH·
50 Zaychikov et al.
The resulting iron(III) is reduced by ascorbate or dithiothreitol back to
iron(II), which can start a new cycle. The use of a negatively charged
[Fe(EDTA)]
2–
complex prevents the iron from interacting electrostatically with
DNA, so the only reactant interacting with DNA is the hydroxyl radical gener-
ated in solution (1).
An alternative source of hydroxyl radicals is potassium peroxonitrite (2).
The radicals are generated via its conjugate acid (ONOOH) when adding a
stable alkaline solution of ONOOK in samples buffered at neutral pH:
ONOOH → NO
2
· + OH·
2NO
2
· → N
2
O
4
N
2
O
4
+ H
2
O → NO
3
–
+ NO
2
–
+ 2H
+
The exact manner in which hydroxyl radicals act on DNA is still not known.
The radicals are thought to abstract an H-atom from the sugar moiety of the
DNA backbone, and secondary reactions of the resulting sugar radical cause
the backbone to break, leaving a gap in one strand of the double helix with
the phosphate groups on either side (3).
1.2. Principle of the Procedure
After formation of the complex of a sequence-specific binding protein and a
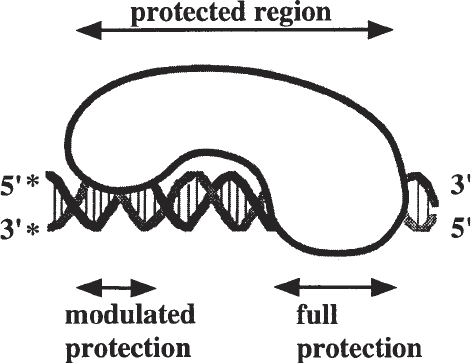
DNA fragment carrying the binding sequence (see Fig. 1), the complex is sub-
Fig. 1. A putative DNA-binding protein interacts with the DNA over three helical
turns. The major portion of the protein interacts with only one side of the DNA over
two helical turns. A minor portion of the protein wraps fully around the DNA. The
asterisk indicates the position of the radioactive label.

Hydroxyl Radical Footprinting 51
jected to hydroxyl radical treatment. Hydroxyl radicals introduce single-base
deletions randomly distributed in the DNA. The concentration of the hydroxyl
radicals is adjusted so that the yield of deletions is less than one per DNA such
that approx 10% of the DNA fragments are affected. Cutting of the DNA is
prevented at those sites on the DNA where the protein is bound. This partially
cut DNA is applied to a sequencing gel. If the DNA is detected by a unique
terminal radioactive label, a DNA ladder is produced that is similar to that
obtained by sequence analysis. Blanks within this regular ladder indicate the
sites where the protein is bound. This footprint becomes more evident if a ref-
erence DNA is included that has been subjected to the same procedure but
without previous protein binding. If complex formation is incomplete (i.e., the
assay contains free DNA), the footprint will be masked by apparent cleavage
within the protein-binding site. This can be avoided by separating the hydroxyl-
radical-treated protein DNA complex from free DNA by nondenaturing gel
electrophoresis (Fig. 2) or by nitrocellulose filter binding before application to
a sequencing gel. Figure 3 shows schematically the footprinting pattern of the
protein DNA complex depicted in Fig. 1.
1.3. Interpretation of the Footprinting Pattern
Figure 3 shows that a footprinting analysis of both DNA strands includes
six DNA ladders (i.e., two DNA sequencing reaction length standards, two
free DNAs as reference, and two complexed DNAs). Blanks in the DNA lad-
der indicate exclusion of radical attack of the DNA because of the presence of
the bound protein. These blanks can be assigned to specific sequence positions
via the length standards.
Fig. 2. Protein-DNA complexes are separated from free DNA by nondenaturing gel
electrophoresis. The two lanes represent the same complex with a single label at one
end of the DNA, at the 3' end, or at the 5' end. Hydroxyl radicals create gaps in the
DNA, which cause a significant retardation of the modified fragments within the gel.
This effect is visible only in the free DNA and is indicated by the shaded shoulder on
the lower band.

52 Zaychikov et al.
The following information can be extracted from the hydroxyl radical
footprinting pattern:
1. The total size of the DNA sequence interacting with the protein can be deter-
mined from the extent of the uncleaved or blank positions.
2. A variation of the intensity of the bands within the interacting sequence reflects
differences in the modes of interaction. By a comparison of the footprints on both
strands, the modes of interaction can often be interpreted:
a. If both strands show a blank at the same region, it indicates that the protein
wraps around the DNA.
b. A blank on only one strand indicates single-strand formation, with one strand
protected by interaction with the protein.
Fig. 3. The bands of the nondenaturing gel in Fig. 2 containing the complex and the
free DNA are eluted and applied (under denaturing conditions) to a sequencing gel.
Lanes 1 and 4 show the free DNA labeled respectively at the 3' and the 5' ends. Lanes
2 and 3 show the DNA recovered from the complex. The complex depicted schemati-
cally in Fig. 1 would result in the footprint displayed in lanes 2 and 3. Lanes G contain
the length standards obtained by a G-specific Maxam–Gilbert sequencing reaction.
Hydroxyl Radical Footprinting 53
c. Modulation of the intensity of the bands with a regular phasing according to
the helix repeat (e.g., 10.3 bp for B-DNA) indicates binding of the protein to
one side of the DNA. This interpretation is supported if the complementary
strand shows the same pattern but with an offset of two or three bases. This
offset is a consequence of the double-helical nature of the DNA, as shown
schematically in Fig. 1.
1.4. Examples of the Application
of Hydroxyl Radicals as Footprinting Probes
1.4.1. Protein DNA Complexes
Numerous Fe
2+
-dependent hydroxyl radical footprinting studies were per-
formed on protein–DNA complexes. Most useful were those studies of protein–
DNA contacts within the transcription machinery.
1. Hydroxyl radicals were used to follow the formation of the transcriptionally
active complex between the DNA-dependent RNA polymerase of E. coli and
T7A1 promoter (4). Temperature-dependent footprinting studies showed that the
transcription initiation complex undergoes three different conformations charac-
terized by a specific “footprint” until a transcription competent complex is
formed. These conformations could be attributed to the so-called closed,
intermediate, and open complex. A bent conformation of DNA in complex with
DNA was concluded from the OH radical probing of the lambda P
R
promoter
complex (5).
2. Site-specific cleavage of the DNA of a RNA polymerase binary complex by both
free and EDTA-chelated Fe
2+
was detected in absence of Mg
2+
ions (6,7). A phe-
nomenon in FeEDTA-dependent OH radical footprints is hypercleavage of DNA
which can be observed in footprints of Mg
2+
-dependent proteins such as E. coli
RNA polymerase (6) or HIV reverse transcriptase (8). The E. coli RNA poly-
merase footprint contains a hyperreactive cleavage spot within the protected
region. This spot could be attributed to a site-specific OH radical cleavage mecha-
nism. This view was supported by footprinting studies using peroxonitrite as an
alternative reagent for generating OH radicals. Using this latter reagent, no
hyperreactive spot was visible in the OH radical footprint of E. coli RNA poly-
merase, indicating that specifically bound FeEDTA probe was responsible for
the hyperreactive cleavage. Free Fe
2+
ion can replace the catalytically active Mg
2+
at the polymerization site of RNA polymerase, being chelated with aspartates of
the active site (7). The chelated Fe
2+
generates OH radicals probably according
to a mechanism that is analogous to the Fenton reaction and causes a strong local
cleavage of both DNA and protein. This hyperreactivity was also observed in
Fe
2+
-dependent OH radical footprints of reverse transcriptase (RT) of the human
immunodeficiency virus (HIV-RT). The catalytically active Mg
2+
of the RNaseH
active site of HIV-RT was replaced by Fe
2+
, leading to site-specific OH radical
cleavage of the DNA. It is interesting to note that no hyperreactive cleavage was
observed at the other Mg
2+
-carrying active site of HIV-RT (i.e., at the polymer-
54 Zaychikov et al.
ization site). These different effects of Mg
2+
/Fe
2+
substitution in the polymeriza-
tion sites of HIV-RT and E. coli RNA polymerase probably reflect a variation of
the redox potentials of the two sites.
3. The movement of E. coli RNA polymerase during mRNA synthesis was followed by prob-
ing a series of specifically arrested transcribing complexes with hydroxyl radicals (9–11).
1.4.2. Antibiotic DNA Complexes
Mithramycin, a small antitumor antibiotic drug, was shown to bind to the
minor groove of GC-rich DNA sequences, thereby protecting only three bases
from hydroxyl radical attack (12).
1.4.3. DNA Structures
The accessibility of bent DNA was studied using hydroxyl radicals. The
bend was induced by A tracts repeated in phase with the helical repeat (13).
Hydroxyl radicals can also be used to measure the number of base pairs per
helical turn along any DNA molecule. The DNA is adsorbed onto crystalline
calcium phosphate before being subjected to radical treatment. From the varia-
tion of the intensity of the bands, the helical periodicity of the DNA can be
directly obtained (14).
1.4.4. RNA Protein Complexes
Splicing-specific ribonucleoprotein complexes were analyzed by hydroxyl
radical treatment. These studies revealed that several regions of the 3'-splice
site of mRNA precursors are not accessible for hydroxyl radicals, for example,
the 3'-intron/exon junction, the polypyrimidine tract, and the site of branch
formation were found to be all inaccessible (15).
1.4.5. RNA Structures
By using hydroxyl radicals, Celander and Cech (16) demonstrated that at least
three magnesium ions are necessary for formation of a catalytically active ribozyme
RNA molecule. By using both FeEDTA and ONOOK-generated hydroxyl radi-
cals, information on higher-order structures of tRNA was obtained (2).
2. Materials
2.1. The Cutting Reaction
Prepare the following solutions separately (see Note 1):
1. 0.1 M dithiotreitol (DTT).
2. 1% Hydrogen peroxide.
3. Iron(II)–EDTA mix: Mix equal volumes of 2 mM ammonium iron(II) sulfate
hexahydrate ((NH
4
)
2
Fe(SO
4
)
2
·6H
2
O) and 4 mM EDTA.
4. Stop mix: 4% glycerol, 0.6 M sodium acetate, 0.1 mg/mL carrier DNA.
Hydroxyl Radical Footprinting 55
2.2. The Sequencing Gel
1. Urea (ultra pure).
2. 20X TBE: 1 M Tris-base, 1 M boric acid, and 20 mM EDTA.
3. Acrylamide solution: 40% acrylamide and 0.66% bis-acrylamide (see Note 2).
4. 10% Sodium persulfate (see Note 3).
5. 10% N,N,N',N'-tetramethylethylene diame (TEMED).
6. Sequencing gel (8%): 21 g urea, 2.5 mL of 20X TBE, and 10 mL of the 40% acrylamide
solution are made up to 50 mL with bidistilled H
2
O and stirred under mild heating until
urea is dissolved. The solution is filtered (filter pore size: 0.2 µm) and degassed for
5 min. Immediately before pouring the solution between the glass plates, add 0.3 mL
of 10% sodium persulfate and 0.3 mL of 10% TEMED.
7. Loading buffer for the sequencing gel (stock solution): 100 mL forma-
mide (deionized), 30 mg xylenecyanol FF, 30 mg bromophenol blue, and
750 mg EDTA.
8. Electrophoresis buffer: 1X TBE.
2.3. The Nondenaturing Gel for DNA Isolation
and Electrophoretic Mobility Shift Assay
1. 20X TBE.
2. Acrylamide solution: 30% acrylamide, and 0.8% bis-acrylamide (see Note 2).
3. 10% Sodium persulfate (see Note 3)
4. 10% TEMED (aqueous solution).
5. 3% Nondenaturing gel: 1.5 mL of 20X TBE, 3 mL of the acrylamide solution,
and 25.5 mL of bidistilled water are mixed and degassed for 5 min. Before pour-
ing the solution between the glass plates add 300 µL of 10% ammonium persulfate
and 300 mL of 10% TEMED.
6. Loading buffer for the nondenaturing gel (stock solution): 50% glycerol and 0.1%
bromophenol blue.
7. Electrophoresis buffer: 1X TBE.
2.4. Other Items
1. Sequencing gel apparatus.
2. Apparatus for nondenaturing gel electrophoresis.
3. Filters for drop dialysis, VS 0.025 µm (Millipore, Bedford, MA).
4. Filtration device (optional).
5. Nitrocellulose filters (BA85, Schleicher & Schüll) (optional).
6. Peristaltic pump (optional).
3. Method
3.1. Establishing the Conditions for Obtaining Optimum Yield
of Specific Protein–DNA Complexes
The method of establishing the conditions for complex formation using the
band-shift assay is described in Chapter 4, see also Chapter 2.
