Moss Tom. DNA-protein interactions: principles and protocols
Подождите немного. Документ загружается.


Assay of Restriction Endonucleases 483
each calculation. The experimental data entered into spreadsheet columns are
counts
complex
and the competitor DNA concentration [D
2
]
t
for each titration point.
3.4. Fluorescence Anisotropy
3.4.1. K
D
Determination. Example: K
D
≈
40 n
M
1. Set the fluorimeter to measure anisotropy in the “single-point polarization” mode.
Each anisotropy value should be measured 10 times (measurement time, 5 s) and
averaged (automatically carried out by the fluorimeter). Set the G-factor to “per
measurement” (see Note 14).
2. An excitation wavelength of 530 nm is used with all the slits on the excitation
monochromator set to 8.
3. Anisotropy is measured in the “L” format through the right photomultiplier with
a 570-nm long-pass filter between the sample and the photomultiplier. Using a
filter rather than a monochromator on the emission side greatly increases the
light intensity at the photomultiplier and so allows measurement of lower con-
centrations of fluorophore.
4. Place a 0.5-mL fluorescence cuvet in the fluorimeter. The cuvet should not be
removed from the fluorimeter or otherwise moved during the experiment. Add
fluorescence binding buffer such that the final volume after step 3 will be
0.5 mL. A measurement temperature of 25°C is used.
5. Add duplex DNA, one strand of which is labeled with hexachlorofluorescein
(Subheading 2.1., item 2) to give a final concentration of 10 nM (see Note 15).
Note the anisotropy.
6. Add EcoRV endonuclease in small aliquots (see Note 16), using a microliter
Hamilton syringe, to cover the range 0–400 nM. Mix thoroughly by gently with-
drawing the contents with a plastic pipet tip and readding to the cuvet. After each
addition, measure the anisotropy.
7. At the beginning (all oligonucleotide free in solution) and end (all oligonucle-
otide bound to endonuclease) of the titration, the fluorescence emission intensity
should be noted (see Note 14).
3.4.2. Data Analysis
1. The fluorescence anisotropy measurements were carried out at [D]
t
≈ K
D
(see
Note 15). Under these circumstances, [E]
f
≠ [E]
t
and so the simplified, hyper-
bolic, form of the binding equation cannot be used. Therefore, data must be fitted
using the full quadratic binding equation (see Note 4). A solution, in terms of
anisotropy, is
A = A
min
+
(
A
max –
–
A
min
)[
×
1
(
([D]
t
+ [E]
t
+ K
D
)–
√[
([D]
t
+ [E]
t
+ K
D
)
2
– (4 [D]
t
[E]
t
)
]
[D]
t
2
where A is the measured anisotropy; A
min
is the anisotropy of free DNA, and is
the A
max
anisotropy of DNA when fully bound to the endonuclease.
2. The data should be fitted to the above equation using GraFit (11). This requires
entry of A and [E]
t
into the spreadsheet provided by the software. [D]
t
is a con-
484 Connolly et al.
stant (i.e., the concentration of oligonucleotide present at the start of the experi-
ment). The titration consists of the addition of multiple aliquots of endonuclease
to the cuvet, resulting in a dilution of the components. Correction should be made
for the dilution of the endonuclease as aliquots are progressively added. Anisot-
ropy, a ratio of fluorescence intensities, does not depend on concentration and is,
therefore, unaffected by dilution. However, as [D]
t
is treated as a constant by this
software it is not possible to compensate for its dilution. Therefore, the final
dilution should not exceed about 10%. This may require endonuclease addition
from several stocks of different concentration. GraFit also requires an estimate of
A
min
, A
max
, and K
D
; suitable values for the first two can be obtained from the
measured anisotropy values at the start (before endonuclease addition) and (after
the final addition of endonuclease) at the end of the experiment. The software
uses nonlinear regression analysis to calculate best-fit values of these three
parameters. Note that the equation given above only holds if the fluorescence emis-
sion intensity of the free and protein-bound DNA are identical (see Note 14).
This is determined in Subheading 3.4.1., step 6.
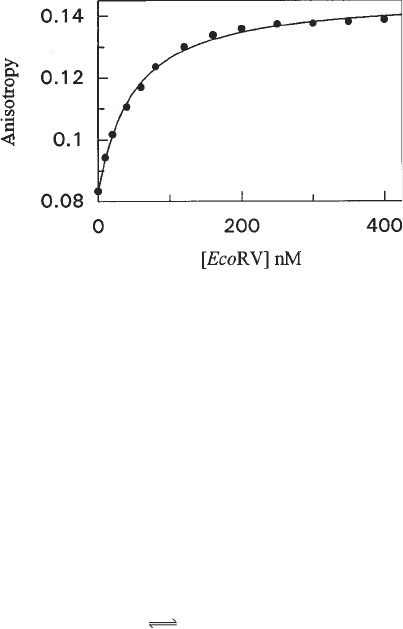
3. A representative titration curve is shown in Fig. 7.
4. Notes
1. With restriction endonucleases, it is often found that the binding of DNA and
Mg
2+
are rapid relative to subsequent events such as conformational changes,
hydrolysis, and product release. This means that rate constants measured under
single-turnover conditions are the same whether the reaction is initiated by add-
ing Mg
2+
to an endonuclease–DNA solution or adding the enzyme to a solution
containing DNA and Mg
2+
. This should, however, be checked by trying both
initiating methods.
2. The K
D
values for the EcoRV endonuclease and the oligonucleotides used in
Subheadings 3.1.1. and 3.1.2. vary between 2 and 40 nM. The DNA concentra-
tion used in Subheadings 3.1.1. and 3.1.2. (25 × K
D
for a K
D
of 40 nM) and an
endonuclease level 10 times higher ensures that ≥95.5% of the nucleic acid
is bound to the protein. Many other concentrations of the two macromolecules
formally meet this requirement. A useful combination, especially for proteins
that are unstable, poorly soluble, and prone to form inactive tetramers and
insoluble higher aggregates when free in solution is [D]
t
= 50 K
D
to100 K
D
and
[E]
t
= 2 × [D]
t
. This results in at least 98% of the DNA being bound and avoids a
large excess of the labile free protein. When bound to DNA, many endonucleases
are both stabilized and protected from aggregation. For substrates with poor K
D
and k
st
(especially so-called “star” sites, where one of the bases in the recognition
sequence is replaced by another natural base), dissociation rate constants are
much greater than the cleavage rate constants. Therefore, multiple protein
dissociations and associations occur prior to a productive cleavage event and so
very poor substrates are never really cleaved under true single-turnover condi-
tions. However, high concentrations of protein and nucleic acid ensure that the
rate (k
a
× [DNA][E]) of association is as fast as possible and minimizes the
Assay of Restriction Endonucleases 485
Fig. 7. Representative binding isotherm determined by fluorescence anisotropy
assay. An oligonucleotide composed of the complementary 14-mers 5'-Hex-
TCCGGATATCACCT and 5'-AGGTGATATCCGGA was titrated with the EcoRV
endonuclease. The binding buffer contained 50 mM Tris-HCl and 100 mM NaCl and
the experiment was carried out at 25°C. The solid circles are experimental points. The
K
D
value determined from the GraFit fit to the curve is 42 nM.
chances that it contributes to the rate-determining step. Whatever levels of pro-
tein and DNA are selected, the rate constants should be independent of protein
concentration if true single-turnover conditions apply. A useful empirical test is
to measure rates at several protein concentrations to confirm this assumption.
3. For the equilibrium,
Endonuclease + DNA
k
on
Endonuclease–DNA K
D
= k
off/
k
on
k
off
the stability of a protein–DNA complex depends on k
off
. Most proteins have k
on
values of between 10
6
and 10
8
M
–1
s
–1
enabling an estimate of the half-life (0.693/
k
off
) expected for an endonuclease–DNA complex at different K
D
values:
K
D
= 1 µM (K
A
= 1 × 10
6
/M
–1
); half-life between 0.01 and 1 s
K
D
= 1 nM (K
A
= 1 × 10
9
/M
–1
); half-life between 10 s and 10 m
K
D
= 1 pM (K
A
= 1 × 10
12
/M
–1
); half-life between 100 m and 150 h
4.
K
D
= [E]
f
[D]
f
/[ED] = ([E]
t
– [ED]) × ([D]
t
– [ED])/[ED]
Therefore, the fractional saturation of the DNA with protein, [ED]/[D]
t
(the
parameter measured using filter binding, gel shift, or fluorescence anisotropy),
has a quadratic relationship to [E]
t
. However, under the condition [E]
f
= [E]
t
(which occurs if [E]
t
>> [ED]),
K
D
= [E]
t
× ([D]
t
– [ED])/[ED]
Here, [ED]/[D]
t
has a hyperbolic relationship to [E]
t
, simplifying data analysis.
In order to achieve the condition [E]
t
>> [ED] and to obtain a satisfactory titra-
486 Connolly et al.
tion it is also necessary to have [D]
t
< K
D
. DNA concentrations ≤15% K
D
are
sufficient to satisfy the assumption [D]
t
< K
D
and give insignificantly different
K
D
values whether the hyperbolic or quadratic binding equation is used. How-
ever, very low [D]
t
often gives binding curves of very poor quality. This may
result from insufficient ligand stabilization of the protein by the nucleic acid.
Therefore, a [D]
t
in the 5–15% K
D
range is recommended. Concentrations of pro-
tein are chosen so that the midpoint of the titration curve yields a protein con-
centration approx equal to the expected K
D
. Clearly, the K
D
will not be known
prior to experimentation, emphasizing the need for an initial estimation, followed
by a second, more accurate, determination.
5. Filter binding requires passage of the mixture through a nitrocellulose filter and a
brief washing of the filter with buffer to remove excess unbound DNA. The entire
process can be carried out in seconds. Gel retardation takes longer; about
1–2 min to load the samples and the same time to run them into the gel. Running
the gel takes 30–60 min. Therefore, gel retardation can suffer, to a greater extent
than filter binding, from dissociation of the protein–DNA complex. To some degree
the problem is self-diagnostic, because it results in a smeared protein–DNA band
rather than the tightly focused one which arises when no dissociation takes place.
In these cases, K
D
determination may be achieved by measurement of the free-
DNA band (16), but if this approach is used, an alternative method is best used to
check the validity of the result.
6. As an example, consider an unmodified DNA with a K
D
of 0.1 nM and four
modified DNAs with K
D
values that range from 1 nM to 1 mM. Direct equilib-
rium analyses are performed for the unmodified substrate and the modified site
with K
D
= 1 nM. Equilibrium competition analyses are also performed for the
four modified sites. If identical values of K
D
are obtained from both direct and
competition analyses for the modified site with K
D
= 1 nM, the modified site
serves as an internal control for the three modified sites which bind more weakly.
7. The relationship between fluorescence anisotropy, lifetime, and rotation is given
by the Perrin equation (7,8):
A = A
0
/(1 + τ/φ)
where A is the measured anisotropy, A
0
= intrinsic anisotropy (i.e., the anisot-
ropy measured under conditions where no depolarization takes place), τ is the
lifetime of the fluorescence excited state, and φ is the rotational correlation time,
a measure of rotational diffusion that depends on the molar volume and, hence,
the molecular mass attached to the fluorophore. The values of τ and φ must be
balanced: a very short lifetime, relative to the correlation time, would lead to
A = A
0
; a very long lifetime to A = 0. Use of this method requires 0 < A < A
0
. The
τ value of approx 3 ns for hexachlorofluorescein attached to oligonucleotides is
suitable for distinguishing between free and protein-bound oligonucleotides. The
measured anisotropy is given by:
A = (I
||
– I
⊥
)/(
⊥
I
||
+2I
⊥
)
Assay of Restriction Endonucleases 487
I
||
and I
⊥
are the intensities of the parallel and perpendicular components of the
emitted light when parallel excitation is used.
8. The buffer components used for k
st
and K
D
determination will obviously vary
somewhat depending on the enzyme under study and the particular experiment.
However, for most DNA-binding proteins, including EcoRI, EcoRV, and BamHI
restriction endonucleases there is pronounced nonlinearity in the dependence of
log K
D
versus salt concentration (17) at salt concentrations ≤0.1 M, presumably
because at these low salt concentrations, coupled protein–protein equilibria lead-
ing to aggregation (18) become significant. This phenomenon can produce mis-
leading quantitative comparisons and thus salt concentrations below 0.1 M should
be avoided. Bis-Tris propane is useful as it has two pK values (6.8 and 9), allow-
ing pH-dependence studies with a single buffer. However, other buffers, of
appropriate pK can equally well be used. Bovine serum albumin and DTT can
help to stabilize certain restriction endonucleases, especially at low concentra-
tions. With several DNA-binding proteins, including EcoRI endonuclease,
aggregation and ultimately precipitation, in the absence of the stabilizing DNA
substrate, is quite common at salt concentrations below 0.4 M. In these cases,
intermediate dilutions should be made with buffers containing 0.6 M NaCl. Five
percent glycerol, in the dilution buffers, also aids stability.
9. The “classic” gel-shift buffer, used to prepare and run nondenaturing polyacryla-
mide gel is 0.089 M Tris-borate, pH 8.0, containing 1 mM EDTA (19). Many
protein–DNA complexes, particularly those of high affinity, are stable in this
buffer. Therefore, it is often possible to incubate enzyme and DNA in a selected
binding buffer and then carry out gel retardation in Tris-borate electrophoresis
buffer without disturbing the equilibrium initially set up. However, the binding
of restriction endonucleases to DNA depends on pH, salt concentration (NaCl or
KCl), and divalent cations (Ca
2+
, a Mg
2+
mimic that does not allow hydrolysis,
often considerably strengthens binding) (18,20,21). Ideally it would be best to
use identical incubation and electrophoresis buffers. We have obtained satisfac-
tory gel retardation data using Tris-borate at pH 7.5. Replacing the EDTA with
up to 5 mM CaCl
2
is also not detrimental. However, many buffers are not suitable
for electrophoresis, giving poorly resolved or smeared bands, especially if NaCl
or KCl is added.
10. Gel retardation requires that the sample applied to the gel be denser than the
electrophoresis buffer and so sinks into the well. The presence of 3% glycerol in
the binding reactions ensures efficient sinking of the sample (18). If this method
is used, it should be checked that glycerol does not perturb K
D
values (e.g., by carry-
ing out filter binding ± glycerol. An alternative involves addition of half a volume
(i.e., in this case, 5 µL to the 10 µL binding reaction) of 25% (w/v) sucrose solution
following incubation and immediately prior to loading on the gel. This dilutes the
sample, and in cases of weak binding, the associated fast k
off
rates may lead to some
dissociation. However, if the binding is strong, the small dilution factor is unlikely
to be a problem. It is best not to add the bromophenol blue marker to the solution
of enzyme and DNA, as the dye can diminish (18) binding.
488 Connolly et al.
11. Phosphorimaging is the best way to quantitate the amount of radioactivity in
bands on gels. Phosphorimaging systems are now fairly standard in most labora-
tories. As an alternative autoradiography followed by scanning of the bands may
be used, but it is more cumbersome and less accurate.
12. Fitting to the full kinetic model is actually the least robust for the unmodified enzyme-
oligonucleotide pair because the rate constants are so similar numerically that the
program has difficulty partitioning among them. Thus, many closely spaced, and
accurate, time-points should be taken and this will usually require rapid quench.
13. The total concentration of endonuclease and radiolabeled reference DNA (D
1
)
are chosen to be at least fourfold over the predicted equilibrium dissociation con-
stant (K
1
) for their interaction. [E]
t
and [D
1
]
t
in the range K
1
to 30 K
1
have been
tested; the K
2
(equilibrium dissociation constant for competitor DNA) is not
affected significantly.
14. The change in anisotropy is only strictly related to the molecular mass of the
fluorophore and hence the amount of oligonucleotide complexed with endonu-
clease, if the following two criteria are met. First, the detector response to the
parallel and perpendicular polarized light must be identical. The G-factor mea-
sures this response, which is automatically corrected for, at each reading, with
the fluorimeter used here. Second, the quantum yield of the free oligonucleotide
and the enzyme-bound oligonucleotide must be the same (i.e., under identical
conditions, the free and bound oligonucleotide should emit the same amount of
light). If the quantum yield of the free and bound oligonucleotide varies the equa-
tion used to fit the data will not be valid (7,22,23). In the example shown in
Fig. 7, the emission intensity of the free and bound oligonucleotide varies less
than 10%. However, the use of other hexachlorofluorescein oligonucleotides and
different buffers with the EcoRV endonuclease often results in large intensity
changes (>10%). In these cases, fluorescence anisotropy cannot be used unless
corrections are made (7,22,23).
15. Good results have been obtained using a DNA concentration about equal to the
K
D
and varying protein concentrations from 0 to (20–30) K
D
. However, lower
concentrations of DNA can also be used, although 1 nM represents the detection
limit. DNA levels much higher than the K
D
should be avoided, as this promotes
stoichiometric titration (where each protein molecule added binds to the nucleic
acid), conditions under which accurate K
D
evaluation is not possible.
16. Purified endonucleases are often stored in buffers containing glycerol. Fluores-
cence anisotropy is very sensitive to viscosity and the addition of large amounts
of glycerol during the titration adversely effects the measurement. Therefore,
intermediate dilutions should be made with buffers lacking glycerol. This usually
ensures that the amount of glycerol added to the fluorescence cuvet is negligible
(≤2%). In the case of weak binding, when an enzyme may have to be added
directly from the storage buffer, dialysis is recommended to remove the glycerol.
Acknowledgments
The Newcastle group would like to thank G. Baldwin and S. Halford
(Bristol) for the use of their quench-flow apparatus and help with the data
Assay of Restriction Endonucleases 489
shown in Fig. 4. B.A.C. is supported by the UK BBSRC and MRC. L.J.J. is
supported by a grant (GM-29207) from the National Institutes of Health (USA).
B.A.C. and L.J.J. have a Wellcome Trust biomedical research collaboration grant.
References
1. Roberts, R. J. and Macelis, D. Rebase; the restriction enzyme database. http://
www. neb. com/rebase/rebase. html
2. Jen-Jacobson, L. (1995) Structural-perturbation approaches to thermodynamics
of site-specific protein-DNA interactions. Methods Enzymol. 259, 305–344.
3. Jen-Jacobson, L. (1997) Protein-DNA recognition complexes: conservation of
structure and binding energy in the transition state. Biopolymers 44, 153–180.
4. Lesser, D. R., Kurpiewski, M. R., Waters, T., Connolly, B. A., and Jen-Jacobson,
L. (1993) Facilitated distortion of the DNA site enhances EcoRI endonuclease
DNA interactions. Proc. Natl. Acad. Sci. USA 90, 7546–7552.
5. Kurpiewski, M. R., Koziolkiewicz, M., Wilk, A., Stec, W. J., and Jen-Jacobson,
L. (1996) Chiral phosphorothioates as probes of protein interactions with indi-
vidual DNA phosphoryl oxygens: essential interactions of the EcoRI endonuclease
with the phosphate at pGAATTC. Biochemistry 35, 8846–8854.
6. Lin, S. Y. and Riggs, A. D. (1972) lac Repressor binding to nonoperator DNA:
detailed studies and a comparison of equilibrium and rate competition methods. J.
Mol. Biol. 72, 671–690.
7. James, D. M and Sawyer, W. H. (1995) Fluorescence anisotropy applied to bimo-
lecular interactions. Methods Enzymol. 246, 283–300.
8. Hill, J. J. and Roger, C. A. (1997) Fluorescence approaches to the study of pro-
tein-nucleic acid complexes. Methods Enzymol. 278, 390–416.
9. Powell, L. M, Connolly, B. A., and Drained, D. T. F. (1998) The DNA binding
characteristics of trimeric EcoKI methyltransferase and its partially assembled
dimeric form detected by fluorescence polarization and DNA footprinting. J. Mol.
Biol. 283, 947–961.
10. Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989) Molecular Cloning. A Labo-
ratory Manual, 2nd ed., Cold Spring Harbor Laboratory, Cold Spring Harbor,
NY, pp. 5.56–5.87 and 11.31–11.39.
11. GraFit (1998) Version 4.05, Erithacus Software, Staines, UK.
12. Scientist (1995) Version 2.0, MicroMath Software Inc., Salt Lake City, UT.
13. Sigma Plot (1996) Version 4.0 SPSS Inc, Chicago, IL.
14. Lesser, D. R., Kurpiewski, M. R., and Jen-Jacobson, L. (1990) The energetic basis of
specificity in the EcoRI endonuclease–DNA interaction. Science 250, 776–786.
15. Jen-Jacobson, L., Lesser, D. R., and Kurpiewski, M. R. (1991) DNA sequence
discrimination by the EcoRI endonuclease, in Nucleic Acids and Molecular
Biology, Vol. 5 (Eckstein, F. and Lilley, D. M. J., eds.), Springer-Verlag, NY, pp.
141–170.
16. Carey, J. (1991) Gel retardation. Methods Enzymol. 208, 103–118.
17. Record, M. T., Jr., Anderson, C. F., and Lohman, T. M. (1978) Thermodynamic
analysis of ion effects on the binding and conformational equilibria of proteins
490 Connolly et al.
and nucleic acids: the roles of ion association or release, screening, and ion effects
on water activity. Quart. Rev. Biophys. 11, 103–178.
18. Engler, L. E., Welch, K. K., and Jen-Jacobson, L. (1997) Specific binding by the
EcoRV restriction endonuclease to its recognition site GATATC. J. Mol. Biol.
269, 82–101.
19. Revzin, A. (1987) Gel electrophoresis assays for DNA–protein interactions
BioTechniques 7, 346–355.
20. Taylor, J. D. and Halford, S. E. (1989) Discrimination between DNA sequences
by the EcoRV restriction endonuclease. Biochemistry 28, 6198–6207.
21. Vipond, I. B. and Halford, S. E. (1995) Specific DNA recognition by EcoRV
restriction endonuclease induced by Ca
2+
ions. Biochemistry 34, 1113–1119.
22. Eftink, M. R. (1997) Fluorescence methods for studying equilibrium macromol-
ecule-ligand interactions. Methods Enzymol. 278, 221–257.
23. Gutfreund, H. (1995) The role of light in kinetic investigations, in Kinetics for the
Life Sciences Receptors, Transmitters and Catalysts. Cambridge University Press,
Cambridge, pp. 291–295.

Intrinsic Fluorescence 491
491
From:
Methods in Molecular Biology, vol. 148: DNA–Protein Interactions: Principles and Protocols, 2nd ed.
Edited by: T. Moss © Humana Press Inc., Totowa, NJ
33
Analysis of DNA–Protein Interactions
by Intrinsic Fluorescence
Mark L. Carpenter,Anthony W. Oliver, and G. Geoff Kneale
1. Introduction
Changes in the fluorescence emission spectrum of a protein upon binding to
DNA can often be used to determine the stoichiometry of binding and equilib-
rium binding constants; in some cases the data can also give an indication of
the location of particular residues within the protein. The experiments are gen-
erally quick and easy to perform, requiring only small quantities of material
(1). Spectroscopic techniques allow one to measure binding at equilibrium
(unlike, for example, gel retardation assays and other separation techniques
that are strictly nonequilibrium methods). Fluorescence is one of the most sen-
sitive of spectroscopic techniques, allowing the low concentrations (typically
in the nanomolar to micromolar range) required for estimation of binding con-
stants for many protein–DNA interactions. Considerable care, however, needs
to be exercised in the experiment itself and in the interpretation of results. The
fundamental principles of fluorescence are discussed briefly in the remainder
of the Introduction.
A molecule that has been electronically excited with ultraviolet (UV)/vis-
ible light can lose some of the excess energy gained and return to its ground
state by a number of processes. In two of these, fluorescence and phosphores-
cence, this is achieved by emission of light. Phosphorescence is rarely observed
from molecules at room temperature and will not be considered further.
Although electrons can be excited to a number of higher-energy states, fluores-
cence emission in most cases only occurs from the first vibrational level of the
first excited state. This has two implications for the measurement of fluores-
cence emission spectra. First, some of the energy initially absorbed is lost prior
to emission, which means that the light emitted will be of longer wavelength
492 Carpenter, Oliver, and Kneale
(i.e., lower energy) than that absorbed. This is known as the Stoke’s shift. Sec-
ond, the emission spectrum and therefore the wavelength of maximum fluores-
cence will be independent of the precise wavelength used to excite the
molecule. Thus, for tyrosine, the wavelength of the fluorescence maximum is
observed around 305 nm, regardless of whether excitation is at the absorption
maximum (approx 278 nm) or elsewhere in the absorption band. Of course, the
fluorescence intensity will change as a consequence of the difference in the
amount of light absorbed at these two wavelengths.
The fraction of light emitted as fluorescence compared to that initially
absorbed is termed the quantum yield. The value of the quantum yield for a
particular fluorophore will depend on a number of environmental factors such
as temperature, solvent, and the presence of other molecules that may enhance
or diminish the probability of other processes deactivating the excited state.
The deactivation or quenching of fluorescence by another molecule, either
through collisional encounters or the formation of excited-state complexes,
forms the basis of many of the fluorescence studies on protein–DNA interactions.
The study of protein–nucleic acid interactions is greatly simplified by the
fact that all detectable fluorescence arises from the protein, all four of the natu-
rally occurring DNA bases being nonfluorescent by comparison. Tyrosine and
tryptophan residues account for almost all the fluorescence found in proteins.
As a general rule, when both residues are present, the emission spectrum will
be dominated by tryptophan, unless the ratio of tyrosines to tryptophans is very
high. The quantum yield of a tyrosine residue in a protein compared to that
observed in free solution is generally very low, illustrating the susceptibility of
tyrosine to quenching. Tryptophan residues are highly sensitive to the polarity
of the surrounding solvent, which affects the energy levels of the first excited
state with the result that the emission maximum for tryptophan can range from
330 nm in a hydrophobic environment to 355 nm in water. Thus, in proteins
containing only one tryptophan, the general environment surrounding the resi-
due can be ascertained. Tryptophan fluorescence, like that of tyrosine, can be
quenched by a number of molecules, including DNA. Unlike tyrosine, the emis-
sion maximum can also change if tryptophan is involved in the interaction and
this can also be used to monitor DNA binding (2).
The extent to which the fluorescence of a protein is quenched by DNA is
proportional to the concentration of quencher. As quenching is the result of the
formation of a complex between the protein and the DNA, the extent of quench-
ing is proportional to the amount of bound protein. Thus by determining the
extent to which the protein fluorescence is quenched when fully bound to DNA
(i.e., at saturation), the fraction of bound and free protein at any point in a
titration can be determined. From these data, the stoichiometry and binding
constant of the interaction can often be obtained. Note that to establish an
