Moss Tom. DNA-protein interactions: principles and protocols
Подождите немного. Документ загружается.

Assay of Restriction Endonucleases 473
9. Determine the amount of radioactivity present in each substrate and product band
using a phosphorimager (see Note 11).
10. From the data obtained from the phosphorimager, determine the percentage of
the two substrate and the two product strands present at each time-point.
3.1.2. Rapid Hydrolysis (t
1/2
15 s or Less
≈
k
st
Values Faster
Than 3 min
–1
; K
D
Between 2 and 40 n
M
) (e.g., with Wild-Type
Eco
RV
and Cognate GATATC Recognition Sites
1. Fill the three syringes of the quenched flow apparatus with the following:
a. Reaction syringe 1: 0.1 mL of 2 µM radiolabeled DNA in 10 mM HEPES,
pH 7.5, 100 mM NaCl and 10 mM MgCl
2
.
b. Reaction syringe 2; 0.1 mL of 20 µM EcoRV endonuclease in 10 mM HEPES,
pH 7.5, 100 mM NaCl, and 10 mM MgCl
2
.
c. Quench syringe 3; 0.1 mL of 0.3 M EDTA.
2. Set the apparatus to mix the 0.1 mL of the oligonucleotide and enzyme solution
(final concentrations of each 1 µM and 10 µM, respectively) and to quench the
reaction at the first time-point (0.051 s) by the addition of the 0.1 mL of the
EDTA solution.
3. Keep the quenched sample on ice.
4. Repeat for each subsequent time-point (in this case, 19 further points between
0.094 and 20 s) (see Fig. 4).
5. As a zero time-point, 0.1 mL of the oligonucleotide solution manually mixed
with 0.1 mL of 10 mM HEPES, pH 7.5, 100 mM NaCl, and 10 mM MgCl
2
, and
0.1 mL of 0.3 M EDTA can be used.
6. Add 5 µL of each of the quenched samples to 5 µL of stop solution (Subheading
2.2., item 6).
7. Proceed with the analysis by gel electrophoresis and phosphoroimaging (Sub-
heading 3.1.1., step 6–9).
3.1.3. Data Analysis; k
st
Determination
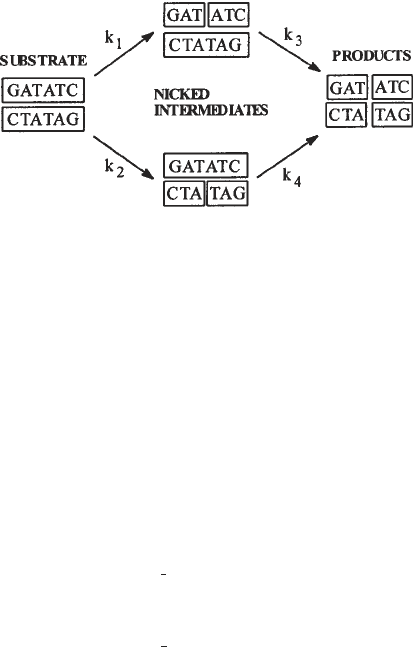
1. Cleavage of a duplex oligonucleotide by a restriction endonuclease involves par-
allel sequential reactions (Fig. 3), giving intermediates in which one strand is
nicked, and described by four rate constants (14,15). In some instances, the reac-
tion scheme can be simplified. This is usually true for wild-type endonucleases
acting on their natural, unmodified, target sequences. Here, both strands are effi-
ciently cut in a concerted reaction and nicked intermediates do not accumulate.
In this case, the cutting of each of the two substrate strands (or the accumulation
of the two radiolabeled products) can be described by a single rate constant with
an acceptable degree of accuracy.
2. If simplification is possible, fit the data to an equation describing a single expo-
nential using GraFit (11). This equation is supplied within GraFit and almost all
biological kinetic software packages. The design of the oligonucleotides used
(Fig. 1) means that the cutting of each strand can be evaluated individually. GraFit
requires time and the percentage of substrate or product present at these times to
474 Connolly et al.
Fig. 3. Parallel sequential cutting of a double-stranded oligodeoxynucleotide by the
EcoRV endonuclease.
be entered into a spreadsheet and to use these data to evaluate k
st
. Concerted
cutting of both strands implies that the k
st
for each of them should be, within the
limits of experimental error, identical. An example is shown in Fig. 4.
3. With modified oligonucleotides or enzymes altered by mutagenesis, cutting often
becomes inefficient and nicked intermediates accumulate. Altering any one point
in a palindromic recognition site produces structural asymmetry, leading to dif-
ferent values of k
1
and k
2
. In such cases, the following equations are used to
obtain values of the four rate constants that describe the unsimplified reaction
scheme (Fig. 3):
k
2
P
1
= S
1
[1 – e
λt
–
—
(e
–λt
– e
k4t
)]
k
4
– λ
k
1
P
2
= S
2
[1 – e
λt
–
—
(e
–λt
– e
k3t
)]
k
3
– λ
where λ = k
1
+ k
2
and S
1
and S
2
are the percentages of total radioactivity in each
DNA substrate strand at t = 0. If end labeling of the two strands is equal, then S
1
= S
2
= 50%. The ability to separate the two DNA substrate strands permits nor-
malizing for “differential labeling” in the event that the two strands have been
radiolabeled with slightly different efficiencies (e.g., if S
1
= 40% and S
2
= 60%,
these values should be entered into the equations). If the radioactivity of the two
strands is markedly different, the program has more difficulty finding the best fit.
This is especially the case when a strand that has an intrinsically faster cleavage
rate constant has lower specific radioactivity than the other strand. Therefore, it
is preferable to attempt to label both strands to the same specific activity.
Reaction times (t) and the amount of products (P
1
and P
2
, expressed as per-
centage of total radioactivity for each time point) are entered into the Scientist
(12) software spreadsheet. A plot of the experimental and “fitted” values of P
1
and P
2
is generated along with numerical values of the rate constants k
1
to k
4
.
Such fits are most statistically robust when the uncertainties in the independent
Assay of Restriction Endonucleases 475
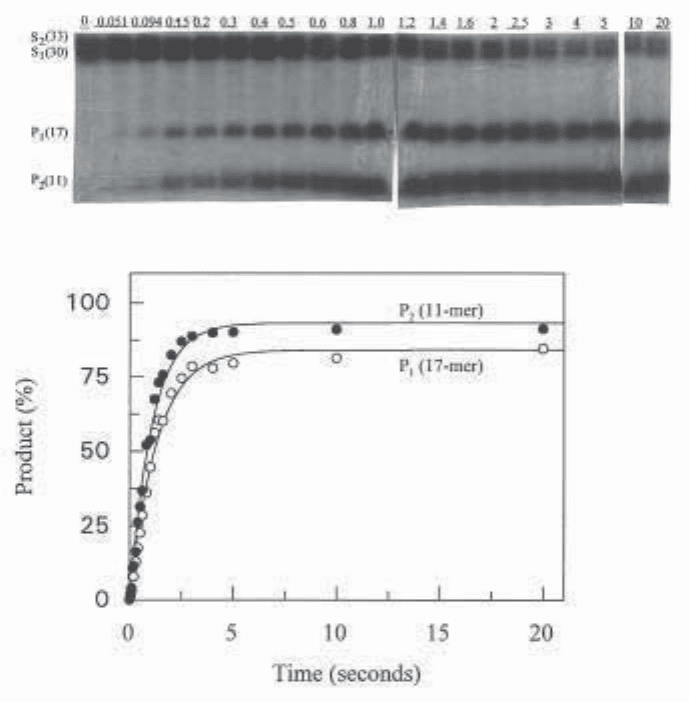
Fig. 4. Cleavage of the duplex oligonucleotide (1 µM) produced by mixing
5'-AAAGTCTGTGGATATCCAAGTGGCTACCGT-*ddA and 5'-CCCCCACGG-
TAGCCACTTGGATATCCACAGACT-*ddA (Fig. 1) with EcoRV endonuclease
(10 µM) using a quench-flow apparatus. The top part shows an autoradiograph of
denaturing PAGE analysis of the two substrates and the two labeled products present
after various mixing times (given on top of the gel lanes in s). The bottom part shows
fits for the formation of each product using the simplified model (i.e., formation of
each product is fitted using a single exponential with Grafit [11]). It should be noted
that data were obtained by phosphorimaging of the gel, and the autoradiograph is pre-
sented for illustrative purposes only. The k
st
values found were 0.94 s
–1
and 0.77 s
–1
for the production of P
2
and P
1
respectively. Fits to the full parallel sequential model
(figure 3) using Scientist (12) (not shown) gave a k
2
(for P
2
and production) of 0.65 s
–1
and a k
1
(for P
1
production ) of 0.51 s
–1
. Thus, in the case of wild-type EcoRV with its
cognate GATATC sequence, the simplified model gives rates that are very similar to
the rigorously correct full model.
476 Connolly et al.
variable (time) are much smaller than the uncertainties in the dependent variable
(here, each radioactivity measurement in a product) and when the number of data
points is sufficiently high to provide a good sampling of experimental uncertain-
ties. The “goodness of fit” of the experimental data to the fitted curve is judged
by the individual reaction point residuals, which should vary no more than 10%.
We have found that a rigorous test of the robustness of the fit to the above equa-
tions is to measure the second strand cleavage rate constants for “nicked” con-
structs, assembled from three individual fragments. When using the full
parallel-sequential model, it is good practice to apply the same calculation method
to reference reactions with the normal enzyme–substrate pair. A comparison of
the rate constants obtained from the “full” and “simplified” calculation methods
then tests the validity of using the simplified-model fit for the cleavage rate con-
stants k
1
and k
2
for the reference pair (see Note 12).
3.2. Filter Binding
3.2.1. K
D
Determination: Direct Binding–Titration of DNA
with Protein. Example: K
D
= 0.66 n
M
K
A
= 1/K
D
= 1.5
×
10
9
M
–1
1. Presoak nitrocellulose membrane filters in filter buffer at the same salt concen-
tration and pH used in the binding reaction. Place in the vacuum manifold.
2. Prepare a solution of 0.5 nM radiolabeled duplex DNA by diluting the radiola-
beled stock DNA solution (100 nM) with binding buffer at the desired salt con-
centration and pH. Add 10 µL of radiolabeled duplex oligonucleotide (0.5 nM) to
each of 10 microcentrifuge tubes; the final concentration of radiolabeled duplex
DNA should be 0.05 nM after step 3 (see Note 4). Add the appropriate volume of
binding buffer such that the total volume of the reaction after step 3 would be
100 µL. Equilibrate on ice for 5 min.
3. To each of the above microcentrifuge tubes add restriction endonuclease to give
final concentrations ranging from 0.05 nM to 5 nM (i.e., the midpoint protein
concentration should be approx equal to the K
D
; see Note 4); equilibrate reac-
tions on ice for 5 min more. During an experimental series, the protein stock
should always be kept on ice and any intermediate dilutions of the stock that are
not used directly in the final reactions are made using enzyme dilution buffer
(salt type and pH always match the experimental conditions). The final dilution
to give the sample used in the reaction should be made with binding buffer (no
glycerol) at the appropriate salt concentration (see Note 8). The salt derived from
the diluted enzyme stock and the DNA source is always accounted for in design-
ing the experiment.
4. Set up two “blank” tubes containing DNA, binding buffer, but no enzyme. One
blank tube will be filtered to obtain R
B
(background counts) and the other used to
determine R
T
(the input counts).
5. Also set up a tube containing DNA, binding buffer, and enzyme at a concentra-
tion 100-fold the K
D
(here, 50 nM enzyme). This tube will be filtered to obtain
R
max
, which represents the total available DNA for binding.
Assay of Restriction Endonucleases 477
6. Transfer the reaction tubes, the two blank tube, and the R
max
tubes to the desired
temperature (e.g., 25°C) and incubate for 30 min.
7. Pipet 85 µL from each reaction tube, the R
B
blank, and the R
max
tube onto a
presoaked 25 mm nitrocellulose filter in the vacuum filtration manifold with
the vacuum applied. The vacuum (flow rate about 0.5 mL/10 s) is applied
continuously throughout the experiment to pull the reaction aliquot through
the filter.
8. As quickly as possible, wash each filter with 350 µL of filter buffer to remove
trapped free DNA.
9. Place each filter in a liquid-scintillation vial and add 2.5 mL of liquid-scintilla-
tion fluid. Count in a liquid scintillation counter.
10. Pipet 85 µL from the R
T
blank tube onto a nitrocellulose filter. Do not wash, but
place the filter directly into a liquid-scintillation vial, add 2.5 mL of liquid-scin-
tillation fluid, and count.
3.2.2. K
D
Determination: Direct Binding–Titration of Protein
with DNA Example: K
D
= 40 p
M
, K
A
= 1/K
D
= 2.5
×
10
10
M
–1
1. Presoak nitrocellulose membrane filters (Subheading 3.2.1., step 1).
2. Prepare a 4 nM stock solution of radiolabeled DNA. For a 4 nM stock DNA
solution, the ratio of radiolabeled DNA to unlabeled DNA is typically 1:3 or 1:4,
depending on the specific activity of the radiolabeled DNA.
3. To each of 10 microcentrifuge tubes, add radiolabeled DNA to give final concen-
trations ranging from 10 pM to 600 pM. (the midpoint DNA concentration should
be approx equal to the K
D
, see Note 4).
4. Add the appropriate volume of binding buffer plus salt at the desired pH such that
the total volume of the reaction after step 6 will be 100 µL. Transfer to ice and
equilibrate for 5 min.
5. Prepare a solution (40 pM) of restriction endonuclease, at the desired salt concen-
tration and pH, by making intermediate dilutions of the stock enzyme solution
with enzyme dilution buffer and the final dilution (to give the 40 pM solution)
with binding buffer as described in Subheading 3.2.1., step 3.
6. To the 10 reaction tubes prepared above, add 10 µL enzyme to give a final con-
centration of 4 pM. Keep on ice for an additional 5 min.
7. Set up 10 “blank” reaction tubes (each with total volume 100 µL) that correspond
to the reaction tubes prepared in steps 3 and 4 (i.e., 100 µL total volume; radiola-
beled DNA concentrations ranging from 10 pM to 600 pM) except that no enzyme
is added.
8. Transfer the reaction and blank tubes to the desired temperature (e.g., 25°C) and
incubate for 30 min.
9. Remove 85 µL from each tube and carry out filter binding as described in Sub-
heading 3.2.1., steps 5–7. As the DNA concentration varies in this experiment, a
different blank (with a DNA concentration that corresponds to its reaction part-
ner) is required for each reaction point.

478 Connolly et al.
3.2.3. Data Analysis: Direct Binding
1. For “normal titrations,” where DNA is titrated with protein, values of K
obs
are
obtained by nonlinear least-squares fits to a single-site binding isotherm:
[ED]
=
K
A
[E]
f
[D]
t
1 + K
A
[E]
f
Note that here K
obs
is K
A
and not K
D
(see the footnote in Subheading 1.). The
actual equation entered into SigmaPlot is (this titration is carried out under con-
ditions [E]
f
≈ [E]
t
, see Note 4):
R
F
– R
B
= MAX
K
A
[E]
t
1 + K
A
[E]
t
The experimental data entered into the spreadsheet are the enzyme concentra-
tion, [E]
t
, for each titration point and R
F
(counts representing complex retained
on the filter) for each titration point; R
B
(counts obtained when radiolabeled DNA
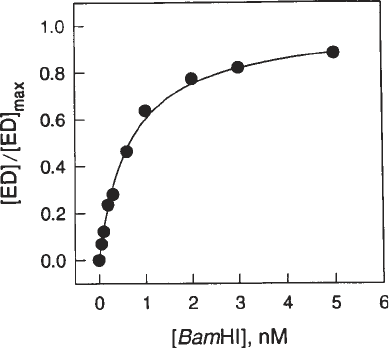
is filtered without enzyme). A plot of the experimental counts (circles) and ideal-
ized counts (line) versus [E]
t
is generated as shown in Fig. 5. In this figure, [ED]/
[ED]
max
i.e., (R
F
– R
B
)/MAX is plotted against [E]
t
. The nonlinear least squares
best fit to the equation will give both K
A
and MAX. The parameter MAX is the
asymptote of the binding isotherm and represents the theoretical maximum counts
retained by the filter. This theoretical MAX can be checked against the experi-
mentally determined R
max
(see Subheading 3.2.1., step 5). The retention effi-
ciency, MAX/(R
T
– R
B
) or R
max
/(R
T
– R
B
), is a good index of reproducibility for
each set of conditions (enzyme, DNA, salt, pH, temperature). The three enzymes
(EcoRI, BamHI, and EcoRV) have different retention efficiencies.
2. In the case of “reverse titration” (i.e., when protein is titrated with DNA), values
for K
A
are obtained essentially as above, except that here the equation for a single-
site binding isotherm is
[ED]
=
K
A
[D]
f
K
A
[D]
t
[E]
t
1 + K
A
[D]
f
and R
F
– R
B
= MAX
(
1 + K
A
[D]
t
)
Because the concentration of enzyme is held constant and titrated with DNA, [E]
t
should be 5–10% of the K
D
value (see Note 4). This is in contrast to a “normal
titration” (DNA held constant and titrated with protein), where [D]
t
is 5–10% of
the K
D
.
3.2.4. K
D
Determination: Competitive Equilibrium Binding. Examples:
K
D
of Reference DNA = 0.25 n
M
; K
D
of Competitor DNA = 0.69 n
M K
D
of reference DNA = 0.25 n
M
; K
D
of competitor DNA = 0.81
µM
1. Presoak nitrocellulose membrane filters as in Subheading 3.2.1., step 1.
2. To each of 10 microcentrifuge tubes, add 10 µL of radiolabeled reference spe-
cific DNA from an 10 nM stock solution to give a final concentration of 1 nM
(see Note 12).
Assay of Restriction Endonucleases 479
Fig. 5. Representative binding isotherm determined by the direct equilibrium filter-
binding assay. The specific BamHI substrate 5'-CGCGGGCGGCGGATCCGGGCGGGC
was titrated with BamHI endonuclease. The binding buffer contained 0.14 M potas-
sium acetate at pH 7.3 and the experiment was carried out at 25°C. Solid circles are
experimental points. The fitted Sigmaplot curve gives a K
A
value of 1.51 × 10
9
/M.
3. Add the appropriate volume of unlabeled competitor DNA such that the final
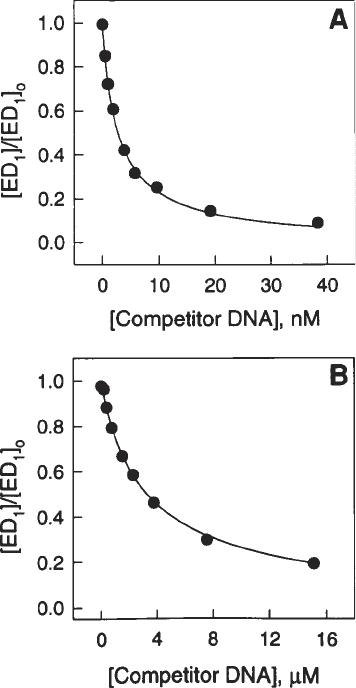
concentrations of competitor DNA will vary between 0 and 40 nM for an expected
K
D
of competitor DNA ≈1 nM (see Fig. 6A). If the K
D
expected for the competi-
tor DNA is much higher (e.g., approx 1 µM), then the final concentrations of
unlabeled competitor DNA should vary in the range from 0 to 20 µM (see
Fig. 6B). Add the appropriate volume of binding buffer (at desired salt concen-
tration and pH) such that the total volume of the reaction after step 4 will be
100 µL. Place tubes on ice for at least 10 min.
4. To each of the above 10 microcentrifuge tubes, add restriction endonuclease to
give a final concentration of 0.8 nM, taking into account the considerations
detailed in Subheading 3.2.1., step 3 for enzyme dilution (see Notes 8 and 12).
Keep on ice for 5 min or more.
5. Set up two additional tubes: a “blank” tube containing radiolabeled DNA, bind-
ing buffer, but no competitive DNA and no enzyme to give R
B
and a tube to
obtain the R
max
(i.e., maximum counts retainable by the filter) containing radio-
labeled DNA, binding buffer, and a final concentration of 80 nM enzyme (no
competitive DNA). The R
max
tube should also be chilled on ice.
6. Transfer all reactions to the desired temperature (e.g., 25°C) and equilibrate for
30 min.
7. Remove 85 µL from each tube and carry out filter binding as described in Sub-
heading 3.2.1., steps 5–7. Background counts (R
B
) are obtained by filtering the
tube containing radiolabeled DNA in the absence of protein and competitor DNA.
480 Connolly et al.
Fig. 6. Representative equilibrium-competition curves for the interaction of BamHI
endonuclease with specific and nonspecific sites. A 40-base-pair specific “snapback”
substrate 5'-TGGGTGGGATCCCACCCACCCCCTGGGTGGGATTCCACCC was
used as a radiolabeled probe for both competition curves. The binding buffer con-
tained 0.14 M potassium acetate at pH 7.3 and the experiment was carried out at 25°C.
Solid circles are experimental points. Test unlabeled specific competitors were (a) the
same specific substrate used for the direct binding assay in Fig. 4 and (b) a nonspecific
site (CCTAGG) embedded in the same flanking context. The K
D
value determined
from the Sigmaplot fit to curve A is 0.69 nM (K
A
= 1.45 × 10
9
M
–1
) and to curve B is
0.81 mM (K
A
= 1.23 × 10
6
/M
–1
).
Corrected reaction counts are obtained by subtracting the background counts from
each reaction. The tube containing radiolabeled DNA and 80 nM enzyme is used
to determine the R
max
value (see step 5).
Assay of Restriction Endonucleases 481
3.2.5. Data Analysis: Competitive Equilibrium Titration
Results are fitted, using SigmaPlot, to a binding isotherm as previously
described (6) using the equation
[ED
1
] =
[E]
t
([D
1
]
t
– [ED
1
])
K
1
(
1 +
[D
2
]
t
)
+ ([D
1
]
t
– [ED
1
])
K
2
where [E]
t
= total enzyme concentration, [D
1
]
t
= total radiolabeled reference
DNA concentration, [D
2
]
t
= total unlabeled competitor DNA concentration,
K
1
= dissociation constant for the radiolabeled reference DNA; K
2
= disso-
ciation constant for the unlabeled competitor DNA. Solving the equation for
[ED
1
] yields
[ED
1
] =
1
{
K
1
+
K
1
[D
2
]
t
+ [E]
t
+ [D
1
]
t
–
冪
[
(K
1
+
K
1
[D
2
]
t
+ [E
1
]
t
+ [D
1
]
t
)
2
– 4[D
1
]
t
[E
1
]
t
]}
2
K
2
K
2
where
[ED
1
] + [D
1
]
t
R
F
– R
B
R
max
–R
B
The dissociation constant K
1
for the reference DNA is always determined by
direct equilibrium binding at the start of a competition experiment. R
max
,
obtained at saturating concentrations of protein and without competitor DNA
(see Subheading 3.2.4., step 5) represents the maximum available [D
1
]
t
. The
known values for K
1
, [D
1
]
t
, [E], R
max
, and R
B
(see Subheading 3.2.4., step 5),
are entered into the SigmaPlot software at the start of each calculation. The
experimental data entered into spreadsheet columns are the corrected counts
(counts retained on the filter for each titration point minus background counts;
R
F
– R
B
) and the competitor DNA concentration [D
2
]
t
for each point. The curve
generated by plotting the corrected counts as a function of increasing concen-
trations of competitor DNA ([D
2
]
t
) is fitted to the best value for the equilibrium
dissociation constant K
2
using SigmaPlot nonlinear regression analysis.
Figure 6 presents the results as a plot of the ratio of [ED
1
]/[ED
1
]
0
where [ED
1
]
0
is the concentration of enzyme–DNA complex obtained in the absence of com-
petitor. The [ED
1
]
0
found in this experiment should not be significantly differ-
ent from the [ED
1
]
0
calculated for a direct binding experiment.
3.3. Gel Retardation
3.3.1. K
D
Determination: Direct Binding–Titration of DNA with Protein
1. Set up and equilibrate binding reactions as described in Subheading 3.2.1., steps
2–4. The final reaction tubes should additionally contain 3% (v/v) glycerol (see
Note 10). A “blank” should contain all the components except endonuclease.
Smaller samples (typically 10 µL) are used in gel retardation than are used for

482 Connolly et al.
filter binding (85 µL in the examples given above). Therefore, if only gel retarda-
tion is being carried out, the final volume should be 10 µL. Alternatively, it can
be very useful to make up a binding reaction and split into aliquots for simulta-
neous analysis by both filter binding and gel retardation (either direct or compe-
tition titration).
2. Following incubation, load the aliquots of the samples, typically 10 µL, into the
wells of 10% nondenaturing polyacryalmide gels (see Subheading 2.4., items 1
and 2 and Note 9).
3. Run the gels at a constant power of 20–25 W, with cooling, until a bromophenol
blue marker (see Subheading 2.4., item 5 and Note 10) in an adjacent lane, not
containing the experimental samples, reaches the gel front (about 1 h).
4. Remove the gel from the electrophoresis apparatus and seal in plastic using a
vacuum bag sealer.
5. Determine the amount of radioactivity present in the bands that correspond to the
free and bound DNA using a phosphorimager (see Note 11).
3.3.2. K
D
Determination: Competitive Equilibrium Binding
1. Set up equilibrium competition reactions according to the steps outlined in Sub-
heading 3.2.4., steps 2–4, except that the binding buffer for all reactions should
include 3% (v/v) glycerol (see Note 10). If only gel retardation is being carried
out smaller volumes (see Subheading 3.3.1., step 1) (e.g., 10 µL should be used. A
“blank” should contain all the components except endonuclease and competitor DNA.
2. Perform gel retardation analysis and phosphorimaging as described in Subhead-
ing 3.3.1., steps 1–5.
3.3.3. Data Analysis: Gel Retardation
1. For direct titration it is possible to determine K
obs
in an analogous manner to that
described in Subheading 3.2.3. In this case, however,
[ED]/[D]
t
= Counts
complex
/Counts
free
+ Counts
complex
= fraction of counts
complex
where counts
complex
and counts
free
are the counts in the shifted (enzyme-DNA)
and unshifted (free DNA) bands, respectively. The experimental data entered
into the spreadsheet are fraction of counts
complex
and the enzyme concentration
[E]
t
for each titration point.
2. For competition binding the equations given in Subheading 3.2.5. are applicable.
In this case,
[ED
1
] = [D
1
]
t
counts
complex
counts
max
where counts
complex
and counts
max
are, respectively, the counts in the shifted band
for each titration point at a particular competitor DNA concentration and for the
control using large amounts of enzyme and no competitor DNA (see Subhead-
ing 3.2.4., step 5), where all the DNA is shifted into the complex. The known
values for K
1
, [D
1
]
t
, [E]
t
, and counts
max
are entered into SigmaPlot at the start of
