Moss Tom. DNA-protein interactions: principles and protocols
Подождите немного. Документ загружается.

Site-Directed DNA Photoaffinity Labeling 369
8. Column fractions are analyzed by TLC as described in Subheading 3.1.1. The R
f
of daeCTP is 0.221 (3).
9. Fractions containing product are pooled and concentrated to 10–12 mM. The aryl
azido or phenyl diazirine (Fig. 1) are coupled to daeCTP as discussed in Sub-
heading 3.1.1. for 5-aa-dUTP. Tether length versions of AB–dCTP have also
been synthesized with similar lengths of tether to those for the other tether-length
nucleotides discussed (Fig. 1), and the R
f
values of these range from 0.067 for
ABG–dCTP to 0.078 for ABG
3
–dCTP.
3.2. Immobilized DNA Templates
pTZ1 plasmid DNA is used for the synthesis of the SUP4 tRNA
Tyr
DNA
photoaffinity probes.
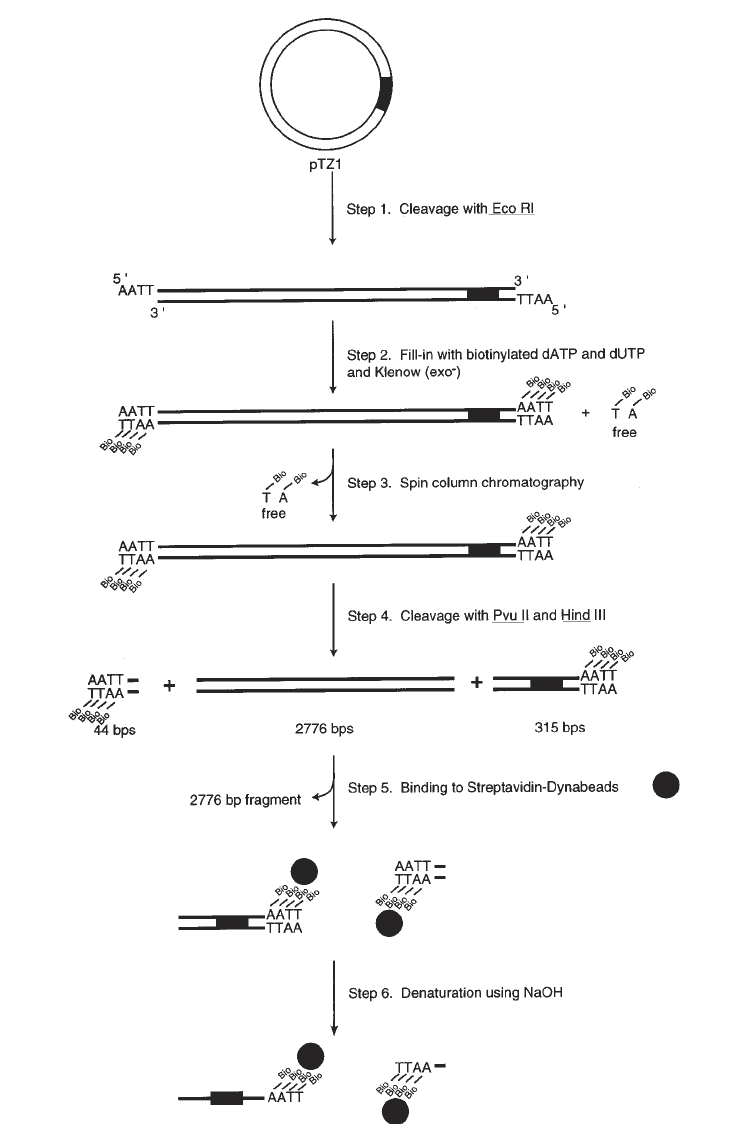
1. DNA is biotinylated by initially digesting 200 pmol of pTZ1 plasmid with either
HindIII for nontranscribed strand templates or EcoRI for transcribed strand tem-
plates (see Note 6) (Fig. 2, step 1).
2. The 5' overhangs are biotinylated by the incorporation of Bio-11–dUTP and Bio-
14–dATP (Sigma Chemical Co.) using the exonuclease-free version of the Klenow
fragment of DNA Polymerase I (Amersham/Pharmacia). The 200-µL reaction con-
tains 200 pmol of linearized pTZ1, 20 µM Bio-14–dATP, dCTP, and dGTP, 25 µM
Bio-11–dUTP, and 150 U of Klenow fragment in buffer A (Fig. 2, step 2).
3. Unincorporated dNTPs are removed by spin-column chromatography (11) with a
2.5-mL Sephacryl S-200 spin column (Pharmacia) equilibrated in buffer B
(Fig. 2, step 3). Aliquots of the samples are removed before and after the spin
column to quantitate recovery.
4. Biotinylated DNA is precipitated by the addition of 2.5 vol of ethanol and placing
the sample at –20°C overnight.
5. The sample is then resuspended and digested with EcoRI and RsaI for non-
transcribed strand templates or HindIII and Pvu2 for transcribed strand templates
to generate a 315-base-pair biotinylated DNA fragment containing the SUP4
tRNA
Tyr
gene (Fig. 2, step 4).
Biotinylated DNA (40 pmol) is bound to Dynabeads M-280 Streptavidin
(Dynal) in the following procedure:
1. Washing Dynabeads
a. The supernatant from 200 µL Streptavidin Dynabeads (10 mg/mL) is removed
using a MagneSphere
®
Technology magnetic separation stand (Promega) and
the beads are resuspended in 200 µL PBS + 0.1 mg/mL BSA.
b. The beads are washed one time with 200 µL buffer C, and resuspended in
400 µL 2X B/W at 5 mg/mL (see Note 7).
2. Binding Reaction
a. A reaction is assembled consisting of 400 µL of the 5-mg/mL washed
Dynabeads and 400 µL of the 0.4-pmol/µL biotinylated DNA (see Note 8).
370 Persinger and Bartholomew

Site-Directed DNA Photoaffinity Labeling 371
b. The reaction is mixed by gentle vortexing every 5 min during a 45-min incu-
bation at 37°C (see Note 9).
c. Buffer is removed using a magnetic stand and analyzed for binding efficiency
by agarose gel electrophoresis (Fig. 3, lanes 2 and 5).
d. Beads are washed with 200 µL of 0.5X buffer C and the washes saved for
further analysis (Fig. 3, lanes 3 and 6).
e. The beads are resuspended in 100 µL of 0.5X buffer C and stored at 4°C
(Fig. 2, step 5) for extended periods of time.
3. Stripping off nonbiotinylated DNA strand
a. Supernatant is removed from the beads and the beads are resuspended in 20 µL
of 0.1 M NaOH.
b. The sample is incubated with occasional vortexing for 10 min at room tem-
perature.
c. The supernatant is removed and beads are washed one time with 50 µL of 0.1 M
NaOH and washed one time with sterile deionized water (Fig. 2, step 6).
4. Dephosphorylating DNA Beads
a. DNA beads are washed three times with 50 µL buffer D and resuspended in
100 µL buffer D.
b. Five units of shrimp alkaline phosphatase (Amersham/Pharmacia) is added to
the reaction and incubated at 37°C for 60 min.
c. Another 5 units of enzyme is added and incubated for an additional 60 min.
d. Beads are washed three times with 50 µL of TE (pH 8.0) + 0.1% SDS.
e. The beads are washed three times with 50 µL PBS + 0.1 mg/mL BSA and resus-
pended in 100 µL PBS + 0.1 mg/mL BSA (final concentration, 0.2 pmol/µL).
f. The remaining enzyme is heat inactivated by incubating the reaction at 65°C
for 15 min and gently vortexing every 5 min. The single-stranded DNA beads
can be stored for several months up to 1 yr at 4°C.
3.3. DNA Probe Synthesis
Immobilized DNA templates made by the previous procedure (Subheading
3.2.) are used to construct DNA photoaffinity probes on either the transcribed
or nontranscribed strand of the gene in the following procedure.
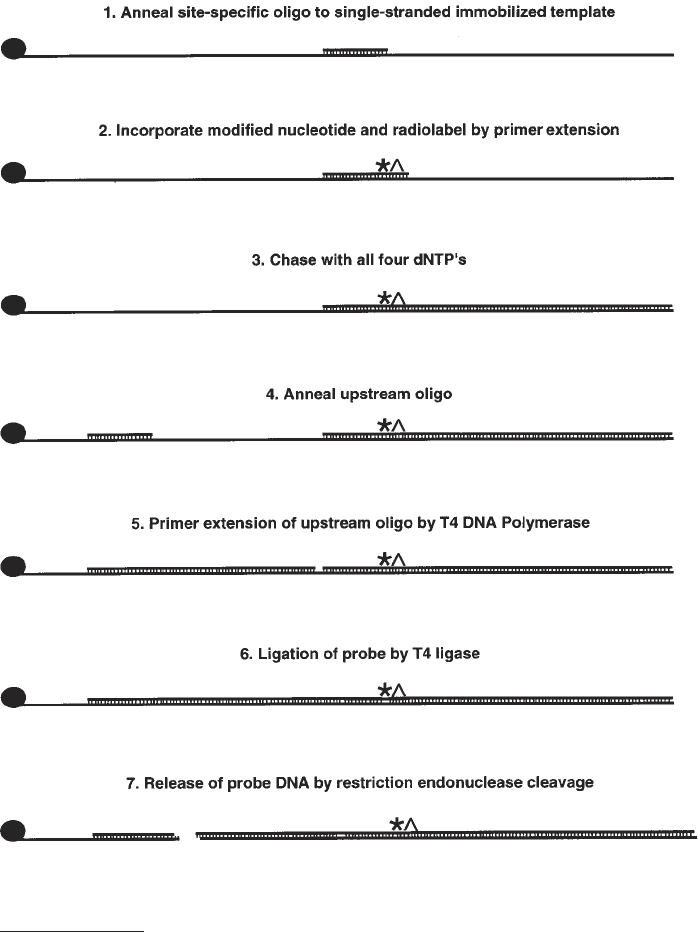
1. First primer extension. Remove 1 pmol of immobilized template and place in a
1.5-mL microcentrifuge tube.
2. Wash the beads three times with buffer D and resuspend in 10 µL of buffer D.
3. To each reaction add 5 µL of sterile deionized water, 1 µL of 2 mg/mL BSA, 2 µL
buffer E, and 2 µL of 2 pmol/µL site-specific oligonucleotide (see Note 10).
4. Reactions are vortexed and heated at 70°C for 3 min and vortexed gently again
before placing at 37°C for 30 min to allow the oligonucleotide primer to anneal
Fig. 2. (previous page) Diagrammatic representation DNA template preparation for
solid-phase DNA probe synthesis.
372 Persinger and Bartholomew
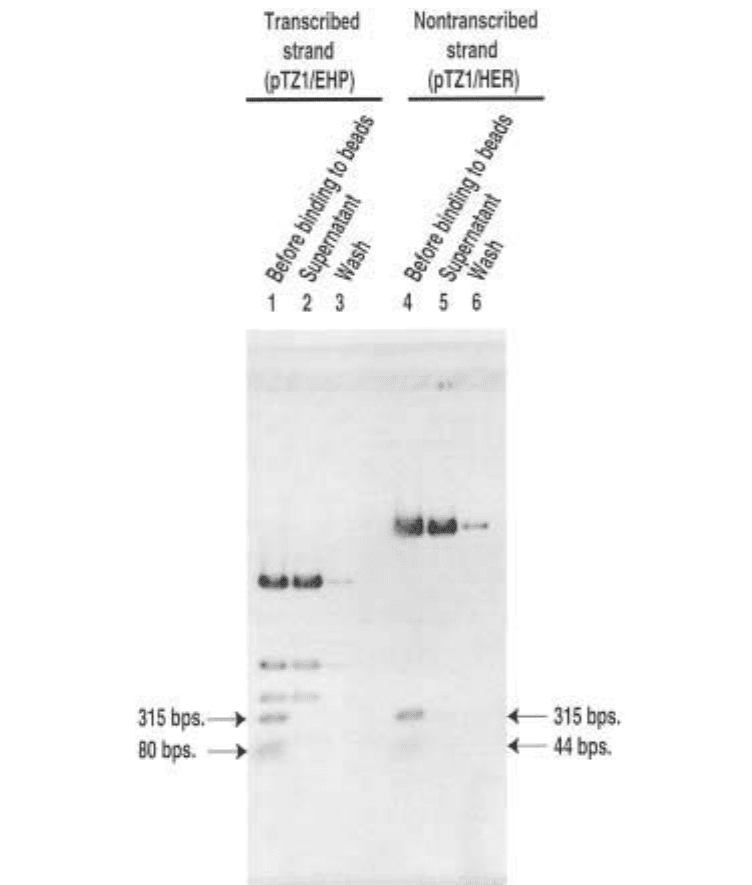
Fig. 3. Agarose gel analysis of DNA bead preparation. Lanes 1 and 4 are
biotinylated pTZ1 DNA cut with EcoRI, HindIII, and PvuII (pTZ1/EHP) used for the
modification of the transcribed strand, or HindIII, EcoRI, and RsaI (pTZ/HER) used
for the modification of the nontranscribed strand before binding to Streptavidin
Dynabeads. The biotinylated 315-base-pair fragments contain the SUP4 tRNA
Tyr
gene.
The 80- and 40-base-pair fragments are also biotinylated, but are not used as a DNA
template. Lanes 2 and 5 are taken from the supernatant of the binding reaction of DNA
with Streptavidin Dynabeads after incubation. The abseπnce of the 315-base-pair frag-
ments in these samples shows efficient binding of probe DNA to Dynabeads. Lanes 3
and 6 are samples from a final wash of the beads after binding.
Site-Directed DNA Photoaffinity Labeling 373
(Fig. 4, step 1). During all incubations, samples must be vortexed gently every
5 min to keep the beads resuspended.
5. To the reaction add 1 µL of 100 µM modified nucleotide described in Subhead-
ing 3, an α-[P
32
]-labeled nucleotide, and 0.25 U of the exonuclease-free version of
the Klenow fragment of DNA polymerase I (Amersham/Pharmacia) (see Note 11)
and incubate at 37°C for 5 min (see Note 12) (Fig. 4, step 2 and Fig. 5, lanes 1–4).
6. Next, full-length extension is done by the addition of dNTPs to a final concentra-
tion of 0.5 mM and incubation for 5 min at 37°C (Fig. 4, step 3 and Fig. 5, lanes 5–8).
7. Klenow fragment and dNTPs are removed by washing the beads three times with
50 µL of TE (pH. 8.0) + 0.1% SDS and two times each with 50 µL buffer D.
8. Beads are resuspended in 20 µL of buffer D.
9. Three microliters of 3 pmol/µL upstream oligonucleotide, 1 µL of 2 mg/mL BSA,
and 1 µL of buffer E are added to a 20 µL reaction containing the DNA beads.
10. The sample is vortexed gently every 5 min and incubated at 37°C for 30 min
(Fig. 4, step 4).
11. The upstream oligonucleotide is extended by the addition of 3 µL 5 mM dNTPs
and 3 units of T4 DNA polymerase (see Note 11) and incubation for 10 min at
37°C (Fig. 4, step 5 and Fig. 5, lanes 9–12).
12. The upstream strand is ligated to the site-specific primer by the addition of 1 µL
of 10 mM ATP and 5 U of T4 DNA ligase and incubated at 37°C for 60 min
(Fig. 4, step 6 and Fig. 5, lanes 13–16).
13. Beads are washed two times with 50 µL TE (pH 8.0) + 0.1% SDS, two times with
50 µL of buffer D, and resuspended in 20 µL of buffer D.
14. Next, the reaction is heated at 65°C for 15 min to inactivate any residual enzyme.
15. The DNA probe is released from the bead by the addition of 12–20 U of specific
restriction enzyme (see Notes 11 and 13) to cut at a site between the SUP4
tRNA
Tyr
gene and the attachment site (Fig. 4, step 7 and Fig. 5, lanes 17–20).
16. After restriction enzyme digestion, the probe is washed from the beads in a series
of three washes of 50 µL each with buffer D, which are pooled in a fresh
microcentrifuge tube.
17. The sample is extracted with phenol:chloroform (1:1) followed by extraction
with chloroform.
18. The DNA probe is ethanol precipitated by the addition of 1/10 vol of 10 M lithium
chloride and 2.5 vol of ethanol.
19. Samples are placed at –20°C overnight.
20. Samples are spun down at maximum speed in microfuge at 4°C for 30 min.
21. The supernatant is decanted and the pellet is allowed to dry.
22. Samples are resuspended in TE (pH 8.0) + 0.05% Tween-20 at a final concentra-
tion of 2–10 fmol/µL and stored at 4°C.
23. After resuspension of the probes, 1 µL is removed for analysis on a 4% native
acrylamide gel, 20 cm × 20 cm × 0.8 mm (Fig. 6).
3.4. DNA Photoaffinity Labeling
Transcription complexes were formed on probe DNA using the 500-mM
KCl fraction from Bio-Rex 70 chromatography of the S-100 extract, made from
374 Persinger and Bartholomew
Fig. 4. Schematic representation of solid-phase DNA probe synthesis.
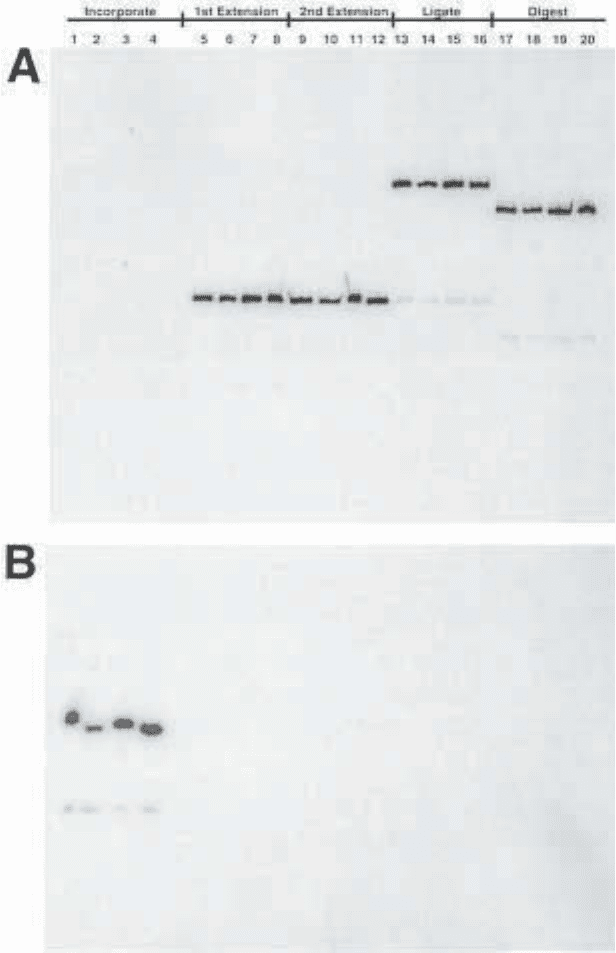
Fig. 5. (opposite page) Analysis of DNA probe synthesis modified at bps +11
on 10% urea PAGE. Incorporation of BP–dUMP, FAB–dUMP, DB–dUMP, and
AB–dUMP and [α-
32
P] dATP (lanes 1–4). Full-length extension of oligonucleotide
primers in the presence of all four dNTPs (lanes 5–8). Upstream oligo annealed to
template and extended to site-specific primer by T4 DNA polymerase in the presence
Site-Directed DNA Photoaffinity Labeling 375
of all four dNTPs (lanes 9–12). Ligation of upstream extension product to site-specific
oligonucleotide by T4 DNA ligase (lanes 13–16). Digestion of the probe DNA to release it
from the Streptavidin Dynabeads by BamHI (lanes 17–20).

376 Persinger and Bartholomew
Saccharomyces cerevisiae strain BJ926 (BR500) (3). In addition, transcription
complexes were also formed using recombinant TFIIIB and partially purified
TFIIIC (12). TFIIIC was obtained from the flowthrough fractions of Ni-NTA
chromatography of His-tagged RNA Pol III (3).
1. A typical 20-µL photoaffinity-labeling reaction contains 4 µL of buffer H, 9 µL
of buffer I, 1 µL of 500 ng/µL pLNG-56 or pTZ1, see Note 14, linearized with
EcoRI, 1 µL of a 2 fmol/µL DNA probe, and 1–4 µL of BR-500 extract (see Note 15)
and adjusted to 20 µL with deionized water. Optimal protein concentration for
transcription activity and photoaffinity labeling is determined by multiple round tran-
scription assays using pTZ1 plasmid DNA and labeled ribonucleotides (3).
2. The photoaffinity-labeling reaction is incubated at 25°C for 30 min for assembly
of complexes onto the probe DNA.
3. The sample is irradiated at this point to crosslink the assembled complex or a
stalled ternary complex can be formed by the addition of a 3 NTP mix. Com-
plexes containing only TFIIIB are formed by addition of heparin to a final con-
centration of 100 µg/mL to the assembled transcription complex for release of
TFIIIC and Pol III from DNA.
4. After irradiation, the DNA probe is enzymatically digested in two steps to leave
a small radioactive tag covalently attached to the crosslinked protein.
5. The first step of digestion is by the addition of 2.3 µL of 0.5 mg/mL DNase I
(Gibco Life Technologies) to a 21-µL reaction and incubation at 25°C for 10 min
(see Note 16).
6. Immediately add 1 µL of 10% SDS to each sample and incubate at 90°C for
3 min and then place on ice for 5 min.
7. Next, 2 µL of the zinc acetate solution and 1 µL of 20 U/µL S1 nuclease (Gibco
Life Technologies) is added, and samples are incubated at 37°C for 10 min.
Fig. 6. Analysis of DNA probes modified at bps +11 on a 4% nondenaturing
acrylamide gel.
Site-Directed DNA Photoaffinity Labeling 377
8. The reaction is stopped by the addition of 1 µL of 0.5 M Tris base to adjust the pH
to approx 7.0 and 7 µL of 5X DB buffer.
9. The sample is heated at 90°C for 3 min and cooled on ice for 5 min prior to being
loaded on to a 4–20% SDS-PAGE that is 20 × 20 cm × 0.8 mm.
10. After electrophoresis, the gel is dried with a slab gel dryer with heating at 80°C
and vacuum for 2 h, and it is exposed to film for autoradiography or to a
phosphorimager screen (see Note 17) (Fig. 7).
3.5. Peptide Mapping
1. Large-scale photoaffinity-labeling reactions for peptide mapping experiments are
as described in Subheading 3.4., except everything is scaled up to a final reac-
tion volume of 2 mL (see Note 18).
2. Samples are irradiated in a multichannel pipettor tray instead of the original
sample tube in order to keep the depth of the sample the same as a standard 20-µL
labeling reaction.
3. DNase I and S1 nuclease digestion is done as described in Subheading 3.4. with
volumes scaled up to the appropriate amounts for the increased reaction size.
4. Next, the samples are concentrated by ultrafiltration using a Centricon 30
(Millipore) to lower the sample volume and allow for loading on a 0.8-mm-thick
8% SDS polyacrylamide gel with a 1.4-cm well.
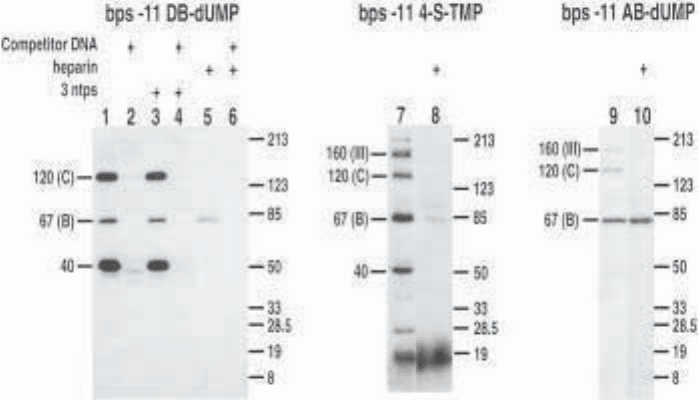
Fig. 7. Comparison of three photoreactive moieties incorporated at bps –11. Sub-
units labeled in both preinitiation (lanes 1, 7, and 9) and heparin-stripped complexes
(lanes 5, 8, and 10) differ from one photoreactive group to the next. Examples of
TFIIIC-specific competition (lanes 2, 4, and 6).
378 Persinger and Bartholomew
5. Photoaffinity-labeled BRF was excised from the gel and electrophoretically
eluted using a Bio-Rad Model 422 Electro-Eluter for 4 h at 10 mA per gel slice
into a volatile buffer consisting of 50 mM ammonium bicarbonate and 0.1% SDS.
6. The eluate is dried down by vacuum centrifugation, resuspended in 200 µL ster-
ile deionized water, and dried down again.
7. Gel-purified BRF is treated with 25 µL 70% formic acid and 1.4% diphenylamine
at 70°C for 20 min to further digest DNA and to cleave the protein at Asp-Pro
sites (Fig. 8).
8. The sample was extracted five times with an equal volume of water-saturated
ethyl ether (fresh).
9. Next, samples are evaporated to dryness by vacuum centrifugation, resuspended
in sterile deionized water, and dried down again.
10. The pellet is resuspended in 40 µL of 2% SDS and 0.1 mM of 2-mercaptoethanol.
11. Proteins are cleaved with cyanogen bromide by the addition of 1 µL of 1 M
hydrochloric acid and 1 µL of 1 M cyanogen bromide in acetronitrile to a 15-µL
sample or addition of formic acid to a final concentration of 70% and 1 µL of 1 M
cyanogen bromide in acetonitrile for a more complete digestion.
12. Samples are incubated at 25°C for 10 min or 2 h.
13. Samples are resolved on a 10–20% tricine gel (13). After electrophoresis the gel
is stained by Coomassie R-250 staining, dried, and placed on autoradiography
film to visualize radiolabeled fragments (see Note 19) (Fig. 8).
4. Notes
1. We have found that differing brands of TLC plates cause products to migrate
somewhat differently and may result in different observed Rf values.
2. We store concentrated stocks of modified nucleotides wrapped in foil at –80°C.
Only small diluted stocks are stored at –20°C wrapped in foil to help protect the
major stock from inadvertent photolysis.
3. Photoreactive nucleotides are tested to determine the range of nucleotide concen-
trations that can be used to incorporate the nucleotides by primer extension and
can be compared to the unmodified nucleotide. An aliquot of each sample can be
irradiated and the DNA will crosslink the Klenow fragment of DNA polymerase
I and cause the labeled oligonucleotide to have a much slower electrophoretic
mobility. Some of the oligonucleotide does not get crosslinked to DNA poly-
merase, but is visibly photolyzed as evident by smearing of the free oligonucle-
otide band.
4. Another question we have addressed is whether a modified nucleotide at a given
position affects normal DNA–protein interactions in that region. This can be done
for transcription complexes by gel shift analysis or performing transcription
assays on wild type DNA and probe DNA to determine if DNA modification
affects the level of transcription.
5. Our lab has now prepared and is characterizing a modified dATP analog to
increase the number of DNA sites that can be modified (M. Zofall, manu-
script in preparation).
