Lewin Benjamin (ed.) Genes IX
Подождите немного. Документ загружается.


18L
urleuorqlolalaH
puP
sraluequl
Jo
suolllv
aql
llolg
srolelnsul
tI'62
'urleuorql0leleq
J0
peards
otll
lsulp6e
siaurpq
aphold {eu slolelnsul
o
'sull
pue'slalualts
'sreluequa
uolJ slle#e 6uqearpeuL.ro 6uLlenqre
fue;o
aDessed
lrolq
ol alqe
a.lp srolelnsul r
u
r.lPruoJ
r.{lojalaH
pue
sjaluPr{ul
Jo
suoLllv
aqt
llolE
srolelnsul
'suolpunJ
Jleql
lnoqe
uolleurJoJul
palrPlJp
JAPq
]J^ 1ou
op eM
lnq
'sJruosoelJnu
Jo
^lqluasseal
pue
l.lqruassesrp er{l uI
pJ^loAuI
osle Jre .daql asneraq
z(lqeqord
'paqIrJsuPJl
Suraq are
leql
suo6ar ur
urteuroJqr;o
Llrr8atul
Jql urelureu
o1
parrnbar
eJe sropPJ
Jaqlo
'rtedar
pue
uoIlPJIIdar
y51q
8ur
-pnlJur
'pareldsrp
aq.{eu srruosoJlJnu
qJIqM
ur suort)pJJ JJqlo JoJ
partnbar
oslp st
If VC
'sseJoJd
aql ataldruor
o1
parrnbar
Jq
lsnlx
sJol
-JpJ
Jeqlo
'aru,{zua
Jql
puqaq
Surlqurassear
sr
leqt
euosoepnu
e ot
1l
ppe
ot sdlrq
uJql
pue
aseraru,{.1od
yNU
Jo
luoJJ
uI aruosoalf,nu
e
ruoJJ
gZH-VZHSrqJelap
Jf,Vd
qllqM
uI
'i
ir'f,{
.
Hil*l*
ur uMoqs
Iapou
Jq1 s1sa33ns
slqJ,
'seuolsIq
aJo)
ruorJ seuosoJlJnu
Jo
uorleruJoJ
slsISSe
1I
esnPJeq
'uollduf,suert
JJIIe sJruosoepnu
;o
.,i.lqruassear
aqt ur
pJ^lo^ul
eq osle .{eru
I)vg
'uoltdr:ls
-uer1
Suunp srJrrreDo
SuDeldsrp JoJ
rusrueqJJlu
e
;o
tred
sI
J)yd
teql
stsaSSns
slqJ
'sreulp
grH-YzHlsol
e^eq
leqt
,,sJruosexeq,,
01 sauroso
-el)nu
suJluof,
lI'lJitA.
tzl uotldt.rrsuerl
Surrnq
'sJeurp
szH-vzH
JSol ol ruJql SJSnPJ
u'saruos
-oelrnu
palelosl
ol
prppe
sI
Jf,v{
urqM
'saua8
a^rlJe
Jo
urleruoJqf,
Jql
qtIM
pelel)osse
s1
t1
'sa1o,{.re>1ne
IIe
uI
panrasuoJ
IIaM
Jre
teqt
slrunqns olvrl
Jo
stsrsuo)
IIVC
('uoll
-drnsuen
1o
aseqd
uotte8uolJ eqt
Suunp
Llert
-;oads
tl
qlIM
sJlelJossp
lnq
'Jseralu,{1od
y51g
yo
ged
lou
sl
tI)
'rotrpJ
uotle8uola
uotldtnsuerl
p
a>lrl sJneqeq
qJIqM
'(ffVg)
uorldursuBrl
ulletuoJqJ
sJlelIIIJpJ
pJIIeJ
ulJlord
e sartnbar
l
aseraru[1od
VXU
f,qo4n zz uotldtnsuerJ
'uoltduJsupJl
3ur,,u.o11oy
uroJeJ
ueql
tsnur
sJeuretJo aI{1
'1I
pulqJq
VNq
pJ{eu
Sutneal
prole
oJ'pareldsrp
eq
lsnu
tl
Jo
peeqe
sreluepo
Juolsrq aql
'uotte8uola
Surrnp
Sunue.tpe
anup
-uor
ot
r
rod
'seuosoJpnu
,{q
papadumrn
VN11
Jo
r{JlJJls
uoqs
e uo
stsarpuLs
VNg
sue6
eserauru(1od
VNU
leqt
sueJur
srq;
'(.raloruord
eqt
tP
pJ8ueq3
ag Lery uorlezrue8ro
JlrrosoelJnN
't'0€
uoIlJJS
aas) arnlrnrls
urleruoJqf, a8ueqr
01
dJv
;o
srsr{.
-1orpz{q
,{q
pateraua8
^d8raue JSn
qlr-qM
pue
slolJeJ
uorldulsuerl
,{.q
palrnnal eJB
]eql
saxaldruor
Suqaporuar
sattnbar
sJal[elJo
aql
Jo IPnoIueJ
aq1
'(arnpruts
ulleluorq3
ut
a8ueq)
TJJIJJU
salrS
e^rtrsuasradlg
esevN(
'gI'62
uollJeS
aas)
vNq
ruor;
pareldslp uJaq aAPq
sJJlrrelf,o
auolsle
JSneJJq'eseVN(
ol eAIlIsuJSradr{q
are
teqt
sJlls,{q
pa4reru JJP sJalourord
azltpy
'uo1l
-eltrur
lp lsaq
pezIJaDeJPqJ
uJJq seq ssarord
aq1
'uorlduJsueJl;o
sa8els
IIe
roJ
luaurarrnbar
La1 e sr
VNO
ruoJJ
seluosoJlJnu
Suneldstq
'uo4d
grsuell
laUP saulosoallnu
olut
olquasseor
ol souolstq
aql loJ
pue
uoqoulsuelJ
6uunp slauepo
areldstp
o1
aseraur\1od
vNU
roJ
q+oq
pallnbar ele slollPJ
r\re11ouy
r
srollPl
lPtlads
arrnbau
Alquasseau
puP
luaualPlds!o
auosoallnN
'ruroJ
papuexa
up
uI
tslsred
Ieru
YNCI
eql
pue
'pareldstp
,{lluauerurad
aq
,{eru JeIuPlJo
eql
uaql-lsJlJ
Jql
purq
Jq
Alal er
pauLu r
sJnu
|
1
uo)
ase.ra urz(1od
JJqloup
asnef,aq
'aldtuexa
roJ-elqPIIeAe
lou
sl
VNo
aqr
g
'(aserarudlod
VNU
qtltt
slJeluoJ
Suqeru
.dq
,,>1req
passed,, sI
rauleDo
aqt sdeq
-red
'VNq1
aqt
Suop
spaarord
IIJS1I
ueql
JZIs
ra8rel
uana;o
rralqo
ue
se sluJuodruor
8utso1
ro
3urp1o;un
lnoqtlm
'qSnoqr
'YNC
qtlm
1reluor
ur€lJJ
plnoJ JJruelJo
ue
,lroq alzznd
P suIeuJJ
1I
'VN(
eqt
qlIM
Deluo)
tsol
f,1ptol JJAJU
JJIUPIJo
eql
teql-elqeqord
sdeqlad
ro-alqtssod
st
11)
'araql
seqJplleeJ
rJrupl)o
eql'alqPIIP^P
sI
aseraru.{1od
eqt
pulqJq
VNq1
eql
;1
'sassar8
-ord
1r
se
sreuleDo
auolsJr1
sareldsrp
aseraru,{1od
vNU
teql
asoddns
01 sI
Iepou
Surz[run
aq1
'uoperlldar
tnoqtlm
patsnlpe
Jq uP)
sJurosoJlf,nu
Jql
Jo
suoulsod
aqt
teqt
tutod
3rnlsa
-Jatur
aqt
sJ>lpru
lpseJ
sIqI'(alalduor lou
sl
1I
q8noqlp) sarnuru
ual
ulqtr,tn
sreadde
Stmroutsod
'peqs{qplsaer
st uorssardal
uaq6'8u1uot1tsod
IeuosoelJnu
aqt
sr{orlsap
uotldtDsuert
1eql
stsaS
-3ns
urnl
q
sIqJ'aseqd
ur
pazrueSro
raEuol
ou
JJe
tnq
Llrsuap
euPS
aql
1e lUaSard
ale
sJruoso
-alJnu
leql
seleJlpul
sIqJ'saluosoalf,nu
Jo
ueDPd
pauoprsod aq1 sareldar
rearus
praua8 e'passardxa
sr aua8
eqt
ueq6
'suor8ar
,€
Jql
q
1so1
st 3ut
-uortrsod
laua8 aqt
ssoJJe
eJuelslp
luerqtu8ts
e
ro;
raloruord
eql
ruoJJ
panue8ro aJe
1eql
sJrxos
-oalJnu;o
uraged
e s,{.e1dsrp
aua8
aql
l.1pt1tu1
'aua3
aq1;o
pue
,E
aq1
le
JIIS UOIIJIJ1SaJ
e 01 e^Ile
-lar
salls
a8erreap
JrrrurcXe
ol
aseJpnu
IeJJolorJIru
Sursn
Lq
peuIuPXa
sr Suruorllsod
'raloruord
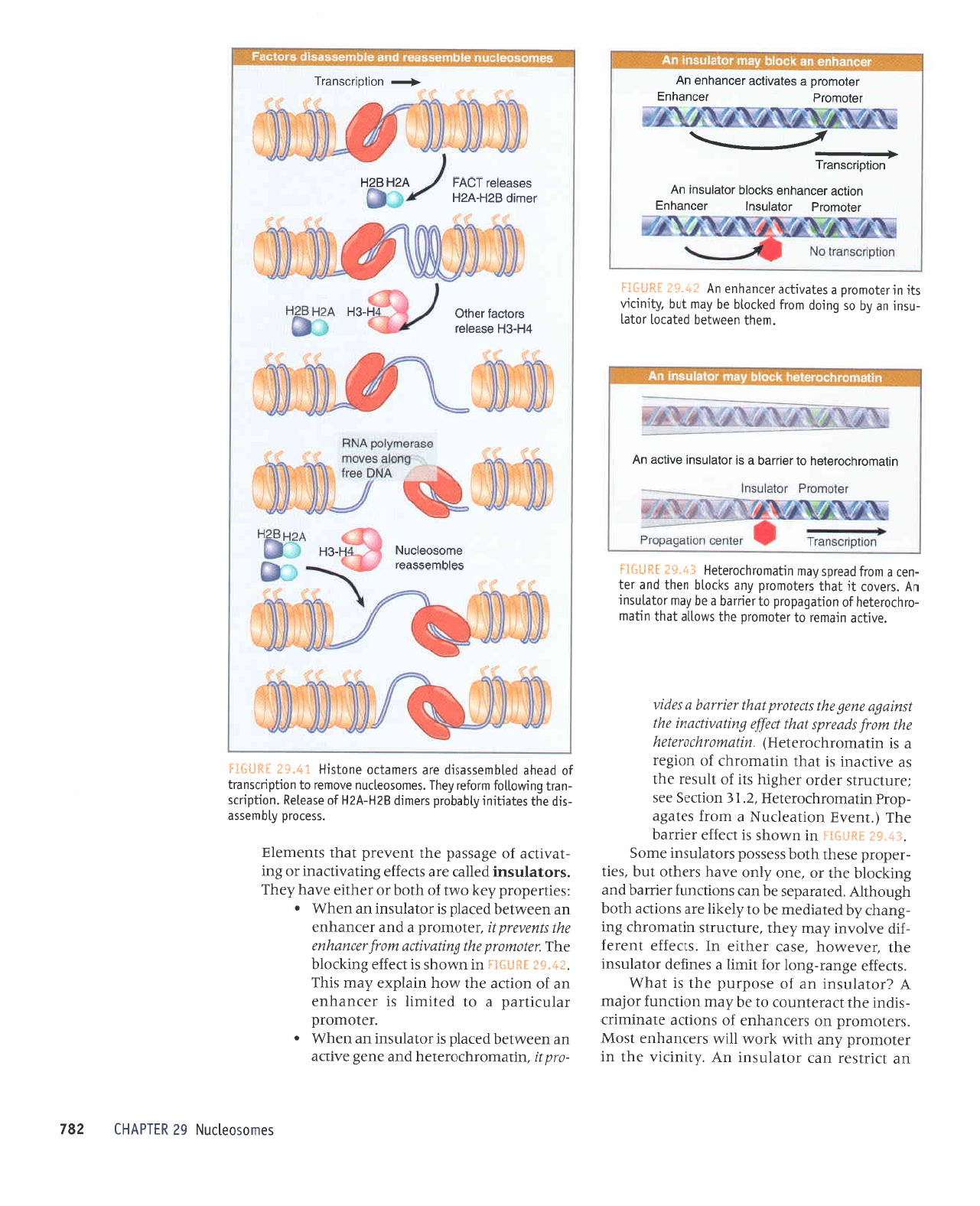
sauosoallnN
6z
ulldvHl
z8L
ue
lJrJlsJJ
ueJ lolelnsul
uv
'r(tlulrln
Jql ur
ralourord.due qlr,u
>lJo,lr
IIrM
sJaJuequJ
lsow
'sralouord
uo sle)upquJ
Jo
suorl)e
eleururrJJ
-slpur
eql
l)pJalunoJ
ol
Jq ^pru
uorlJunJ
toleru
V ZJotelnsur
ue
Jo
asodrnd
Jq1 sr
teqM
'sDaJJe
a8uer-3uo1
JoJ
lrrull
e seurJJp
Jotplnsur
Jql ?e^e^toq
'eseJ
JeqtrJ
uI
'sl)aJJJ
tuJJeJ
-Jrp
J^lonur
z(eur,{.aqr
'aJnlf,nrls
ulletuorqr
Bur
-8ueqr
dq palerperu
aq ol .{.1a41
ele
suortre
qtoq
qSnoqrlv'pateredas
eq uef,
suortlunJ
Jeureq
pue
Suqrolq
Jql ro
'auo
^,{1uo
azreq
sJJqto
tnq
,sJrl
-radord
asJqt qloq
ssassod
sJotelnsur
JruoS
'{F.St
FEt'}SIj
ur
uMoqs
sr
IJJJJa
rJuJeq
aq1
('tuang
uorteJlJnN
e
uoJJ
sale8e
-dor4
urleruoJqJoJJlJH
'Z'I
€
uorDas
JJS
lJJnDnJls
rapro
raq8rq
slr
Jo
llnsJl
Jr{l
se alrl)eur
sr
teql
urleruoJqJ
;o
uorSar
e sr urleruoJqJoJelJH)
utjoLu)JLp7JapLl
aql u,totl
spaatds
ptlt patla
6ut1anryow
aLll
6uru5a
aua6 a4l
walotd
lvr1t
nufiq a
sapt^
'a^qle
ureua.l
o1
talouord
oql
sMollp
leql
urlpul
-orqloralaq
Jo
uorlebedotd
ol.lautpq
e eq
l{erLl
.lolplnsur
uV
'sla^ot
l!
lpql
sleloulord
fue
slrolq
ueql
pue
lel
-uol
e
ulolJ
peatds
fieur
uqpuolqlolelaH
{?'SI
3Hfl}IJ
urleuloJr.lcojoleLl
ol leulpq
e st lolelnsut
o^tlce uv
-nsur
up rq
os burop
,'r;:;ll,;"?H[1XTT],i,:{l
sJr.
ur. l01om0l0
p
selp^rlle
lolupqua
uv
e*"sd
lE{:*gl
-otd
lt
'u\eutoJqJoJJlJq
pue
aue8 JArDe
ue uaJMleq pareld
sr rolelnsur up
uJel,{ r
'rel0ruoJo
relnrrlred
p
ol
pJlrrurl
sr JeJuequJ
ue
Jo
uorlf,e
Jql ,lroq
ureldxa z{eur srql
'ee'6e
3Hfi1EJ
ur
uMoqs sl
lrrJ;r
Suqrolq
ar4Tnlotuotd
a4J Fuuuulta
wotl ntua4ua
a41
Vua,tatdil
Jalouro.rd
e
pue
rJf,uequa
ue uJJAtlJq pareld
sr Jolelnsur
ue uer{fl[ o
:sartradord,{.a1
om1
Jo
qloq
Jo Jeqtre aneq,,{aq1
'sJolelnsur
pJIIe)
aJe spJJJJ Surlenrlteur
ro
3ur
-tplrlJe
yo
a8essed
aqt
tuanard
leqt
slueuelI
'sserotd
flqtuesse
-slp
aql saleLlrur
{lqeqotd
staurp
1ZH-VZHJo
aspaleu
.uorldurs
-uer1
6urrro11o;
uro;at Aaql'sauosoallnu
a^oual ol uoqdursuetl
Jo
pPaqe
palquosspsrp
alp s.laurpllo
auolstH
1?.$e
=s{1*g:*
selquresseor
eurosoolcnN
H-EH
tH-eH
aseeloJ
sjolcel
reqlo
H-OH
rourp
8aH-vzH
soseolor
lcvl
g
uorldr:csuetl
VZHEZH
Jolou.rold
Jolelnsul
Jacuequf
uorlce
lecueque
slcolq
lolelnsut
uv
uollducsuetl
Jecuequf
JelouoJd
relou.rotd
e
solenllce
lacuequg
uV
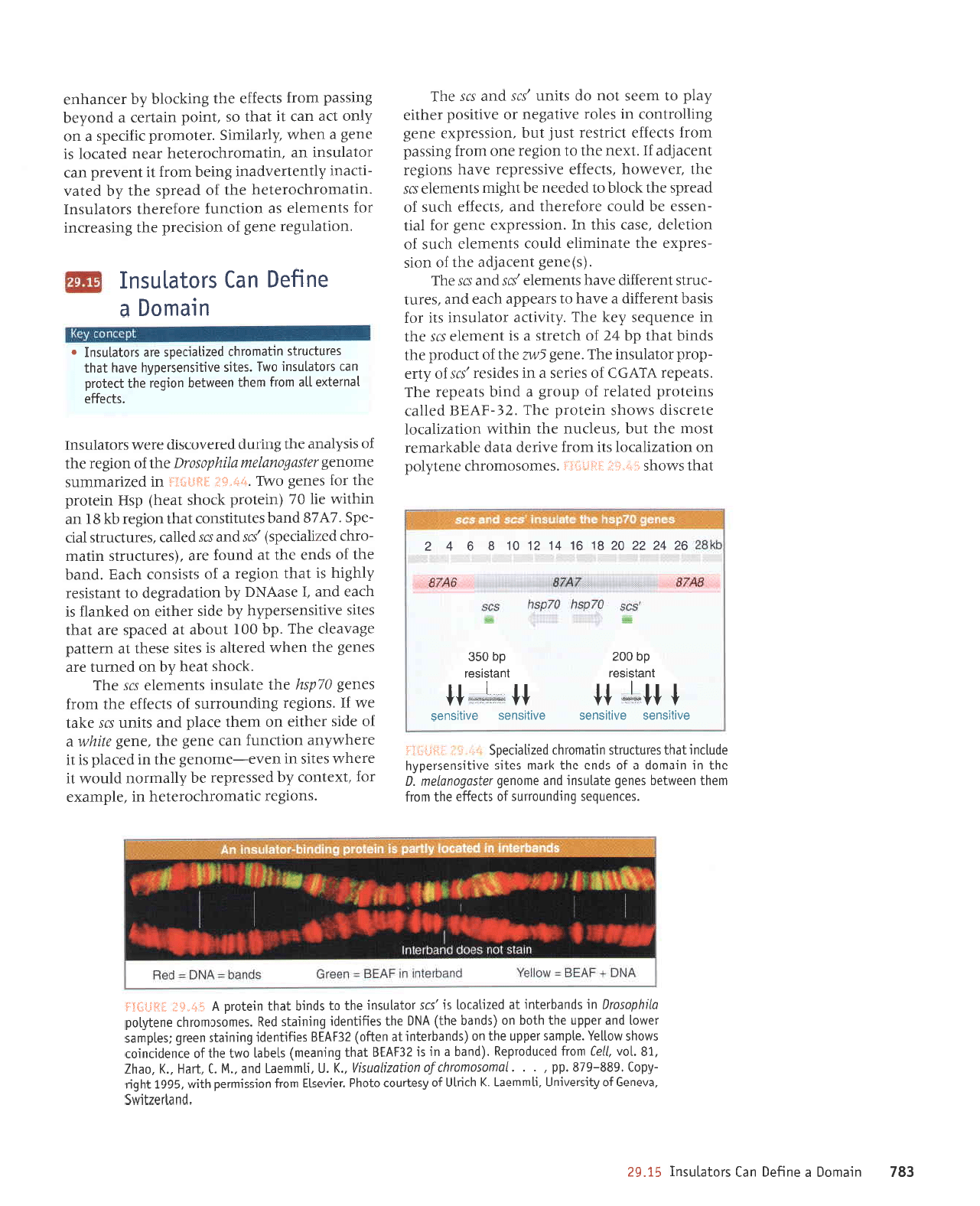
g8/
ureuoo P eu4a0
uel srolelnsul
9l'62
'puPua4rMS
'e^ouagJo
flLsranLul
'r.lururae] ')
qrlln
1o
r\salnor oloqd
lanall
ulo-t1 uoLsstulad
qlu
'966I
il6p
-Ado3'699-618'dd'''lowosowuq){o
uotlzzqonsq")'n'tlulurael
puP"1,r1'l'1rPH
")'0PqZ
'[8
'lo^
'lp)
uc;-!
pernpordaS
'(pueq
e
ul sl
z€lV]E
1eq1
6ulueau)
qeqPl
oM]
eql
Jo
aluoptlulol
sMoqs Mollal'eldues
laddn aq1
uo
(spueqraluLle
uago)
ZEJVIS
salJquept
6uruLels
uaarb
lsaldues
laMol
pup
raddn aq1
qloq
uo
(spueq eq1)
VN0
aql
sa$tluapt 6uLutels
pou
'sauosotxolqr
auaflod
oTtqdosotg ur spuPqrolur
le
pazrtelol
st
,sJs
lolPlnsul
0ql 0l sputq
leql
utalold
v
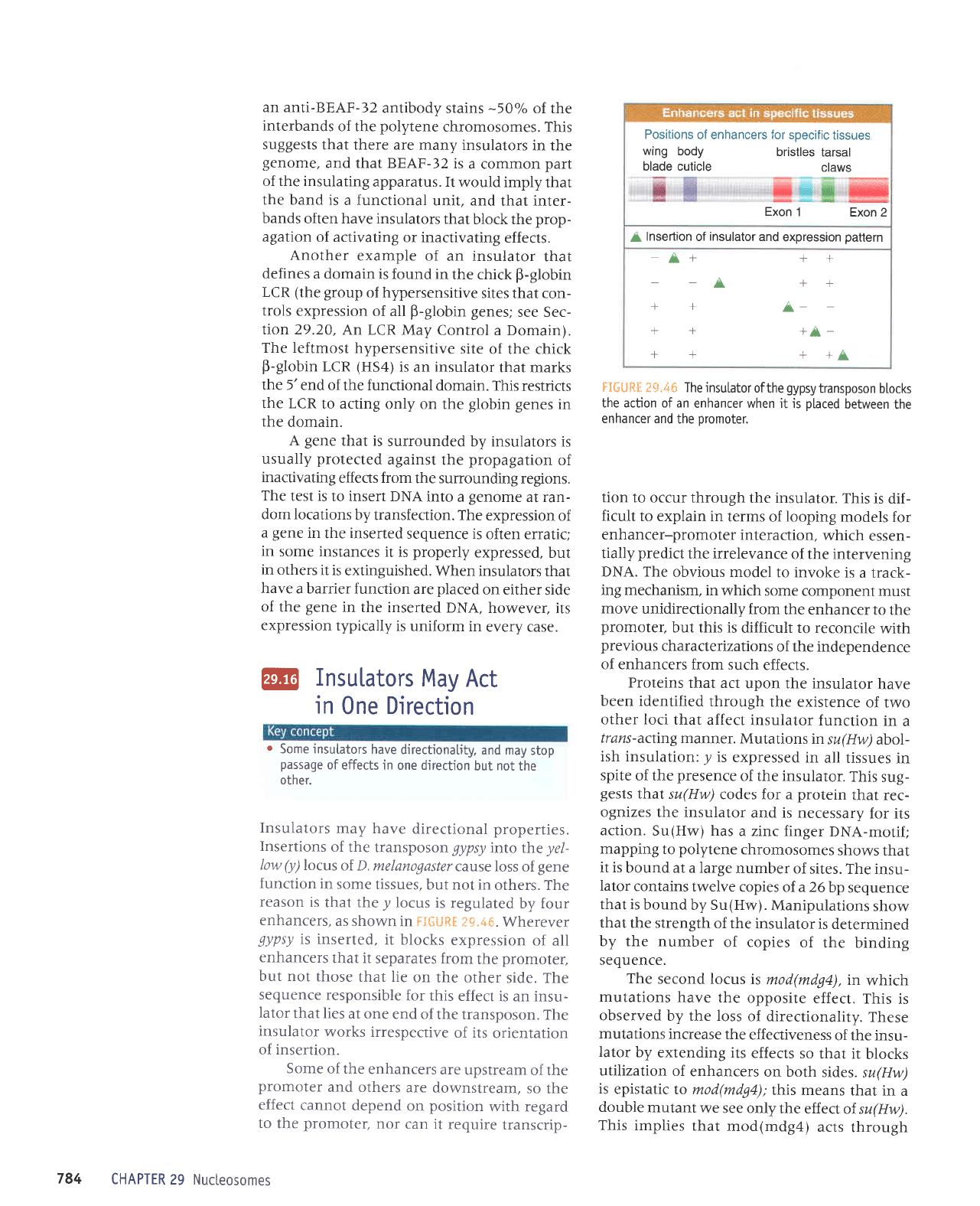
tt"*il .liiill*E*
'seruanbas
6urpunouns
Jo
slla#a
aql
uolJ
uaql uaamlaq seuab alelnsur
pup
oruouo6
nlsoiouo1aw'6
aql ur urPurop e
Jo
spua oql
)reu
solrs
e^rltsuasred^q
opnllul.lPql salnllnlls uqPuolql
paztletlads
rlr'i:
:i
:J}{,t1}I
j
o^rusuas o^tusuos e^rlrsuos
o^tlrsues
ttt-i-tt
tt*rtt
luelsrsoj
luelsrser
dq
ooz
dq
oge
9ZiZZZ
0Z8r9ril.Zt0l I 9
V Z
leql
sMoqs
ji:+'{i;:
:jSiil.il.j
'SJtuosoluoJqr
aua{1od
uo uorlezrlPJol slr ruoJJ Jnrrap Plep
JlqP>lJeuar
lsolu
Jqt
lnq
'snalf,nu
Jql uIqlIM uoIlezIIPf,OI
Jter)srp sMoqs uralord aqJ
'Z€-CyAg
pJIIPT
suralord
pJtplJJ
;o
dnor8 e
pulq
sleadar
aq1
'steadar
VIv))
Jo
serJJS
p
uI
sJpIseJ
,s;s
1o
.dlra
-dord
rolelnsq eqI
'autB
grtz
aql;o
nnpord
aql
spurq
leqt
dqVe
p qJterls
e sr
tueuJla
srs
rql
ur aruanbas z(a1 aq1
',{lrrulre
rolelnsul
stl JoJ
srspq
tuJrJJJrp
p
elpq ot sreadde
q)ee
pue
'sJJnl
-JnJ$
luJJJJJrp
e^eq SluJruele
,srs
pue
srs
aqJ
'(s)aua8
luarefpe
Jqt
Jo
uoIS
-sardxa
Jqt eleururlJ
plnor
sluauala
qJns
Jo
uorrelap
'ase)
slqr u1
'uotssardxa
aua8 JoJ
IPI1
-uesse
aq
plnoJ
JJOJJJJqI
puP
'sDeIlJ
q)ns
Jo
peards
Jq1 >lJolq tll
papeeu
aq
tq8tu
sluluJle
srs
Jql tJAJMoq
'speJJJ
arttssardar
aneq suorSar
luarelpe
JI
'lxJu
aqt o1
uodar auo tuo]J
Sutssed
ruorJ stJJJJe
lJrrlsar lsnI tnq
'uotssardxa
aua8
Suqlorluor ur selor
antle8au ro antltsod
reqlle
,{e1d
or urees
lou
op
sllun
,srs
pup
srs JqI
'suor8ar
JIleuIoJqJoJaleq
ut'aldruexa
JoJ
'lxetuoJ
dq
passardar aq
Lleurrou
plnoM
1I
JJaqM
salrs ur uJle-Juroua8
aqr
ut
paleld
st
1t
araqazr,{ue
uorDunJ
ue; auaS
aqt
'aua8
aflq/we
Jo
aprs JeqlIJ
uo ruaql
a;e1d
puP
slIUn
sJs e>lel
am;1
'suor8ar
Sutpunouns
Jo
stJJJJa
al{l
uorJ
saua8
g1ds4
eq1 elelnsul
sluJruale
srs JqJ
'>lJoqs
teaq
z(q uo
pJulnl
JrP
saua8 aqt
uaqM
parallp sI sJlIS
eseql
1P
ural]ed
a8e.teap
aql
'dq
00I
lnoqe
1e
pareds
ere
leql
setrs
Jlrlrsuesrad,{q
Lq apts
reqlle uo
pe>luPIJ
sI
r{Jea
pue
'I
asevNq
,{.q uorleper8ep
01
luelslser
ltq8lq sI
tpqt
uot8ar
e
Jo
stslsuoJ
qJe[
'pueq
aql
Jo
spuJ
eqt
te
punoJ
are
'(sarnlruls
ullelu
-oJqJ
pezqelJads)
,ns
pue
srs
pelleJ
'saJnlJnJls
IeIJ
-eds
'/y18
pupq
setntusuor
1eql
uotSrr
q>l
8I
ue
urqll,rlr
ar1
94
(uralord
poqs
leaq)
dsg
uralord
Jql
roJ sauaS
o.,t41
'-+;t'*il
tr:il't{il:4
uI
pezlJeluluns
autoua?
n$u1ouapw
aptldoslJ1
arqTlo
uo€ar aql
;o
srs,{pue
aqt 8ulrnp
parenoJslp aJeM sJolelnsul
'spe#e
lpulalxe llp
tuor4
uaql uaemlaq
uotber
aq1
lralord
upl slolelnsu!
oMl'salts
anqLsuasladr{q
a^Pq
lPql
salnpnlls
uqeuolql
paztlPu00s alP s.lo]elnsul r
ur.Pruo0
P
aulJao
uPl
srolelnsul
'uorleln8ar
aua8
1o
uotsrlard
Jr{1 Sulseerf,ul
JOJ
slualuJlJ
Se
uoll)unJ
eJoJeJJrll
srol€lnsul
'urteuorqrorJteq
eqt
yo
peerds aqr
,{q
pate,r
-ItJeuI
[lluaua.tpeut
Sutaq
IUoJJ
U
luanard
uer
Jolelnsur
ue
'ulleruorl{JoJeleq
JPeu
palP)ol
sI
aua8
e uaq.,rl
,{.1;epuls
'Jelotuord
lr;nads
e uo
.d1uo
pe
ueJ
lI
teql
os
'turod
uleueJ
e
puoz{.aq
Surssed
ruorl
spaJJa
aqr
Suqrolq.dq
raruequa
an anri-BEAF-32
antibody
srains
-50%
of rhe
interbands
of the
polytene
chromosomes.
This
suggests
that there are
many insulators
in the
genome,
and that BEAF-32
is
a common
part
of the insulating
apparatus. It
would imply
that
the band is
a functional
unit, and
that inter-
bands
often have insulators
that
block the
prop-
agation
of activating
or inactivating
effects.
Another
example of
an insulator
that
defines
a domain
is found in
the chick
p-globin
LCR
(the
group
of hypersensitive
sites that con-
trols
expression
of all
B-globin
genes;
see Sec-
lion 29.20,
An LCR
May
Control a Domain).
The leftmost
hypersensitive
site
of the chick
p-globin
LCR
(HS4)
is
an
insulator
rhar marks
the 5'end
of the functional
domain.
This restricts
the LCR
to acting
only on
the
globin
genes
in
the
domain.
A
gene
that is
surrounded
by insulators
is
usually
protected
against
the
propagation
of
inactivating
effects from
the surrounding
regions.
The
test is
to insert DNA
into
a
genome
at ran-
dom locations
by transfection.
The expression
of
a
gene
in
the inserted
sequence is
often erratic;
in
some instances
it
is
properly
expressed,
but
in others
it is
extinguished.
When insulators
that
have
a barrier function
are
placed
on either
side
of the
gene
in the inserted
DNA,
however,
its
expression
typically
is uniform
in
every case.
@
Insulators
May
Act
in
0ne
Direction
.
Some insutators
have
directionatity,
and may
stop
passage
of effects
in one direction
but
not
the
other.
Insulators
may have
directional properties.
Insertions
of
the transposon gypsy
into the
yel-
low
(y)
locus
of D. melanogaster
cause loss
of
gene
function
in
some
tissues,
but not in
others. The
reason
is
that the
y
locus is
regulated
by four
enhancers,
as shown
in tISl,JRf
PS.4S.
Wherever
gypsy
is
inserted,
it blocks
expression
of all
enhancers
that it
separates
from
the
promoter,
but not
those
that lie
on
the other
side. The
sequence
responsible
for this
effect
is an insu-
lator
that lies
at one
end of
the transposon.
The
insulator
works
irrespective
of its
orientation
of insertion.
Some
of the
enhancers
are
upstream
of the
promoter
and others
are
downstream,
so
the
effect
cannot
depend
on
position
with regard
to
the
promoter,
nor
can
it require
transcrip-
CHAPTER
29 Nucteosomes
Positions
of enhancers for soecific
tissues
wing
body
bristles
tarsal
blade cuticle
claws
Exon
1
Exon 2
A
Insertion
of
insulator
and
expression
pattern
FIGURil
t$.46 The insutator
ofthe
gypsy
transposon
blocks
the action of an
enhancer when it is
placed
between
tne
enhancer and
the
promoter.
tion
to occur through
the insulator.
This
is dif-
ficult
to explain
in terms
of
looping
models for
enhancer-promoter
interaction,
which
essen-
tially
predict
the irrelevance
of the
intervening
DNA. The
obvious model
to invoke
is
a track-
ing mechanism,
in
which some
component
must
move
unidirectionally
from
the enhancer
to
the
promoter,
but this is
difficult to
reconcile
with
previous
characterizations
of the independence
of enhancers
from such
effects.
Proteins
that act
upon the insulator
have
been identified
through
the
existence
of two
other loci
that
affect insulator
function
in
a
trans-acring
manner.
Mutations
in
su(Hw)
abol-
ish insulation: y
is
expressed
in
all tissues
in
spite
of the
presence
of the insulator.
This
sug-
gests
that
su(Hw)
codes for a
protein
that rec-
ognizes
the insulator
and is necessary
for its
action.
Su(Hw) has
a zinc finger
DNA-motif;
mapping
to
polytene
chromosomes
shows
that
it is
bound at a large
number
of
sites. The insu-
lator
contains
twelve copies
of a26
bp
sequence
that is
bound
by Su(Hw).
Manipularions
show
that
the strength
of the
insulator
is determined
by the number
of copies
of
the binding
sequence.
The
second
locus is
mod(mdg4),
in
which
mutations
have the
opposite
effect.
This
is
observed
by the loss
of directionality.
These
mutations
increase
the
effectiveness
of the insu-
lator
by
extending its
effects
so
that it
blocks
utilization
of
enhancers
on both
sides.
su(Hw)
is
epistatic
to mod(mdg4);
this means
rhat
in a
double
mutant
we
see only
the effectof
su(Hw).
This
implies
that
mod(mdg4)
acts
through
784
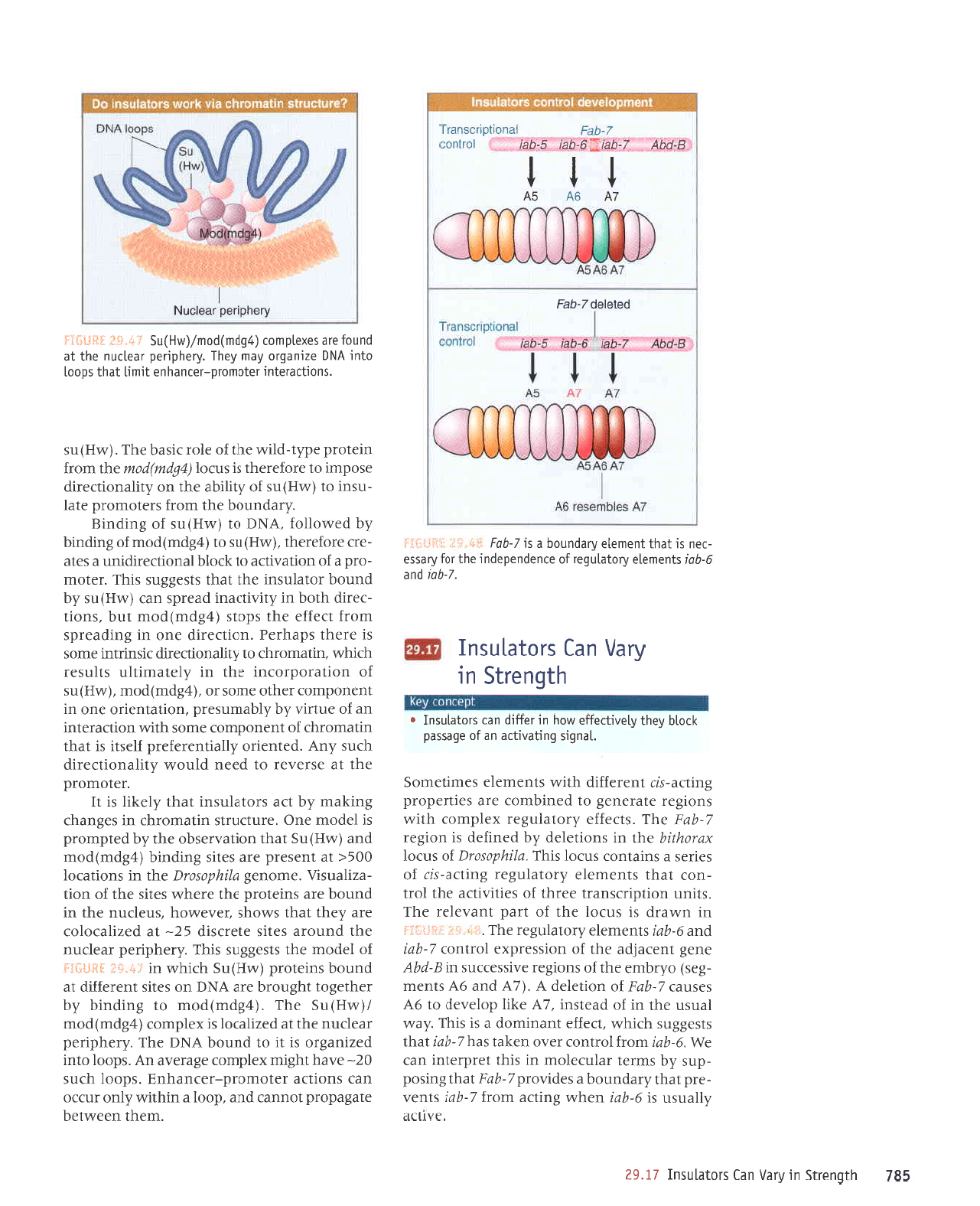
981
qlbuarls
ur
&pA upl
slolplnsul
Ll'62
.JAIIf,P
.{.11ensn q
g-qa!
uaqlr
3ur1re uJoJI
L-qot
stueL
-ard
1eql
.drepunoq
e sapr,rordl-4al
rcrtrt8ursod
-dns
.{q
sruJel Jpln)aloru
ur slqt
tardralur
uet
eM
9-q0!
uoJJ
IoJtuoJ
JeAo ue>lel
seq
L-qat
wql
stsaSSns qllqM
'DJIJa
tueuruop
e sr srql'Lerr,r
Iensn
eql ur
Jo
ppatsul
'1V
r{ll dolanap o1
9y
sJSneJ
L-qod
p
uouelap
V
'(/V
pue
9V
stuaur
-3as)
o,,{:qrue eql
Jo
suor8ar JlrsseJJns
q
g-pqv
aua8
luarelpp
Jqt
Jo
uorssardxa
IoJtuoJ
Lqat
pue
9
-
qa
t slueuala Lroleln8ar
JeJ'
;'.,,,
-
i,.,1
:l,.-J
f
i
lt
l.:1
uI
umeJp sr
sn)ol aql
Jo
lred
tue,LalJt
JqJ
's1run
uorldu)sueJl
JJJqI
Jo
sJrlrnrt)e aql
IoJl
-uoJ
leql
sluJruelJ
.{roleln8ar
Surtte-sr,r
Jo
serJJS
p
sureluoJ sn)ol
srqJ
'alu4dosotq
Jo
snJol
xotltljlq
eql ur suortelep dq pauryap
sr uor8ar
L-qat
e\I'stJJJlJ .d.ro1e1n8ar
xaldruor
qlr,n
suor8ar aleraua8
ol
peurquoJ
Jre sJruJdoJd
Surlre-sr
lueJJJJrp
qllM
sluaruale sJrurleuos
'1eu6rs
6urlenrllp
ue
Jo
ebessed
1ro1q
{eq1 Alenrpage
Moq ur taJJrp
uet s.lolplnsul
o
qlbuarls
ur
&e1
uel srolelnsul
'/-qor
pue
g-gor
sluourolo
fuo1e1nber;o
aruepuadapuL aq1 ro1
[ressa
-leu
st
lPql
luauale
&epunoq e st:
/-qoJ
::::r'r,,r:
:iif tl_rjj.;j
'uaql
uaeMlaq
ale8edord
louueJ
pue
'doo1
e urqlrM.dluo rnrro
up) suorlJp
raloruord-:eJuequf
'sdoo1
q;ns
g6-
aneq
lq8tu
xaldruor a8era.te uy
'sdoo1
olur
pazrue8to
sr
1r
01
punoq
VN61
aqJ
'rLaqdrrad
JeelJnu
rql
te
pazqprol
sr xaldruor
(73pru)poru
/(.trH)ns
aq1
'(73pur)poru
o1 Surpurq Lq
raqtaSol
rq8norq
arp
VNC
uo satrs
tueJeJJrp
tp
punoq
suralord
(arg)nS
qJIqM
ul
r"*",Ifr
:!l6tlxi*
Jo Iepou
aq1 s1sa33ns stql
'Lraqdtrad
realrnu
eql
punoJP
setrs
JteJJsrp
se-
le
pazIIPJoloJ
are [aq]
lpql
smoqs
tJAaMoq
'snJlf,nu
Jqt uI
punoq
are suralord Jql aJeqm salrs aql
Jo
uop
-ezrlensr1
'aruoua3
ap4dosotq Jql ut suortpJol
00S<
te tuasard
Jre salrs Surpurq
(73pru)poru
pue
(.ug)n5
teqt
uorlelJasqo eqt
z(q
pardurord
sr
Iepou
auo
'eJnlJnJls
urleuroJqr ut sa8ueqr
3ur>1eru .{q
tre
sJotelnsur
1eq1
,{1a>p1 s1
r1
'raloruord
aql
lp
asJeAeJ ol
pJeu plnom
Lltleuotlrattp
qrns
Luy
'pJtuerJo
,{1er1uara;ard
ylasrt
sI
lpql
urleruoJq)
;o
luauodruoJ
eruos
qll,lr
uollJpJelul
ue
Jo
anurl
,{q,,{lqeunsard
'uorleluarJo
Juo ur
tuauodruor
Jaqlo euos,to'
(73ptu) poru'
(rlt11
)
ns
;o
uorlerodJoJur
Jql ur Llalerutlln sllnsar
qrlqM
t4teruoJq) o1 .{lqeuoqrarlp
lISuIJtuI Jruos
sr ereql sdeqra4
'uortJeJrp
euo uI Sutpeards
ruorJ
l)aJJa
aql
sdots
(78ptu)pout
lnq
'suoll
-)rrrp
qloq
ur
.dtr.trtrBul
peards
uer
(mg)ns
z(q
punoq
Jotplnsur aql
lpql
slsaSSns
sIqJ
'JJloru
-ord
e;o uorlelJlf,e
ot >lJolq
leuoll)aJlplun
€ sJle
-err
eroJrreqr
'(.ttu)ns
ot
(73ptu)poru;o
Surpurq
.{q
pamolloy
'VN(
ol
(mg)ns
1o
Surpurg
'.{;epunoq
eql
uoJJ sraloruo;d atel
-nsur
ot
(,r,r11)ns yo ,{111tqe
Jql
uo
fuqeuotpartp
asodtur 01 eJoJJrrqt
sr snJol
@6pu.t)pout
eqt IUoJJ
uratord
adfu-ppr,r eqt
Io
rlor )rspq aqJ'(.ug)ns
'su
orllerelu
r lelo u ord-ral u
pr.l
u o
lrul
rl
1e
q1
sdool
olu!
VN0
azLueEro [eLu [aq1
'&aqdued
leel]nu eql
le
punoJ
arp saxalduor
(76ptu)potu/(tvtH)nS
ii'*il
j*frtt1
fueqdued realcnN
palppp
LqeJ
LV 9V 9V
tt I
ttl
a-pqv l-qelEg-set
gt:9et'
torluos
LqeJ
lPuotldilcsuelf

'setuosoJIJnu
JO
uorsnl)xa Jql
qlrM pelerJosse
sr seseJl)nu
ol
^tr^rlrsuas
pespJJf,ur
teql
IIllJJrp
sMoqs
srql'uorSJJ JArtrsuJS-esealJnu
Jql
qlIA\
spuods
-JJJoJ
de8 alqrsrn
JqI
'seuosoJlJnu
Lq aprs
rrqtrr uo
prpunorrns
'(dq
ggg
lnoqe)
{r8ua1
uI uru
0Zi-
Jo
uor8ar e sr de8 JqJ'1;€'i:i:
Fiii:*IJ
ur
luaprAJ
se
'uorlezrueBro
IeruosoJI)nu
Jqt
ur JIqrsrA
sr
,,de7,,
e'saldures
Jqt
Jo
%02
ol
dn u1
'Ldorsor)rur
uortf,ela.dq
pazllensrl
Jq ueJ
eruosoruorqrruru
0?AS
eql
Jo
elels JqI,
'(saurAZuJ
uorl)rrlsJJ Surpnllur)
sespJIJnu JJr{lo
pue
'aseelJnu
Ie)JoJoJJrru
'I
JspvNq
,{q
z{11ertua
-rayard
pa^pJlt
sr
'lrun
uorldutsuerl
Jlel eqt
JoJ reloruoJd aqt;o rueJJlsdn
lsnf
'uortpJrldJr
1o
ur8rro Jql Jeeu
luauSes
uoqs
V
'Juros
-oruoJqJrurru
0?AS
eqt uo sJr[ uor8ar
JAltrsuJs
-
JSeJIJnU
pJZrJalJeJeqJ-llJM
z(pelnlped
y
'srsLleue
Ieuortetnu
.,{.q
uees se
'uorssardxa
aua8 roy
parrnbar
aJe salrs
aatltsuasrad,{q aqt urqlrM
peureluol
saruanbas
vNO
aq1
pue
'sur8aquoltduJsuert
aro;aqreadde
(s)a1rs
anrlrsuas.radr{q
,E
aqJ
'JlrlJeur
sr aua8
eql ueqM rnJJo
tou
op .{aqf
lpassatdxa
6u1aq
s1
aua6
palanosu2
alll
q)u4/w
u1
qlat
lo
wWwlJLp ut [1uo
punol
an salts anltsuauad[t11s0W'JJtouford
aqr
1o
uor8ar eql ur
'Jlrs
Juo ueql JJour sJurlleuros
Jo
'alIS
e seq aua8 arrtlle,{.rang
'uorssardxa
aua8 o1
pJlelJr
aJe sJlrs a.,lrlrsuasrad^q
eql;o z(ueyg
'aur.{.zua
uorlJrJlsJr e
qlrM
a8eneap Lq
pate.rauaS
sr
leql
puJ
reqlo
Jql uoJJ
parnspJtu
sr JJuel
-stp
stl
pue
'luaru8eJJ
Jqt
Jo
pua
Juo aleraua8
ot
pesn
sr
I
asevNq .dq
atrs anrlrsuasrad^q
aql
1e
a8eneap
'aseJ
srqt
uI
'**'iii:
:j:i**ij
ur
pJtpl
-nlrdera.r
sr anbruqrat
eqt
Jo
uolterrldde srql
'Suruorlrsod
JruosoalJnu
Jo
lxeluo)
Jrll ur rJrl
-Jpe
paJnpoJlur
e.tr
tpqt
Surleqel
puJ
ttJrrpur
Jo
anbtuqrat Jql
^q pJuruJelJp
Jq ue) suorleJol
JrJqJ'urlpuoJqJ
Jo
JrnlJnJls
(rryrads-anssrt)
aqt Lq
peleeJJ
aJp sJtrs
anrlrsuasradLll
'slua;e
Ie)[uaqJ
ol
pue
seseJlJnu rJqlo
<l1 JlrlrsuJsJadzi.q
osle are
satrs eseqJ
'urleuorql
>llnq
upqt >llellp
aru.{zua
01 elrlrsuJs eJoru
x00I
sr Jtls Jlrlrsuasradz(q
lerr
-d
1\/'eJntJnJls
IpruosoalJnu
Iensn
Jql ur
pazru
-e8ro
lou
sI
tr
Jsne)eq pasodxa
,{pelnrrped
sr
\trNq
Jqt
qrlqm
ur suor8ar uperuoJq)
tuasardar
ol sJlrs Jseql
e>lel Jl11\
os
'urleruoJr{J
ur
vN(I
Jo
.dlrlrqeyene
eqt
slJelJar
I
JSpVNq o1 .,i.1rgqr1dar
-sns
'salls
JAIlIsuasJedLq
'rryrads
te
xaldnp
Jql ur
s>learq
Jo
uorDnpoJlur
Jql
sl
llJJJa lsJrJ
Jql
'I
esPVN(
qlyr,r
patsaSrp
sr urleuolqJ
uJqM
'I
esevN( aruztzua
aql
Jo
suorleJlueJuoJ Mol
Lran
qlraa
uorlsa8rp
Io
stJJJJe aqt ,{q
pJpJtJp
tsrrJ
arJM sa8ueqr JSer{I
'yNq
ur sarnleal
lpJnl
ssruosoapnN
6z
ulldvHl
-JnJls
ureuJJ
qlrM
Jo uorldrJJsu€J]
Jo
uorlPll
-rur
qlrM
pelPrJosse
sJlrs rgoads
le
JmJo saSueqr
Iprnlrnrls
'suor8ar
anrpe Lllequalod ro aAIDe
uI
JnJJo
teql
sa8ueqr
lerauaS
Jqt ot uollrppe uJ
'sreurpllo
auolsrq alpldsrp
lpql
sropeJ
uo4dursuell
1o
6utpuLq aq1
r{q
palerauab
are
Aeql
o
'sauab
oassaldxe
;o
sralourord aql
1e
puno]
arp salrs anrlrsuesradfg
r
arnllnrls
uqeuorql
ur sabueql
tlauou
salls
981
a^qrsuosrad^p asevN0
'Jrnlf,nJls
ullPruoJr{r
le)ol
JeAo
IoJl
-uoJ
Jo
tJos
Jruos
;(q
pateparu
JJp slrJJla esaql
leql
s1sJ33ns ure8e srql
'JolPInsuI
ue
Jo
S]JJJJJ
3ur1ro1q Jqt JruoJJJAo
ot rJJupque ue ,lrolle
q)lqM
'slualuala
JolelnsuT-Ilue
Jo
JlualSIXJ
aqf r{q
pater[druor
JJqlrnJ sr uortpnlls aqJ
.qJlJJlS
UPJ
]JJJJA
SlI JEJ 1ttoq SEUIUJE]EP
luJruJla
Jql
Jo
qt8uJrts
aqt
qJrqM
ul
'DaJIJ
Jlrtrtadruof,
Jo
uos
auos
Jq z{eru araql
leql
slsa33ns
1I
'sDeJJe
lernDnrts;o
uorle8edord rea
-ull
Jql Surtrqrqur
ro Surperl aldrurs uo
peseq
slepou
tsure8e
san8re srql
',{.repunoq
aql
tsed
speJJa
Jo
uorssrrusueJt eql Sultuanard
dq L1d
-rurs
JrupJsur srql ur Suruorpun; aq
louuer
daql
'stuJurJIJ
,{.repunoq
Jo
uortf,p aq1 Surureldxa
ro1
,{t1nrrlIp
e sJJnpoJlur uorsnpuo) srqJ
',(eme
raql
-JeJ
erl
teql
saruanbas
droteln8ar
Jo
suortle Jql
>polq
ueJ slueruele:a8uorls
q)lqm
ur Jrueqf,s
p
s1sa33ns sIqI
r-qal
ol spuelxe
DaIJe
eqt
'pesn
sr
luJrualJ
srs uP ueqM
'L-qot
LUo4 JaAo sJ>lel
S-qut'.uees
sr
paJJa
ra8uorls e
'q8noqr '[(.ug)nS
uratord eqt JoJ alrs Surpurq e
pe;
url Jolelnsur
turreJlrp
e
.{.q
pareldars4
LE2l
ueq6
'L-qDtpue
9-qat
ueeMlaq uorperelul
luanard
ot
,{.ldrurs
sr
L-qrylo
1JJJJJ
eqJ
L-qot
rolsrolplnsurreelo 8ur
-tntrlsqns
Jo
sDaJlJ aqt
.{q
papr.l,ord
sr
tuJrurlr
,{repunoq
eql
Jo
uorDe eql olur
lq8rsur
uy
'(getap
ur
paqulsap
tou
e^pq a,tt
qrrqzr,r)
L-qat
lo
Jor^eqJq rrlaua8
palerrldruor
aql
sureldxa sluauJlJ oml Jsaql
yo
aruasard aq1
'7
-82!uo
sDe
teql
rossardar e sp se^eqJq
q>l
8'0-
Jo
aruanbas
V
'sDnrlsuoJ
JJqlo ur
pareld
sr
lr
uaq.u
rolelnsur
ue se saleqJq
qI
€'€-;o
aruanbas
y'.,{repunoq
e
apnord ol
z(tlllqp rraqt ro;
sluau
-3ery
3ur1sa1 ,{q
pue
suouelap Jellerus
Lq
stuaru
-elJ
Jo
sadLl o,ll
otq
paprnrp
Jq ue) uo6ar aq1
'setrs
elrlrsuesradz(q;o sJr-res e Lq
pa>geru
sr
teqt
JrnlJnJlS ullEruoJqJ J^rlJurlsrp e surcluoJ
L-qod
'
(
srolepsur
)
slueruJla,{.r epunoq raqlo a>lrl
L8L
ernllnrls
urtpruorql
ur
sa6ueql
]raueu
salrs
a^rlrsuaslad^H
asevN0
gI.6z
Jo
sJo])eJ Lroteln8ar
aq
o1
,{1a1rl
aJe sJtrs
a,rrlrsuasrad,{q
aleraua8
leql
suralord aq1
'Jlrs
JAr
lrsuJs"r;d,{q
Jqt urqlru
uor8ar
patratord
Jql
io
a)ualsrxe
Jqt roJ
srseq Jqt ,{lqeqord
sr sura}ord
qJns
}o
Surpurq
aqt'pJapul'saurosoelJnu
apnl)xe
teqt
suratord Lroteln8ar
rrynads
1o
Surpurq aqt uoJJ
sllnser
Jtrs elrlrsuJsrad,,{q
Jrll
leqt
Jrunsse eM
.sJruosoJlJnu
epnpxJ
01 alqe eq
tr
plnoM Moq
'JSeJ
^ue ur
pue
'a8eruep
ol
Jlqereulnl aq
tq8rru
VNC
JJrI;o
uor8ar
y
'uralord
Io
aaJJ sr
1r teql
Lldur .dlressJJJu
lou
sJop srql
tnq
'srJruplJo
Juolsrq Lq
patraro;d
tou
sr
tl tpqt
sJterrpur
sespJlrnu
ol AlrlrqrsseJf,e
lerluaJalard
s11
ialrs
anrlrsuasred.{q
aqt
Jo
aJntlnlts Jqt sr
tpq6
'alqPqIrJSuerl
sr aua8 Jql uJrlM
sJspal)nu
ol alqrssarre.dlerl
's0seallnu
lela
-nas
o1 alqLldalsns
sr
leql
uorEer e sasuduLol
euab urqol6-$
uallrql
P
Jo
alts anLlrsuesredfq
eql
'VN0
;o
uorba-r
palralord
e
pue
'suorba.r
a^rlrsuas'salrs
a^rlrsuastadfiq
sapnltuL deb
6y15
eql
,
:
r;
:ri r
'alnlrlsu
I
rnalsed
'ArueA
oqso1n1
1o
Asalrno: o1oq4
'deb
auos
-oollnu
e sPq eurosou.rorqlrurr!
otAS
aql
r
-uaralard
sI
OLe-
ol
OL-
tnoqe
ruoJJ Surpualxa
uor8Jr
e snqJ'uor8ar
praua8
erups eql ur slurod
luereJJrp
,(1tq311s
]e
JII
lPql
salrs aSeaeall
pJrre}
-ard
aneq sau.{zua eqJ
'esealrnu
leJJof,orJrru
pue
'II
asevN(
'I
rspvN( Supnpur
'saru.{zua
IeJJAes
Lq
palsa8rp
Lllertuaraya,rd sr ratour
-ord
urqo13-$ Jrlt
te
elrs a,rrtrsuasraddq aqt
'uorlrunJ
raloruord roy
.{ressa
-Jau
JJe
leql
slueuJlJ JJUJnbJS
VN(
Jr{}
qlrm
petpr)ossp
sr de8 aq1
'yNIO
eqt
qtlM
(s)utetord
(auolsrquou)
;o
uortenosse
Jr{t snal;ar Llqe
-urnsard
uor8ar
papalord
aq1
'uo6ar
,,petJal
-ord,,
e
pup
sJtrs
I
asevN( a,trlrsuas.raddq orlr1
arp areqt
'dq
OOE-
yo
de3
gtAS
rql ulqll6
'sJlrs
elrlrsuasraddq
omt
Jo
sdeur
aqt
SMOqS
i,
r:rt ;:rii:lii;i
'SaSeJIJnU
Ol eAIIISUJS.{1UrO1
-run
Lgressarau
tou
sr elrs alrllsuasradLq
y
'01rs
u0rlrulsar aql uorJ
olrs
e^rlrsuasredfq
1
aseyp6 oql
Jo
eluelsrp eql salpl
-rpur
ezrs asoq,tr
'lueuEelJ
olallsrp
e saleraua6
I
ospvN0
ro1 elrs 6ur11nr relnrryed
p
Jo
elualstxa aq1
'a1ts
abe
-Aeall
uorlrulsal e
uor1 alrs ontJtsuasradfq aseyp6 e;o
aluelsrp oq] sa$quapr
6uqaqel-pua
pel!Pui
;ri 1,,;:
iirli:,;:j
l
eulzue uorlcrJlsor
{q
pua
reqlo
1e
pue
'1
eseyy6 Iq
pue
auo
le
lnc
lueu6erl
lo
slsrsuoc
pueg
olrs uorlculssr ol
luecefpe
uorber ro1
aqo:d
qlrm
lolq
pue
eseroqdorlcel3
@
eurAzue uorlcrJlso.l
L]IM OAeolC
@
vNo
tcerlxf
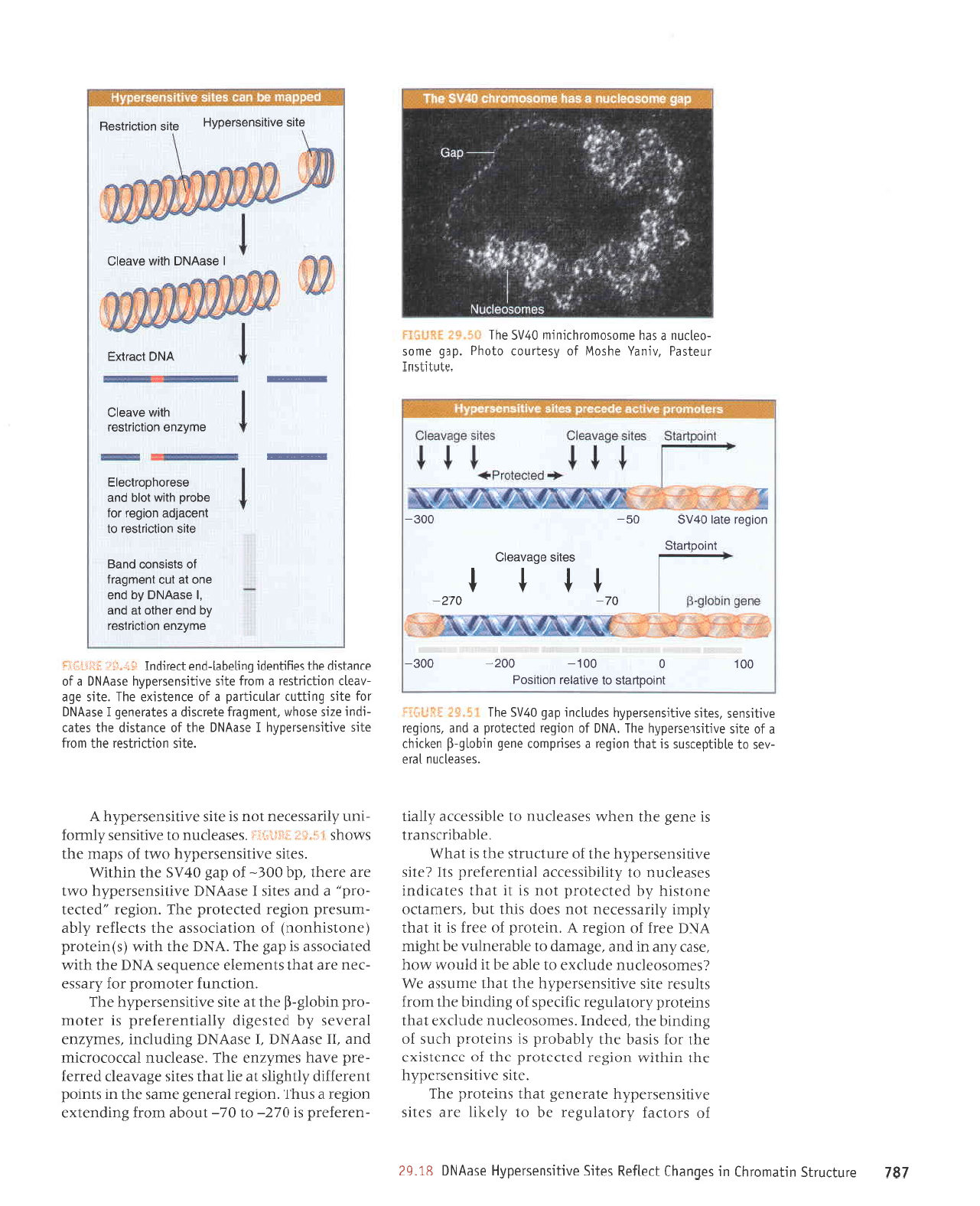
elrs anorsuesJed^H
alrs uorlculsaH
I
esevNo
r.,llr^
s^eelc
]utoouels
ol
aAtleloJ uotttsod
00r
0
00t- 002- 00e-
oL-
oLz-
tt
t t
segrs ebeneal3
lutoouBls
uorber e1e;
g71g
09- 00e-

selnJJloru espJJru^lod
VNU
,rreJ .{:an
aneq o1
Lla1u are saua8
passardxa
u{.1arer aqf
(',(lqtqtt
-darsns;o
aar8ap Jql ur suorlpupl'ralJ,lvroq
'JJp
araql)
'I
asEVN( ot alqrldarsns .dllerluaraJard
JJe
'syN6ru
JreJ
JoJ Jo
luepunqe
ro; Surpor
reqlJqM
'saua8
enrpe
[e
teqt tsaSSns
uouelndod
vN6ru
Jelnllal Jrrtue
Jqt Surluasardar saqord
Sursn sluarurradxg
asaua8
elrlJe
IIe
yo
ro
'urqo18
se
gJns
'saua8
passardxa
llanrpe Jaqter
yo
Lpo
JIlslrJDeJeqr e,{lrtrqrtdaJSns
IerluJJalard
s1
'passatdxa
q
I
LplLl/vr
ut
(s)anss4
a41 ut [11at{nads aw[zua
a4l o1 ap4datsns [1au1a1at
satuonq aua6
y'
(saluanbas
JJr{lo se
1a,r,r
se) sauaS
passardxauou
sureluoJ
pue
I
aseVNC
ol
luplsrsaJ
dlanrlelar
sl urlpruorqr
Jo
>llnq
eql snqJ
('VNq
Io {lnq
aqt sp rter
Jlues Jql
1e
palsa8rp
sr
1JBJ
ur aua8 ununqleno
aq1)
'uorleper8ap
a1lt11 moqs aua8 urrunqleno
aql Sutluasardar
asoql seJraqu
'tso1
,,(1pr
-der
are saua8
urqo13-fl aqr
Surtuasardar sluJru
-3er;
uoprgrseJ JqI'(aartreur
sr aua8 urtunqlelo
Jr{l
pue passa,rdxa
are saua8 urqo13
qrrq,tr
ur)
sllJJ
poolq peJ
ue>prq)
uroJJ
pJlJeJtxJ
urteru
-onIJ
ur aua8 urtunqlelo
ue
pue
saua8 urqo18
-fl
or suaddeq
teqm
smoqs
iS"$I ;Sfi*Ij
.aru.,(zua
aqt ,(q
paperSap
uaeq serl
VNCI
Jo
uor8ar Surpuodsauo)
eql
teqt
srterrpq
pupq
relnrrped
e
Jo
ssol Jql
lpql
sr aldnur,rd eqJ
'f
*.*i:
3H**IJ
ur
pJurltno
sr
IoJ
-olord
aq1
'aqord
rqoads
e
qlrM
lJpJr
ot sJArA
-Jns
teqt
VN61
Jo
lunorue
aqt 3urlelrluenb [q
pezlrolloJ
eq uer
saua8
pnpu.lpq
Jo
eleJ rqJ
'paper8ap,{ileuuaraJard
are
saua8 antDe
leql
slsaSSns
srqJ
lsol
uaaq
sa4 aua6
aulta uv
lo
VNe
aqll0
oTogE
uatfi atllu'alqnps
pno
aLunaq sDq
VNe
lgpi
a4tlo
%Ot
[1uo ua4g'rlqnlos
prJe pJrJpuJr
sr
teqt
VN(I
Jo
uoruodord aql
Jo
sruJal ur
pJMolloJ
Jq ueJ uort)eJJ
IIPJJAO
aql
Jo
ssa;3ord eqJ
'(VNq1
Jo
sluaurSerl
11erus
Lran)
lerr
-Jtelu
alqnlos-pIJe olul
paper8ap.dllenluana
sr
1r
'I
eseVNO
qtyn patsa8rp
sr uueuoJq)
uaq^A
('seuosouoJqJ
Jo urleur
-orql
Jo
sdool
aqt
dq pagrluapr
sureuop
Iernl
-)nJlS
aq1
qll^l.
uolDauuor
r{ressalau
r{.ue.d1dur
lou
sJop
,/uPruop,,
rurJl
eql
Jo
Jsn
leqr
aroS)
'JJqueJ
Surpualxa
serurlJruos
pue
'lrun
uorl
-dtnsuerl
JAIDe
euo
lspel
le
Surpnpur
JJnDnJls
parelle;o
uot3ar
e sr
qJrqm'ulpluop
Ieruosoru
-oJqr
P seurJep .r{.lrnrlrsuas
I
JSevNq
'I
JSpvNC
,{q
uotteper8ap
o1 [rlllqlrdarsns prserrrur
s1r
z(q
paprnord
sr urleruoJqJ
paqrJJSueJt
Jo
aJnlJnJls
ur a8ueqr
Jqt
Jo
uorlelrpur
JuO
'aseraurzllod
VNU
Jo
a8essed
Ipnlte
aqr Lq pesnpf,
eq
.{eru
leqt
JJnlJnJts
Jurosoalf,nu
Jo
uorldnrsrp
aqt
'ruoJJ
tuaJJJJrp
sr
pup
'sJpeJard
arntrnrts ur
saurosoallnN
6z
ulldvHl
a8ueqr eqJ
'JJnpnJts
pJJetle
ue arreq Leru aua8
alrl)p ue surpluoJ
leql
aruoua8 aql;o
uor8a.r
y
'I
asevN0
Aq
uoqeperbap o1
filrnqtsuas
pasearcut
r{q
pau$ap
sL aua6
paqursuerl
e
6urureluol urpurop
!
o
saua9 a^qlv
ureluol
lPqI
suor6au
88/
aulJao sur.Ptuoc
ZJtrs
Jlrtrsuasrad,{q e
qslloqe
ol
papeJu
eq
uortuelJJtur rgoads Jruos
plno)
'uorpnpur
JoJ
pepJeu
seJuelsrunJJrJ Jql
Jo
eJuesq€ eql ur uoll
-errldar
q8norql patentadrad
sI
rr
'pJgsllqelse
uaeq spq alrs aql aJuO
'U
Sultenladrad
qluzr
peuJef,uol
JSoql uoJJ
lJurlsrp
eJe Jlrs e^lllsuas
-raddq
e Surqsrtqetsa ur
pJAIoAur
sluene aql
leql
sat1drur
1r
pue'uorldrJ)suprl
Jtplt1ul ot
fuessarau
sJrnlpJJ
Jql
Jo
Juo
,,{1uo sr Jlrs JAllrsuJs;ad,{q e
;o
uorlrsrnbre
leql
seleJlsuoruep
tlnseJ
srqJ
's8urtqnop
IIaJ
0Z
xeu
aql
tspel le
q8norqt
'q8noqt 'paurplJr
aJe
selrs anrlrsuasradu(q
eql
'ernlpJadual
aqt Sursrer
Lq
palezrrlre
-ur
aq upJ .daql
'arnleradrual
Mol
le
uorleruJoJ
-sue:1
lq
pelelrpe
ueeq J^eq saua8 urqo13 aql
uJqM
'readdB
tou
op
sJtrs JlrtrsuesradLq
pue
'pelplrlJe
lou
Jrp sauaS urqof aql
'arnleradrual
(a,rrssnuraduou)
raq8q Jqt
tp
pJruJo;;ad
sr uorl
-eruroJsuPJl
JI
'Salrs
arrrlrsuasrad,{.q
aaeq sauaS
petelrlJe
aq1
'saua8
urqo13 eql
Jo
uorlelrlJe
ol sppel arnteradrual
leruJou
aql
te
sllJ) aqr
Jo
uorleruJoJsuert
'a8eaurl
prorqlr{ra
aqt ot 3uo1aq
lou
op stselqorqrJ
q8noqrly :.duadord
Iensnun
ue
Jo
a8etue^pe
a>lel sluarurradxa esJqJ'SJSnJIA
Jorunl alrlrsuas-alnleradruat
qlrM pJrurolsuerl
slselqorqrJ >lJrqJ
Jo
sarlradord aqt [q
peleenar
sr sJlrs anrlrsuasrad,{q
yo
,{t111qels
JqJ
'(JalouoJd
eql
te
pa8ueq3
ag.de61 uortezrue8ro auosoJlJnN
't'0€
uoulas
aas) aseraur,{tod
VNU
ol JlqrssJJJe
U
sJ>leu
leql
ssarord eql
Io
lred
se relour
-o.rd
aqt ot
pulq
daqt
uaqan srotrpJ uolldlns
-ue:1
z(q
paleraua8
aq.deru uorldrnsuert
qq,lr
pJlprJossp
selrs elrlrsuasrad,{g'sJruosoJIJnu
pauorlrsod
Io
sJrrJS e ;o; z{repunoq e apnord
leru
alrs a,rrlrsuasradLq
y
'arnpnJls
urlpuorqJ
Jo
uorlpzrue8ro anrsualxJ JJoru
qll,lr pJlerJosse
are Laqt
'sJSpJ
Jruos uI
'JJueJrJruSrs
le.rnlrnrls
Jarpo
qlrm
sJlrs
pue
'sJrJruorluJr 'uoperlldar
;o
sur8rro
'uortdrnsuerl
aleln8a.r
teql
stuau
-ala
Jeqlo
'sraloruord
qlIM patprJosse punoJ
eJp salrs anrlrsuasrad,{.q asneraq
'sad,{.]
snorrerr
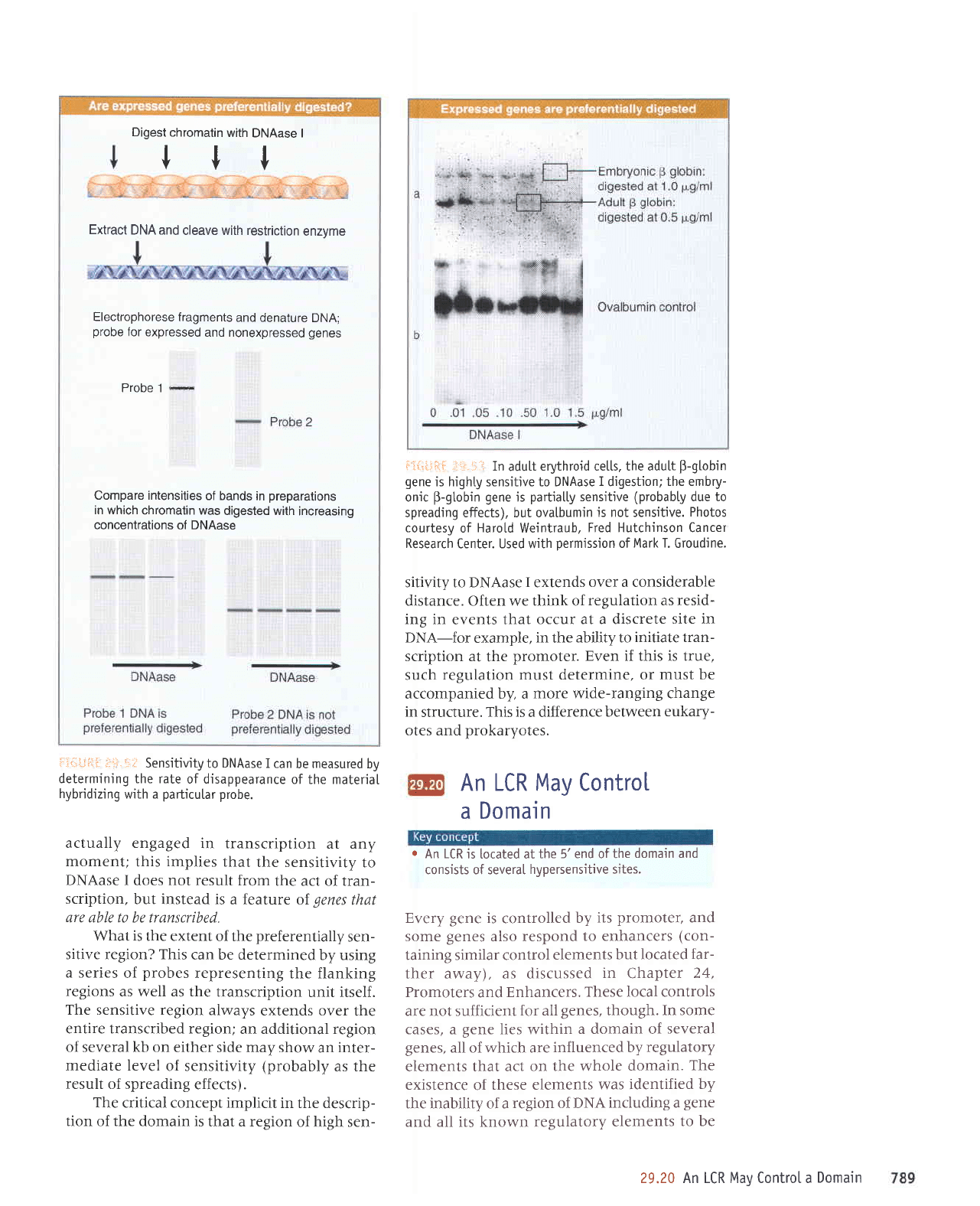
Digest
chromatin
with
DNAase I
Itt
Electrophorese
fragments
and denature
DNA;
probe
lor expressed
and nonexpressed
genes
Probe.l
*
Compare intensities
of bands in
preparations
in which
chromatin was
digested with
increasing
concentrations of DNAase
Extract
DNA and
cleave with restriction
enzvme
U'.i4;11.:i.
iF.:-!;
Sensitivity
to
DNAase
I can
be measured by
determining the rate
of disappearance
of the materiaI
hybridizing
with a
particutar
probe.
actually engaged
in transcription
at any
moment;
this
implies
that
the sensitivity
to
DNAase I does not result
from
the act of tran-
scription,
but instead is
a feature
of
genes
that
are able to be transcribed.
What is
the extent
of the
preferentially
sen-
sitive region? This
can be determined
by using
a series
of
probes
representing
the flanking
regions
as well as the
transcription
unit itself.
The sensitive region
always
extends over
the
entire transcribed region;
an additional region
of several kb on either
side may show
an
inter-
mediate level
of sensitivity
(probably
as the
result of spreading
effects).
The critical concept implicit
in the descrip-
tion of the domain is
that a region
of high sen-
lli.,;.j$if.
':l:i,i::
In
adutt
erythroid cetts. the adutt
B-gtobin
gene
is highLy sensitjve to DNAase
I
digestion;
the embry-
onic
B-gtobin
gene
is
partiatly
sensitive
(probabty
due to
spreading effects), but ovalbumin
js
not sensitjve.
Photos
courtesy
of
Harotd
Weintraub,
Fred Hutchinson Cancer
Research
Center. Used with
permission
of
Mark T.
Groudine.
sitivity to DNAase
I
extends
over a considerable
distance. Often we think of
regulation as resid-
ing in
events that
occur at a discrete
site in
DNA-for
example,
in
the
ability to
initiate tran-
scription at
the
promoter. Even if this is true,
such
regulation must determine, or
must be
accompanied by, a
more wide-ranging change
in
structure. This
is
a difference
between
eukary-
otes and
prokaryotes.
An LCR May Control
a Domain
r
An LCR
is located at the
5' end of the
domain and
consists of severaI
hypersensitive sites.
Every
gene
is
controlled
by
its
promoter,
and
some
genes
also respond
to enhancers
(con-
taining similar control
elements
but located
far-
ther away), as discussed
in
Chapter
24,
Promoters
and
Enhancers.
These local controls
are not sufficient
for all
genes,
though.
In some
cases, a
gene
lies within
a domain of
several
genes,
all of
which
are
influenced by
regulatory
elements that act
on the whole
domain.
The
existence
of
these elements
was identified
by
the inability of a
region of DNA
including a
gene
and all its known regulatory
elements
to be
29.20 An LCR Mav Control a
Domain

5' hypersensitive sites 3' hypersensitive site
HS4 HS1 Globin
genes
60 80
FI*IJRE i9.54
A
gtobin
domain is marked by
hypersen-
sitive sites at either end. The
group
of sites at the 5' side
constitutes the LCR and is essentiaI for the function of atl
oenes in the cluster.
properly
expressed
when
introduced into an
animal
as a transgene.
The best characterized example of a regu-
Iated
gene
cluster is
provided
by the mammalian
B-globin
genes.
Recall from Figure 6.1 that the
o- and
B-globin
genes
in mammals
each exist
as clusters of related
genes
that are expressed at
different times during embryonic and adult
development. These
genes
are
provided
with a
large number
of
regulatory
elements, which
have
been analyzed
in
detail.
In
the case of
the
adult human
B-globin
gene,
regulatory
sequences are located both 5'and 3'to the
gene.The
regulatory
sequences include both
positive
and negative elements in the
promoter
region,
as well as additional
positive
elements
within
and downstream of the
gene.
A human
B-globin
gene
containing all of
these
control
regions,
however, is never
expressed in a transgenic mouse
within an order
of magnitude
of
wild-type levels.
Some further
regulatory
sequence
is required.
Regions that
provide
the additional regulatory function are
identified
by DNAase I hypersensitive sites that
are
found
at the ends
of
the
cluster.
The map
of
FI**qr
f *.54
shows that the
20
kb upstream of
the
e
gene
contains a
group
of five
sites,
and
that there is
a single site 30 kb downstream of
the
p
gene.
Ttansfecting
various constructs into
mouse
erythroleukemia
cells shows that
sequences
between the individual hypersensi-
tive
sites in the 5'region
can be removed with-
out much effect, but
that
removal
of any of the
sites
reduces
the overall level
of expression.
The
5'regulatory
sites are the
primary
reg-
ulators, and the
cluster of hypersensitive sites
is called
the locus control region
(LCR).
We
do not know if
the 3'site has any function. The
LCR is
absolutely required
for expression of
each of the
globin genes
in the cluster. Each
gene
is
then further regulated
by its own spe-
cific controls. Some of these controls
are
autonomous:
Expression oI the e and
y
genes
appears
intrinsic to those loci in conjunction
with
the LCR. Other controls appear to rely
upon
position
in the cluster, which
provides
a
suggestion IhaI
gene
order in a cluster is impor-
tant for regulation.
The entire region containing the
globin
genes,
and extending
well beyond them, con-
stitutes a chromosomal
domain. It shows
increased sensitivity to digestion by
DNAase I
(see
Figure 29.52\. Deletion of the 5'LCR
restores
normal resistance to DNAase over the
whole
region. TWo
models for how
an LCR
works
propose
that its action is required in
order to activate the
promoter,
or alternatively,
is required to increase the rate of transcrip-
tion from the
promoter.
The exact nature of
the
interactions between the LCR and the indi-
vidual
promoters
has not
yet
been fully
defined.
Does this model apply to other
gene
clus-
ters? The cr-globin
locus has
a similar organi-
zation of
genes
that are expressed at different
times,
with
a
group
of hypersensitive sites at
one end of
the cluster, and increased
sensitiv-
ity to DNAase I throughout the region. Only a
small
number
of
other cases are known in
which
an LCR controls a
group
of
genes.
One of
these cases involves
an
LCR
that
controls
genes
on
more
than one chromosome.
The Ts2 LCR coordinately regulates a
group
of
genes
that are spread out over
I20
kb on chro-
mosome I I by interacting with their
promot-
ers.
It
also
interacts
with
the
promoter
of the
IFNygene on chromosome
10.
The two types of
interaction are alternatives that comprise
two
different cell fates, that is, in one
group
of cells
the LCR causes expression
of the
genes
on chro-
mosome
I
l,
whereas
in
the other
group
it
causes
the
gene
on chromosome l0 to
be expressed.
What Constitutes
a Regulatory Domain?
r
A domain may have an
insutator,
an LCR,
a
matrix
attachment
site, and transcription unit(s).
If we
put
together the
various types of
struc-
tures that have been found in
different sys-
tems,
we can think about the
possible
nature
of a chromosomal domain. The
basic feature
of a regulatory
domain is that regulatory
ele-
CHAPTER 29 Nucleosomes
